Introduction
Renal cell carcinoma (RCC) represents one of the
most prevalent malignant tumors within the genitourinary system,
accounting for ~5% of all newly diagnosed cancers in men and 3% in
women (1). Previous statistical
data have shown that, in 2019, the United States recorded ~73,820
new diagnoses of RCC, and 14,770 mortalities were attributed to RCC
(1). Among the diverse subtypes of
RCC, clear cell RCC (ccRCC) has emerged as the predominant form and
is widely regarded as one of the most aggressive malignancies in
the urinary tract, with an annual global mortality rate of ~90,000
cases (2). Cytokine-based and
checkpoint inhibitor immunotherapies have been shown to elicit
robust immune responses through distinct mechanisms, thereby having
a pivotal role in RCC treatment (3,4).
However, despite the major improvements that have been made in
terms of understanding tumor initiation and progression, the
etiological mechanisms underlying RCC remain poorly understood
(5). Due to the high incidence and
mortality rates associated with RCC, it is essential to identify
predictive biomarkers that impact immune responses in patients
diagnosed with this malignancy.
Chemokines, a class of small, secreted proteins, are
recognized for their pivotal regulatory roles in mediating immune
cell trafficking and lymphatic tissue development (6,7). As
the largest subfamily of cytokines, chemokines can be further
classified into four primary subgroups according to the arrangement
of their two cysteine (C) residues in their protein sequences,
namely CC-chemokines, CXC-chemokines, C-chemokines and
CX3C-chemokines (7). Within the
tumor microenvironment (TME), chemokines are expressed by a diverse
array of cell types, encompassing both tumor cells themselves and
other essential components, such as immune cells and stromal cells
(8,9). In response to specific chemokines,
diverse immune cell subsets migrate to the TME, orchestrating the
immune response against tumors in a precisely regulated manner
(8,9). Additionally, chemokines directly
target non-immune cells within the TME, including tumor, stromal
and vascular endothelial cells, which have been shown to modulate
tumor cell proliferation, tumor stemness properties and tumor
invasion and metastasis (10).
Consequently, chemokines exert both direct and indirect effects on
tumor immunity, thereby shaping the immunological and biological
phenotypes of tumors, and subsequently influencing tumor
progression, treatment efficacy and patient prognosis (8,10,11).
CXC motif chemokine ligand 1 (CXCL1) belongs to the
glutamate-leucine-arginine (ELR) + CXC chemokine subfamily that is
distinguished by the conserved amino acid sequence of ELR (12). The role of CXCL1 is mediated through
its interaction with its receptor, CXCR2 (13). To date, a burgeoning body of
evidence has demonstrated that CXCL1 displays an increased level of
expression in a variety of different types of human malignancies,
with CXCL1 serving as a potent signal within the TME that promotes
malignant progression (13). The
present study therefore aimed to investigate the expression
patterns of CXCL1 in RCC tissues, also exploring whether CXCL1, as
a component of the RCC microenvironment, may contribute to the
carcinogenic potential of RCC through exogenous and autocrine
mechanisms. The findings of the present study may help to
facilitate the discovery of valuable biomarkers and potential
therapeutic targets for the clinical diagnosis and molecular
targeted therapy of RCC.
Materials and methods
Bioinformatics analysis
The initial CXCL1 RNA sequencing (RNA-seq) data were
acquired from The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) and Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) databases. The
expression of CXCL1 in tumor tissues, as well as its expression in
tumor tissues with different pathological parameters, was
investigated in the GSE53757 dataset (including RNA-seq data of 72
pairs of normal kidneys and tumor tissues) and the GSE40435 dataset
(including 101 pairs of matched adjacent and tumor tissue RNA-seq
data) (14,15). The STRING database (https://string-preview.org) was utilized to identify
core (hub) genes that are co-expressed with CXCL1, and the
expression levels of these hub genes were subsequently examined
using TCGA database. Furthermore, the tumor immune estimation
resource (TIMER) database (https://cistrome.shinyapps.io/timer/) was used to
assess the association between CXCL1 expression and immune cell
recruitment. To determine the MSI status, MSI scores were
calculated using the program Microsatellite Analysis for
Normal-Tumor InStability (MANTIS) with a default threshold of 0.4,
defining high MSI (MSI-H) as scores above the threshold, and
microsatellite stable (MSS or no apparent MSI) as scores below
it.
Clinical samples and
immunohistochemical analysis
Paraffin-embedded specimens, which had undergone
surgical resection at the First Affiliated Hospital of Jiamusi
University (Jiamusi, China) between February 2023 and January 2025
and were subsequently diagnosed as RCC by the Department of
Pathology, were selected for the present study. A total of 68 pairs
of RCC tissues and their corresponding adjacent non-cancerous
tissues, along with complete clinical data, were randomly chosen
for inclusion in the present study. The clinical characteristics of
these 68 samples are presented in Table
I. The study included patients with histologically confirmed
renal cell carcinoma who underwent partial or radical nephrectomy
and for whom high-quality paired tumor and adjacent non-cancerous
tissue samples were available. All participants had complete
clinicopathological data (age, sex, stage and grade). Exclusion
criteria were prior systemic anti-tumor therapy, previous renal
surgery or biopsy on the same kidney, and a concurrent malignancy.
In brief, the specimens were fixed in 10% neutral-buffered formalin
at room temperature for 24 h, then processed into paraffin blocks
and cut into 5-µm thick sections. These sections were
deparaffinized with 100% xylene at room temperature and
subsequently rehydrated through a graded ethanol series. Antigen
retrieval was carried out in a high-pressure cooker using 0.01 M
citric-acid buffer at 120°C for 10 min. To suppress endogenous
peroxidase activity, slides were incubated in 3%
H2O2 in PBS for 30 min at room temperature.
Non-specific binding sites were blocked by applying 20% normal goat
serum (cat. no. AR0009; Wuhan Boster Biological Technology, Ltd.)
and incubated at 37°C for 1 h. Finally, the sections were incubated
overnight at 4°C with a CXCL1-specific antibody (dilution, 1:100;
cat. no. YT2074; ImmunoWay Biotechnology Company), and subsequently
processed using a StreptAvidin-Biotin Complex Kit (cat. no. SA1022;
Wuhan Boster Biological Technology, Ltd.), in accordance with the
manufacturer's instructions. The sections were subjected to routine
3,3′-diaminobenzidine staining and hematoxylin counterstaining at
room temperature for 15 min, followed by an assessment of CXCL1
protein expression, as determined under an epifluorescence
microscope (Olympus Corporation) by two specified parameters: i)
Intensity of staining, ranging from negative-weak (assigned a score
of 1) to moderate (score of 2), strong (score of 3) and extremely
strong (score of 4); and ii) the proportion of stained cells,
categorized as ≤25% (score of 1), >25% to ≤50% (score of 2),
>50% to ≤75% (score of 3) and >75% (score of 4). The total
score was calculated by generating the sum of the parameters (i)
and (ii). A score of ≤4 was indicative of low expression, whereas a
score of ≥5 was indicative of high expression. The staining
intensity was quantified using Image-Pro Plus 6.0 software (Media
Cybernetics, Inc.).
 | Table I.Clinical characteristics of patients
with clear cell renal carcinoma. |
Table I.
Clinical characteristics of patients
with clear cell renal carcinoma.
| Characteristic | n |
|---|
| Sex |
|
|
Male | 46 |
|
Female | 22 |
| Age, years |
|
|
≥60 | 21 |
|
<60 | 47 |
| Tumor grade |
|
| Grade
1 | 31 |
| Grade
2 | 26 |
| Grade
3 | 11 |
| Clinical stage |
|
| Stage
I | 44 |
| Stage
II | 20 |
| Stage
III | 4 |
Cell lines and cell culture
The human ccRCC cell line 786-O and papillary RCC
(pRCC) cell line CAKI-2 were supplied by the American Type Culture
Collection. The cell lines were maintained in Gibco RPMI-1640
medium (cat. no. 11875093; Thermo Fisher Scientific, Inc.)
supplemented with 10% Gibco fetal bovine serum (FBS; cat. no.
A5670201; Thermo Fisher Scientiffc, Inc.) and 1:100
penicillin/streptomycin (cat. no. PYG0016; Wuhan Boster Biological
Technology, Ltd.). In the present study, the medium containing FBS
and penicillin/streptomycin was referred to as complete medium
(CM).
Cell transfection
Overexpression and RNA-interference strategies were
employed for transfection experiments. Pseudovirus particles
containing either empty lentiviral plasmids (cat. no.
LPP-NEG-Lv201-100) or overexpression lentiviral plasmids (cat. no.
LPP-G0095-Lv201-100) were produced by GeneCopoeia, Inc., whereas
small interfering (si)RNAs were synthesized by HyCyte, Inc. The
sense and antisense sequences specifically designed for
RNA-interference to silence the CXCL1 gene were:
5′-GAUGCUGAACAGUGACAAATT-3′ and 5′-UUUGUCACUGUUCAGCAUCTT-3′;
5′-CCAAGAACAUCCAAAGUGUTT-3′ and 5′-ACACUUUGGAUGUUCUUGGTT-3′; and
5′-GCUGCUCCUGCUCCUGGUATT-3′ and 5′-UACCAGGAGCAGGAGCAGCTT-3′. The
negative-control sense and antisense sequence were
5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′. Cell
transfection was carried out according to the supplier's
instructions. In the overexpression study, a total of
4×104 cells were seeded into each well of a 24-well
plate in 500 µl of serum-free medium. After a 24 h incubation at
37°C under starvation conditions, lentiviral particles were added
directly to the medium at a multiplicity of infection of
10−8. Following a further 5 h incubation at 37°C, an
additional 500 µl of 10% serum-containing medium was added. A total
of 24 h later, the entire medium was replaced with fresh complete
medium. 72 h after the initial viral infection, puromycin was added
to a final concentration of 2 µg/ml for selection. Finally,
downstream experiments were performed after an additional 72 h
incubation to allow sufficient phenotypic expression. In the
RNA-interference study, 1.25 µl siRNA stock solution was diluted
with 50 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.), and
was gently pipetted 3–5 times. Meanwhile, 1.0 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was diluted with 50 µl Opti-MEM, and was gently
pipetted 3–5 times. After standing at room temperature for 5 min,
the transfection reagent and siRNA dilution were mixed and gently
pipetted 3–5 times. Subsequently, they were left to stand at room
temperature for 20 min. Finally, the transfection complex was added
to the 24-well cell plate with cells.
ELISA detection assay
Within each compartment of the 6-well dishes,
5×105 cells were grown in 1 ml serum-free medium.
Following 24 h culture, the supernatant was isolated via
centrifugation at 4°C, 1,000 × g for 5 min, and a Human CXCL1 ELISA
kit (cat. no. EK0722; Wuhan Boster Biological Technology, Ltd.) was
employed to determine and quantify the level of CXCL1 in the
supernatant, following precisely the manufacturer's instructions.
The ELISA experiments were repeated three times.
Cell proliferation detection
assay
Concerning the exogenous study, 786-O and CAKI-2
cells were seeded into each well of a 96-well dish at a density of
3×103 cells/well, utilizing CM supplemented with various
concentrations (0, 2, 5, 10, 20 and 30 ng/ml) of exogenous CXCL1
(cat. no. 300-11-25UG; PeproTech, Inc.; Thermo Fisher Scientific,
Inc.). Concerning the endogenous study, 786-O and CAKI-2 cells with
overexpression of CXCL1, 786-O and CAKI-2 cells with decreased
expression of CXCL1, along with their corresponding control cells
were seeded into each well of a 96-well dish at a density of
3×103 cells/well, and CM served as the growth medium for
all cell types. The cells were subsequently incubated for 72 h.
Subsequently, 10 µl Cell Counting Kit-8 (CCK-8) reagent (cat. no.
AR1160; Wuhan Boster Biological Technology, Ltd.) was added to each
well, and the cells were further incubated for 2 h in the
incubator. Finally, the optical density (OD) values at a wavelength
of 450 nm were read using a microplate reader. The cell
proliferation assay experiments were repeated three times.
Cell migration detection assay
The 786-O and CAKI-2 cells were seeded at a density
of 1×104 cells/well into the top chambers of Transwell
dishes (cat. no. 3428; Corning, Inc.). The top chambers were filled
with serum-free medium containing various concentrations of
exogenous CXCL1, whereas the bottom chambers were maintained with
CM. Concerning the endogenous study, 786-O and CAKI-2 cells with
overexpression of CXCL1, 786-O and CAKI-2 cells with decreased
expression of CXCL1, along with their respective control cells were
seeded at a density of 1×104 cells/well (for
overexpression study) or 3×104 cells/well (for siRNA
study) into the top chambers of the Transwell dishes. The top
chambers were filled with serum-free medium, whereas the bottom
chambers contained CM. The cells were subsequently incubated for 48
h, after which the upper surface of the chambers was gently wiped
with a cotton swab to remove the non-migrated cells. Following
methanol fixation and crystal violet staining at room temperature
for 20 min, the chambers were imaged under an inverted microscope
in order to count the numbers of migrated cells. The cell migration
experiments were repeated three times.
Western blotting assay
Total cellular proteins were extracted and
quantified utilizing RIPA protein lysis buffer (cat. no. P0013B;
Beyotime Institute of Biotechnology) and a BCA protein quantitation
kit (cat. no. AR0146; Wuhan Boster Biological Technology, Ltd.,
respectively. Subsequently, 40 µg total protein was subjected to
12% SDS-PAGE and transferred onto a PVDF membrane (cat. no.
IPVH00010; MilliporeSigma). The membrane was incubated overnight at
4°C with primary antibodies against Bcl-2-associated X protein
(Bax; cat. no. CY5059; Shanghai Abways Biotechnology Co., Ltd.;
1:1,000), B-cell lymphoma-2 (Bcl-2; cat. no. CY5032; Shanghai
Abways Biotechnology Co., Ltd.; 1:1,000), phosphoinositide 3-kinase
(PI3K; cat. no. CY5224; Shanghai Abways Biotechnology Co., Ltd.;
1:1,000), AKT (cat. no. AF6261; Affinity Biosciences; 1:1,000) and
β-actin (cat. no. TA811000; OriGene Technologies, Inc.; 1:5,000).
Subsequently, the membrane was further incubated with horseradish
peroxidase-conjugated secondary antibodies at 37°C for 30 min (cat.
no. BA1055, Wuhan Boster Biological Technology, Ltd.) at a dilution
of 1:10,000 and ECL reagents (cat. no. 34095; Thermo Fisher
Scientific, Inc.), prior to being placed in a Tanon 4200 Fully
Automatic Chemiluminescence/Fluorescence Image Analysis System
(Tanon Science & Technology Co., Ltd.) for visualization and
imaging. Protein quantification was performed with Tanon Image
software (Tanon Science & Technology Co., Ltd.). The western
blotting experiments were repeated three times.
Statistical analysis
The SPSS statistics software, version 22.0 (IBM
Corp.) was employed for statistical analyses. The variabilities in
CXCL1 expression among different groups were compared using the
Mann-Whitney U test. To assess the prognostic value of CXCL3, both
univariate and multivariate Cox regression analyses were performed.
Pearson correlation analysis was employed to assess the
relationship between CXCL1 expression and immune cell recruitment.
In the cellular experiments, comparisons between two groups were
performed with an unpaired two-sample t-test. For comparisons among
three or more groups, a one-way ANOVA was applied. When the one-way
ANOVA revealed a significant overall effect (P<0.05), pairwise
comparisons were conducted using the Tukey's post hoc test.
Results
Expression of CXCL1 mRNA is
upregulated in RCC tissues, and its high expression is associated
with the clinical characteristics of patients with RCC
TCGA database analysis revealed that the expression
level of CXCL1 mRNA in ccRCC or pRCC tissues was significantly
upregulated compared with normal tissues (Fig. 1A). Due to ccRCC being the most
prevalent subtype of RCC, subsequent experiments in the present
study were focused on ccRCC. The analysis of clinical grade
(Fig. 1B) and subtypes (Fig. 1C) revealed that the expression level
of CXCL1 in grade 4 ccRCC was significantly higher compared with
that in the normal tissues and grades 1 and 2 ccRCC, and the ccB
subtype exhibited a notably increased expression level compared
with both normal tissue and the ccA subtype. Additionally, in both
the GSE53757 dataset and the GSE40435 dataset, the expression
levels of CXCL1 mRNA were markedly increased in ccRCC samples
compared with the normal tissues (Fig.
1D) or the paracancerous tissues (Fig. 1E). Moreover, clinical staging
analysis revealed that patients with stage III/IV ccRCC exhibited
significantly higher CXCL1 expression levels compared with those
with stage I/II ccRCC (Fig. 1F).
Similarly, grade 4 tumors exhibited significantly upregulated CXCL1
expression levels compared with grade 1–3 tumors (Fig. 1G). Furthermore, significant
associations were observed between the expression levels of CXCL1
and tumor status (T; Fig. 1H),
lymph node status (N; Fig. 1I) and
metastasis status (M; Fig. 1J). A
prognostic assessment was performed among the different groups
utilizing the log-rank test method, which revealed that high CXCL1
expression was significantly associated with shorter overall
survival (OS) times, (Fig. 1K),
shorter disease-specific survival times (Fig. 1L) and shorter progression-free
survival times (Fig. 1M).
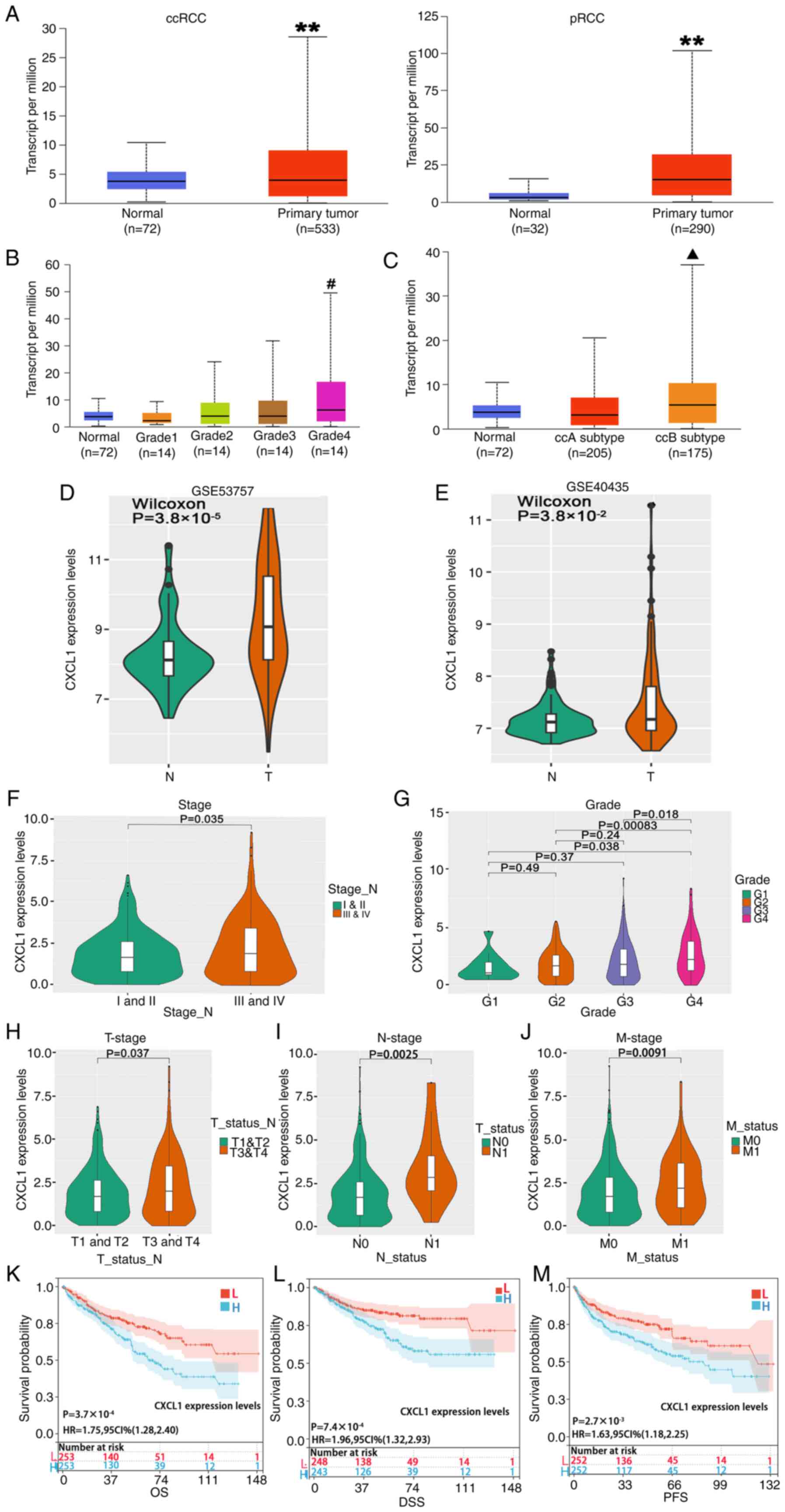 | Figure 1.Increased expression of CXCL1 mRNA
levels is associated with the clinical characteristics of patients
with RCC. (A) The expression profiles of CXCL1 mRNA in normal
tissues compared with ccRCC or pRCC tissues. The different levels
of expression of CXCL1 mRNA within (B) different clinical grades
and (C) subtypes of ccRCC tissues. Expression levels of CXCL1 mRNA
in the (D) GSE53757 and the (E) GSE40435 datasets. The different
levels of expression of CXCL1 mRNA within different (F) clinical
grades and (G) stages of ccRCC tissues. Analysis of the expression
level of CXCL1 within the clinical (H) T-status, (I) N-status and
(J) M-status categories in ccRCC tissues. (K-M) A prognostic
assessment was performed among the various groups. **P<0.01 vs.
normal; #P<0.01 vs. normal, grade 1 and 2;
▲P<0.01 vs. normal and ccA subtype. CXCL1, CXC motif
chemokine ligand 1; RCC, renal cell carcinoma; ccRCC, clear cell
RCC; pRCC, papillary RCC; T, tumor; N, lymph node; M, metastasis;
OS, overall survival; DSS, disease-specific survival; PFS,
progression-free survival. |
CXCL1 expression is associated with
the expression of tumor-associated genes, immune cell recruitment
and microsatellite instability (MSI) in RCC
STRING database analysis disclosed 10 hub genes that
exhibited co-expression with CXCL1 in ccRCC, namely CXCL6, C-C
chemokine receptor type 2 (CCR2), CXCR1, CXCL3, CXCL5, C-C motif
chemokine 11 (CCL11), CXCR2, atypical chemokine receptor 1 (ACKR1),
CXCL2 and CXCR4. TCGA database was employed to assess the
expression levels of these 10 key genes, and the results
demonstrated a significant reduction in the expression of CCL11,
and a notable increase in the expression levels of CCR2, CXCR1,
CXCL3, CXCL5, CXCR2, ACKR1, CXCL2 and CXCR4 (Fig. 2A). A protein-interaction network
analysis that was subsequently constructed based on the CXCL1 gene
revealed that numerous chemokines and their receptors exhibited
strong interactions with CXCL1 (Fig.
2B). Correlation analysis indicated that, excluding CCL11,
CXCL1 exhibited a significant positive correlation with the
expression of the other nine genes (Fig. 2C). TIMER database analysis unveiled
a negative correlation between CXCL1 expression and the
infiltration of B cells, whereas a positive correlation was
observed with the recruitment of CD4+ T cells and
neutrophils (Fig. 2D). The findings
obtained from MSI status study demonstrated a positive correlation
between CXCL1 expression and both the number of mutations (Fig. 2E) and MSI scores (Fig. 2F) in patients with RCC. Furthermore,
patients in the MSI group exhibited significantly higher CXCL1
expression levels compared with those in the MSS group (Fig. 2G).
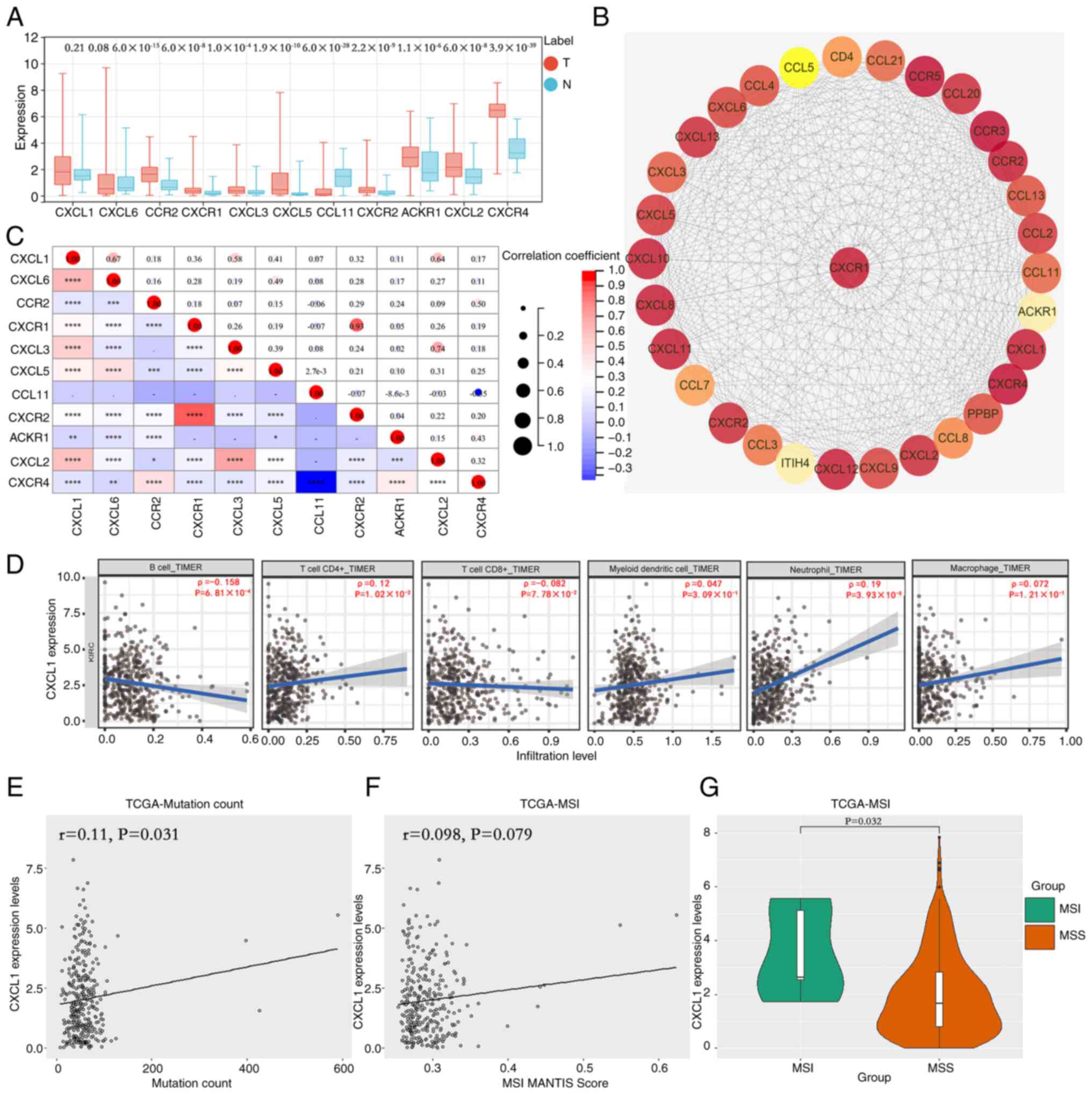 | Figure 2.Expression levels of CXCL1 mRNA are
correlated with the expression of genes implicated in
tumorigenesis, recruitment of immune cells and presence of MSI in
renal cell carcinoma. (A) Identification of 10 hub genes exhibiting
co-expression with CXCL1. (B) Analysis of the protein-interaction
network involving CXCL1. (C) Analysis of the correlation between
CXCL1 and the aforementioned 10 hub genes. (D) Analysis of the
correlation between CXCL1 expression and immune cell recruitment.
The correlation between CXCL1 expression and the (E) number of
mutations is presented, as well as (F) MSI scores. (G) The
expression levels of CXCL1 in the MSI group compared with the MSS
group. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
CXCL, CXC motif chemokine ligand; ACKR1, atypical chemokine
receptor 1; CCR, C-C chemokine receptor type 2; MSI, microsatellite
instability; MSS, microsatellite stable or no apparent MSI; TIMER,
tumor immune estimation resource; TCGA, The Cancer Genome Atlas; T,
tumor; N, normal. |
Expression of CXCL1 protein is
upregulated in RCC tissues, and its elevated expression is
associated with the clinicopathological features of patients with
RCC
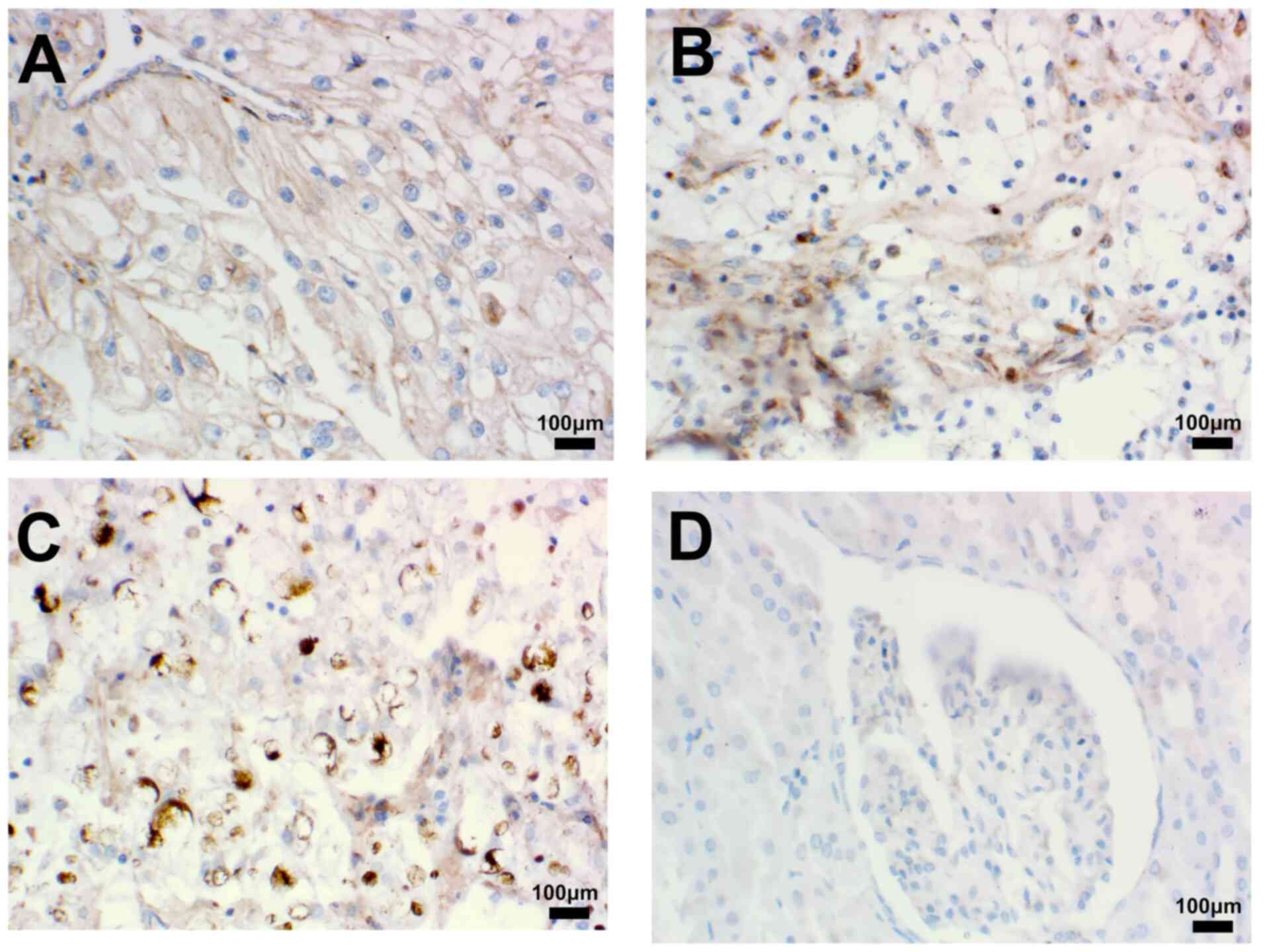
To further elucidate the clinical significance of
CXCL1 in RCC, immunohistochemical experiments were utilized to
assess the expression of the CXCL1 protein. The results obtained
demonstrated that the expression of CXCL1 was significantly
upregulated in RCC tissues compared with paracancerous tissues
(Fig. 3A-D). In addition, the study
of the clinicopathological characteristics of patients with RCC
revealed that the CXCL1 protein level in patients with grade 2–3
RCC was significantly higher compared with that in patients with
grade 1. In addition, the CXCL1 protein expression levels in
patients with stage II/III cancer was significantly higher compared
with that in patients with stage I. However, the present study did
not find any association between the expression level of CXCL1
protein and the age or the sex of the patients (Table II).
 | Table II.Expression of CXCL1 in renal cell
carcinoma and their adjacent tissues. |
Table II.
Expression of CXCL1 in renal cell
carcinoma and their adjacent tissues.
| Characteristic | CXCL1 expression
(mean ± SD) | P-value |
|---|
| Tissue |
| <0.0001 |
|
Adjacent non-cancerous
tissues | 4.485±1.440 |
|
|
Cancerous tissues | 5.485±1.275 |
|
| Sex |
| 0.3836 |
|
Male | 5.391±1.325 |
|
|
Female | 5.682±1.171 |
|
| Age, years |
| 0.9690 |
|
≥60 | 5.489±1.333 |
|
|
<60 | 5.476±1.167 |
|
| Tumor grade |
| 0.0340 |
| Grade
1 | 5.129±1.176 |
|
|
Grade2-3 | 5.784±1.294 |
|
| Clinical stage |
| 0.0384 |
| Stage
I | 5.250±1.296 |
|
| Stage
II/III | 5.917±1.139 |
|
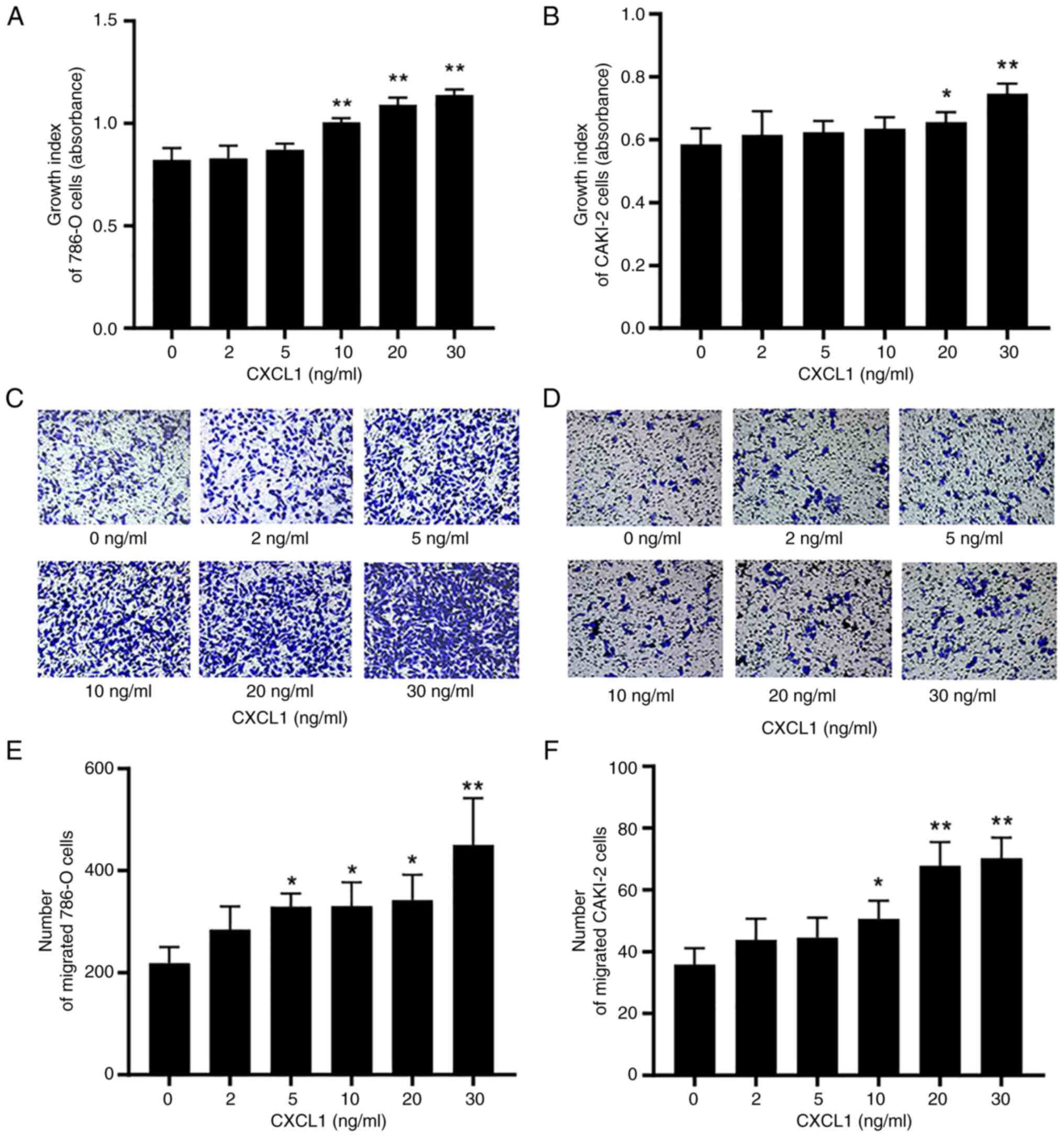
Exogenous CXCL1 contributes to the
malignant behaviors of RCC cells
To assess whether CXCL1 is involved in the tumor
biological behavior of RCC cells through paracrine or endocrine
pathways, relevant studies were performed using exogenous CXCL1.
CCK-8 assay analysis revealed that the proliferation rates of 786-O
and CAKI-2 cells treated with exogenous CXCL1 at concentrations of
10, 20 and 30 ng/ml (for 786-O cells) or 20 and 30 ng/ml (for
CAKI-2 cells) were significantly increased after 72 h in comparison
with the 0 mg/ml treatment group (Fig.
4A and B). Furthermore, the Transwell assay experiments also
demonstrated that the migratory capabilities of the 786-O cells
treated with 5, 10, 20 and 30 ng/ml exogenous CXCL3 and the CAKI-2
cells treated with 10, 20 or 30 ng/ml exogenous CXCL3 after 48 h
were significantly enhanced (Fig.
4C-F).
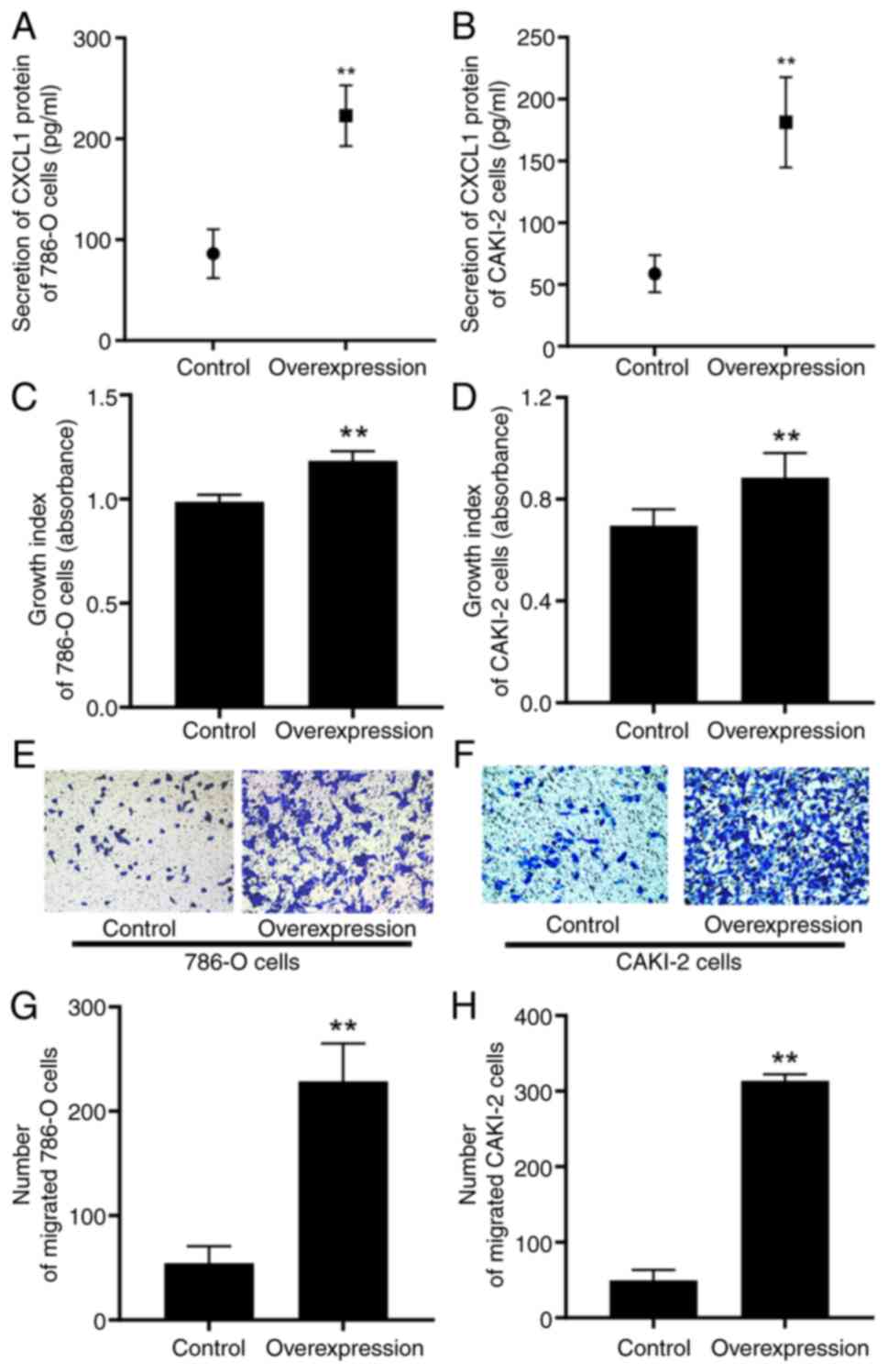
High expression of CXCL1 facilitates
the malignant behaviors of RCC cells
To clarify the involvement of endogenous CXCL1 in
the biological behavior of RCC progression through an autocrine
pathway, lentiviral transfection was used to establish RCC cell
lines with a high expression level of CXCL1. ELISA assay revealed
that, in comparison with their respective control cells, the
secretory levels of CXCL1 protein were significantly increased in
the 786-O and CAKI-2 cells expressing a high level of CXCL1
(Fig. 5A and B). Subsequent CCK-8
and Transwell assays demonstrated that the overexpression of CXCL1
led to significant enhancements of the proliferative and migratory
capabilities of both 786-O and CAKI-2 cells (Fig. 5C-H).
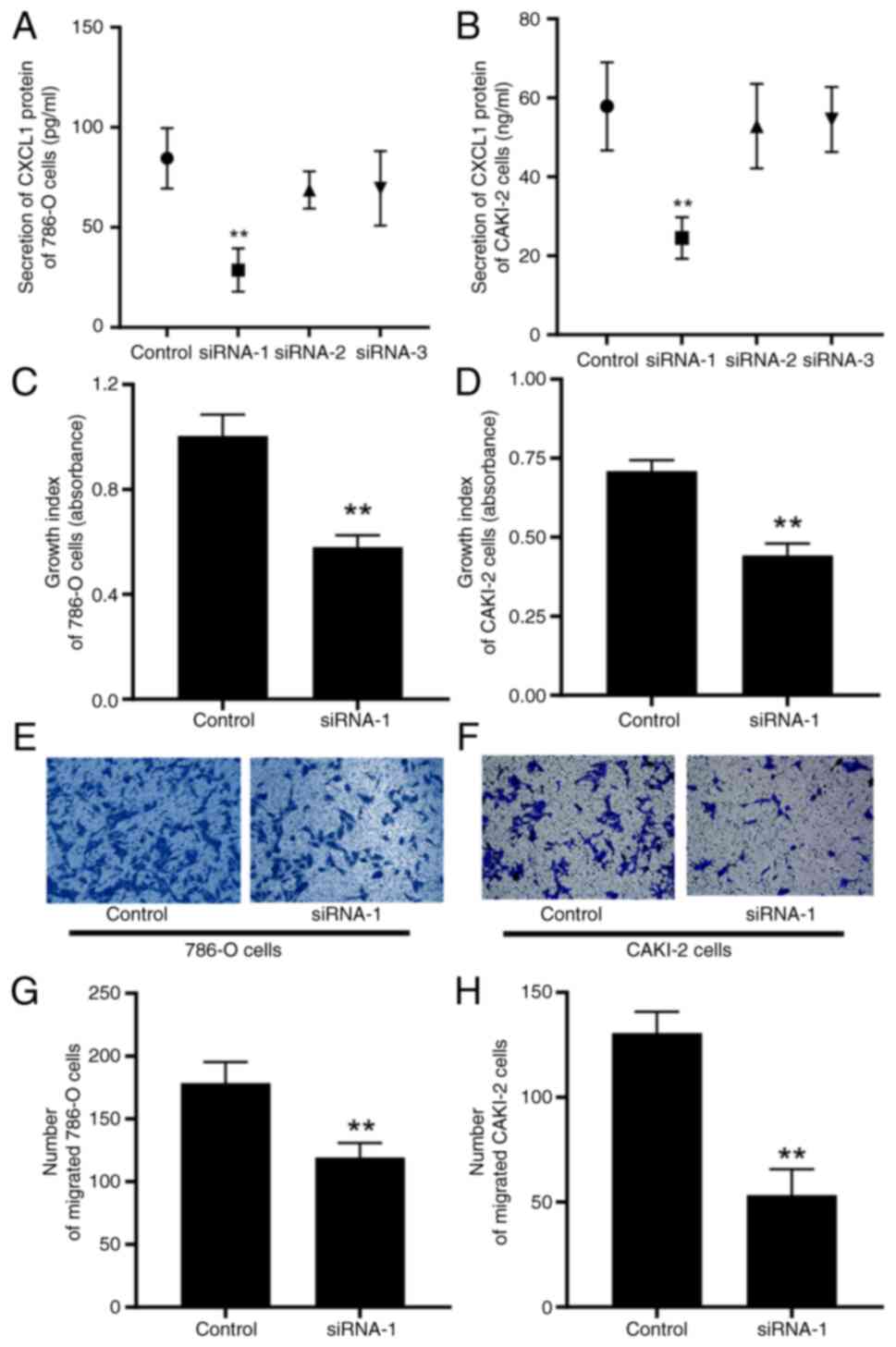
Low expression of CXCL1 impedes the
malignant behaviors of RCC cells
Similarly, in the interference studies, ELISA assay
was used to quantify the secretory protein levels of CXCL1 in the
supernatant, thereby confirming the successful establishment of
786-O and CAKI-2 cells that downregulated CXCL1 at low levels
(Fig. 6A and B). In contrast to the
results observed in the CXCL1 overexpression study, the decreased
expression of CXCL1 led to a marked reduction in both the cell
proliferative and migratory capabilities of the 786-O and CAKI-2
cells (Fig. 6C-H).
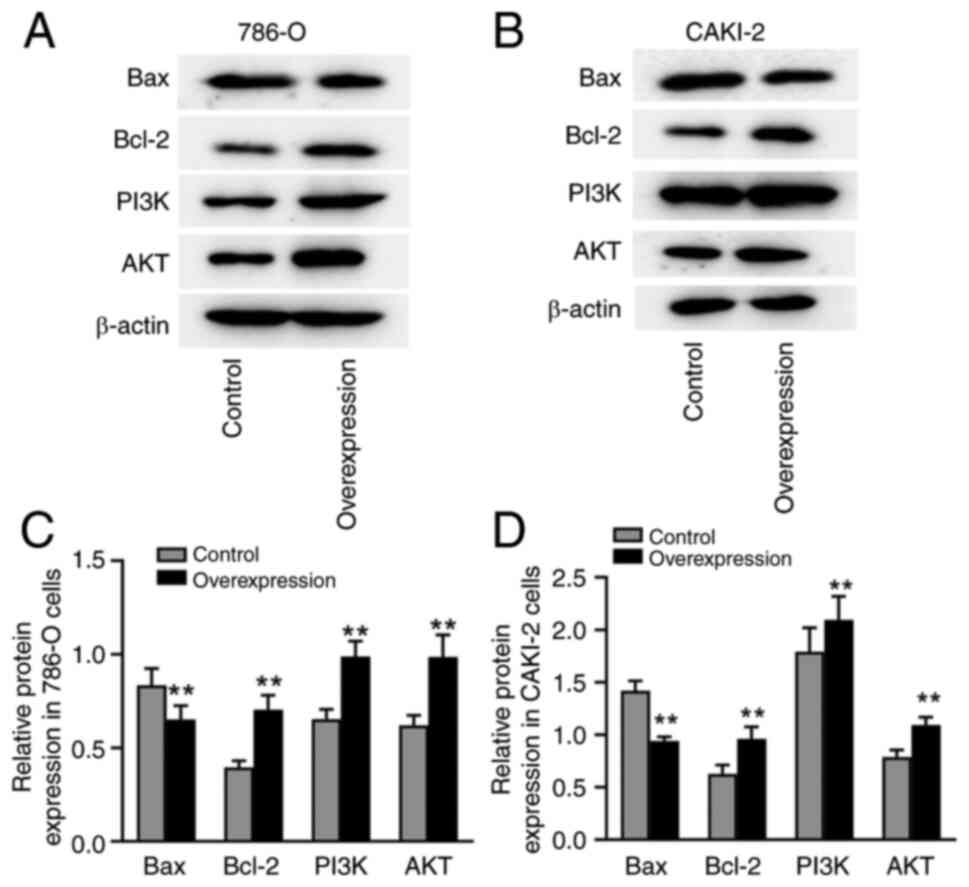
Overexpression of CXCL1 regulates the
expression of PI3K/AKT-associated proteins in RCC cells
Finally, western blotting assays were performed to
investigate the potential underlying mechanisms of CXCL1-mediated
malignant progression in RCC cells with high CXCL1 expression. The
results showed that, in both 786-O and CAKI-2 cells, overexpression
of CXCL1 led to a marked downregulation of the expression Bax, as
well as an upregulation of the expression of PI3K, AKT and Bcl-2
proteins (Fig. 7A-D).
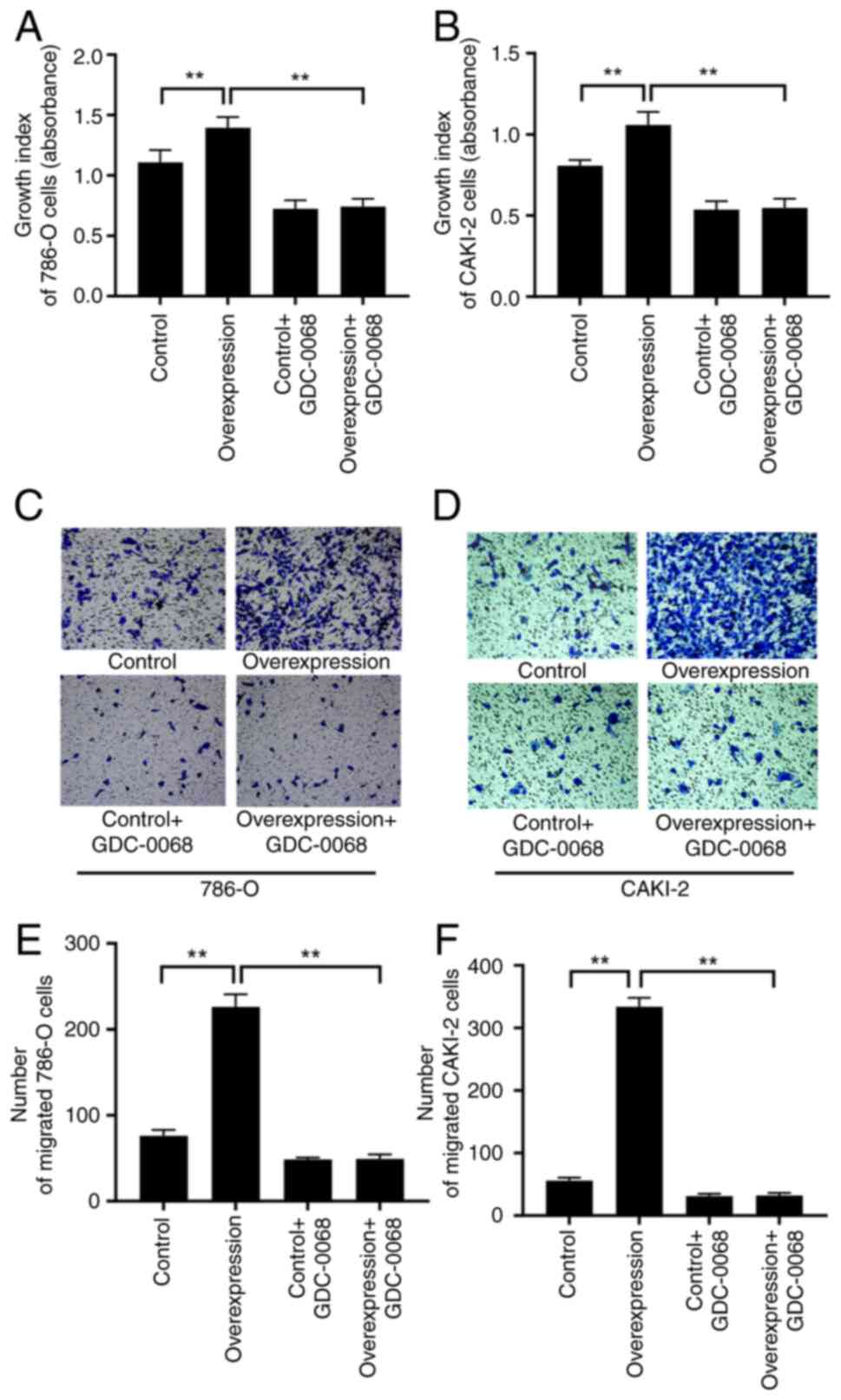
Inhibiting AKT reverses the promoting
effect of a high expression of CXCL1 on the malignant behaviors of
RCC cells
To determine whether the PI3K/AKT pathway regulates
the influence of CXCL1 on the malignant behaviors of RCC cells, a
further experiment was performed, wherein the AKT-specific
inhibitor GDC-0068 was introduced into the cell culture medium to
study 786-O and CAKI-2 cells with CXCL1 overexpression and their
corresponding empty vector-transfected cells. The CCK-8 assay
results revealed that, following GDC-0068 treatment, no significant
differences were observed in the proliferative capability between
the overexpression and control cells (Fig. 8A and B). Similarly, Transwell
(Fig. 8C-F) assays showed that
GDC-0068 could reverse the promoting effects of CXCL1 on migration
in both cell lines.
Discussion
CXCL1, also known as melanoma growth-stimulatory
activity/growth-regulated oncogene a, is a polypeptide that was
originally isolated from human melanoma cells exhibiting
chemotactic properties (16). It
has been proposed that CXCL1 has the capability to induce migration
of immune cells, notably neutrophils, towards sites of inflammation
or infection, thereby participating in immune responses and
inflammatory reactions (17). To
date, accumulating evidence has demonstrated that CXCL1 exhibits
upregulated expression in numerous types of human tumor, and its
upregulation has been shown to be notably associated with advanced
clinical stages, higher grades, distant metastasis and poor
prognosis (13,18). For example, evidence has been
presented to suggest that the expression level of CXCL1 is markedly
upregulated in uterine cervical cancer tissues, and elevated levels
of CXCL1 were found to be notably associated with advanced clinical
stage and decreased survival probabilities (19). Furthermore, previous studies have
revealed a notable association between the expression of CXCL1 and
the histological types of tumors. For example, in the case of
breast cancer, the expression levels of CXCL1 were found to be
markedly increased in tissues of basal-like breast cancer,
mesenchymal-like triple-negative breast cancer and basal-like
triple-negative breast cancer compared with normal tissues
(13). Additionally, a high
expression of CXCL1 was observed in inflammatory breast cancer as
compared with other subtypes of breast cancer (13).
Consistent with the aforementioned findings, the
present study demonstrated a notable upregulation of CXCL1 mRNA and
protein expression in RCC tissues compared with normal and
paracancerous tissues. Notably, high expression of CXCL1 was
strongly associated with advanced stage, higher grade, unfavorable
TNM status and a worse clinical outcome for patients. Moreover, the
findings presented in the current study demonstrated markedly
increased expression of CXCL1 in the ccB subtype of ccRCC compared
with both normal tissues and the ccA subtype. ccRCC comprises two
subtypes defined by distinct gene expression profiles: The ccA
subtype is characterized by the overexpression of genes associated
with hypoxia, angiogenesis, as well as fatty acid and organic acid
metabolism, whereas the unfavorable-outcome ccB subtype features
the overexpression of genes that regulate epithelial-to-mesenchymal
transition, cell cycle progression and wound healing (20). Consequently, the results from the
tissue subtype analysis in the present study also suggest an
association between elevated CXCL1 expression and the malignancy of
ccRCC.
The TME consists not only of tumor cells, but also
stromal cells, immune cells, vasculature, extracellular matrix and
soluble factors, in which the chemokines form a dynamic system that
fulfills a pivotal role in tumor progression (10). It is now widely accepted that the
chemokine system is involved in tumor development via multiple
direct and indirect mechanisms that modulate immune cell
infiltration, angiogenesis, as well as tumor cell proliferation,
invasion, metastasis and stem-like properties (21). The present study on co-expression
hub gene and protein-interaction network analysis revealed that
numerous chemokines and their receptors, including CXCL3, CXCL5,
CXCL8, CXCR1 and CXCR2, exhibit co-expression and interactive
associations with CXCL1 in RCC. Similarly, CXCL3, CXCL5 and CXCL8
(along with CXCL1) belong to the ELR + CXC chemokine subfamily, and
their functions are all mediated by the same CXC receptor, CXCR2. A
number of studies have demonstrated that CXCL3, CXCL5 and CXCL8
serve crucial roles in the oncogenic potential of various types of
tumor (22–25). For example, the investigation of
CXCL3 in prostate cancer in the present study demonstrated that
CXCL3 is highly expressed in tumor tissues, and both the exogenous
administration and overexpression of CXCL3 have been shown to
promote tumor proliferation and migratory capabilities through the
ERK/AKT signaling pathway (26,27).
Within the TME, both tumor cells and
tumor-associated cells are able to release a series of chemokines
that regulate the infiltration and activation of diverse immune
cell types, ultimately influencing the balance between
tumor-promoting and tumor-suppressing activities (8). Among the recruited immune cell
population, several exhibit antitumor activities, including
CD8+ T cells, type 1 T helper (TH1) cells,
multifunctional TH17 cells and natural killer cells. Additionally,
the recruited antigen-presenting cells (APCs), including
macrophages and dendritic cells (DCs), contribute to tumor
regression through activating and amplifying local immune effector
cells (8). On the other hand, other
recruited immune cells, including myeloid-derived suppressor cells,
regulatory T cells, IL-22+ CD4+ TH22 cells,
IL-22+ innate lymphoid cells and plasmacytoid DCs, have
been demonstrated to accelerate tumor progression through fostering
angiogenesis, suppressing antitumor immune responses and
maintaining tumor stemness (8). In
the present study, a negative correlation between CXCL1 expression
and B cell infiltration was demonstrated in RCC, whereas a positive
correlation was observed with the attraction of CD4+ T
cells and neutrophils. It is considered that B cells, possessing
the capacity to generate antibodies, serve an important role as
crucial mediators in humoral immunity (28). Multiple chemokine/receptor systems,
including CCL21/CCR7, CXCL12/CXCR4 and CXCL13/CXCR5, are implicated
in the recruitment of B cells to the TME (29). A previous study has shown that B
cells exhibit antitumor activity through a multitude of mechanisms,
including the direct killing of tumor cells, the production of
specific antibodies against tumor antigens, acting as APCs to
induce T-cell activation and memory T-cell development, and
facilitating the immune responses of CD4+ and
CD8+ T cells (21).
Similarly, although previous studies have established that
neutrophils display antitumor properties through direct cell
toxicity, antibody-dependent cellular cytotoxicity and the
presentation of specific antigens (30), tumor-associated neutrophils notably
present tumor-promoting activities (31).
Subsequently, the functional experiments performed
in the present study showed that both the exogenous administration
and overexpression of CXCL1 led to a marked augmentation of the
proliferative and migratory abilities of RCC cells, whereas
decreased expression of CXCL1 had an inhibitory effect on these
tumorigenic behaviors. CXCL1 has been recognized as an important
autocrine/paracrine molecule in the TME for supporting tumor
progression. For example, silencing CXCL1 has been shown to
markedly reduce tumor proliferation through effectively inducing
apoptosis in hepatocellular carcinoma cells, suggesting that
autocrine signaling networks may serve a role in CXCL1-associated
tumor biology (32). Tumor cells
with higher CXCL1 levels have also been shown to trigger the
infiltration of tumor-associated macrophages (TAMs) and
cancer-associated fibroblasts (CAFs) into the TME. CXCL1 derived
from TAMs and CAFs serves a crucial role in regulating
intercellular adhesion and crosstalk between tumor cells and these
stromal cells, further enhancing tumor proliferation in a paracrine
manner (33). Ultimately, the
findings of the present study have revealed that the high
expression of CXCL1 in RCC cells exerts a strong modulatory role on
the expression patterns of proteins associated with the PI3K
signaling pathway, including Bax, Bcl-2, PI3K and AKT. The PI3K
pathway is a crucial signaling cascade within the body, and an
abundance of evidence has shown that the activation of the PI3K
signaling pathway is one of the most prevalent events in human
cancers, being heavily implicated in the development and
progression of tumors through its role in regulating cell growth,
survival, proliferation and migration (34).
In conclusion, the present study demonstrated that
CXCL1 is upregulated in RCC, and this upregulation is positively
associated with various clinicopathological factors, as well as
tumor-associated chemokine expression and immune cell recruitment.
Furthermore, functional studies indicated that overexpression of
CXCL1 contributes to the malignant behaviors of RCC cells, which
are partly mediated via the PI3K/AKT signaling pathway. However,
although the present study has preliminarily elucidated the role of
CXCL1 in RCC, the authors acknowledge that these findings require
validation through performing in vivo experiments in the
future, and this represents a key limitation of the current
research. Nevertheless, in spite of this limitation, the present
study has not only deepened the understanding of the mechanisms
underlying RCC, but has also offered crucial insights into, and
potential approaches for, the diagnosis and treatment of this
disease, highlighting its notable clinical and translational
implications.
Acknowledgements
Not applicable.
Funding
The study was supported by Grants from Joint Cultivation Project
of the Natural Science Foundation of Heilongjiang, China (grant no.
PL2024H003).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HQ carried out the experimental design and edited
the manuscript. SG and CL performed the cellular and molecular
biology experiments. HX performed the bioinformatic analysis. SG
drafted the initial manuscript. CL carried out immunohistochemical
experiments and statistical analyses. HZ and YW performed cellular
experiments. BW carried out molecular biology experiments. SG and
HZ confirm the authenticity of all the raw data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of School of Clinical Medicine, Jiamusi University
(approval no. 202315; ethical approval period from 2023 to 2025),
and was performed in accordance with the Declaration of Helsinki.
Informed written consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI
|
|
2
|
Baldewijns MM, van Vlodrop IJ, Schouten
LJ, Soetekouw PM, de Bruïne AP and van Engeland M: Genetics and
epigenetics of renal cell cancer. Biochim Biophys Acta.
1785:133–155. 2008.PubMed/NCBI
|
|
3
|
Linehan WM, Schmidt LS, Crooks DR, Wei D,
Srinivasan R, Lang M and Ricketts CJ: The metabolic basis of kidney
cancer. Cancer Discov. 9:1006–1021. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rijnders M, de Wit R, Boormans JL, Lolkema
MPJ and van der Veldt AAM: Systematic review of immune checkpoint
inhibition in urological cancers. Eur Urol. 72:411–423. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lalani AA, McGregor BA, Albiges L,
Choueiri TK, Motzer R, Powles T, Wood C and Bex A: Systemic
treatment of metastatic clear cell renal cell carcinoma in 2018:
Current paradigms, use of immunotherapy, and future directions. Eur
Urol. 75:100–110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rot A and von Andrian UH: Chemokines in
innate and adaptive host defense: Basic chemokinese grammar for
immune cells. Annu Rev Immunol. 22:891–928. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Griffith JW, Sokol CL and Luster AD:
Chemokines and chemokine receptors: Positioning cells for host
defense and immunity. Annu Rev Immunol. 32:659–702. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ozga AJ, Chow MT and Luster AD: Chemokines
and the immune response to cancer. Immunity. 54:859–874. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bule P, Aguiar SI and Aires-Da-Silva Fal
Dias JN: Chemokine-directed tumor microenvironment modulation in
cancer immunotherapy. Int J Mol Sci. 22:98042021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Märkl F, Huynh D, Endres Sal and Kobold S:
Utilizing chemokines in cancer immunotherapy. Trends Cancer.
8:670–682. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O'Hayer KM, Brady DC and Counter CM:
ELR+CXC chemokines and oncogenic Ras-mediated tumorigenesis.
Carcinogenesis. 30:1841–1847. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Korbecki J, Bosiacki M, Barczak K, Łagocka
R, Brodowska A, Chlubek D and Baranowska-Bosiacka I: Involvement in
tumorigenesis and clinical significance of CXCL1 in reproductive
cancers: breast cancer, cervical cancer, endometrial cancer,
ovarian cancer and prostate cancer. Int J Mol Sci. 24:72622023.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao M, Tabuchi H, Nagashima Y, Baba M,
Nakaigawa N, Ishiguro H, Hamada K, Inayama Y, Kishida T, Hattori K,
et al: Gene expression analysis of renal carcinoma: Adipose
differentiation-related protein as a potential diagnostic and
prognostic biomarker for clear-cell renal carcinoma. J Pathol.
205:377–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wozniak MB, Le Calvez-Kelm F,
Abedi-Ardekani B, Byrnes G, Durand G, Carreira C, Michelon J,
Janout V, Holcatova I, Foretova L, et al: Integrative genome-wide
gene expression profiling of clear cell renal cell carcinoma in
Czech Republic and in the United States. PLoS One. 8:e578862013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Richmond A and Thomas HG: Melanoma growth
stimulatory activity: Isolation from human melanoma tumors and
characterization of tissue distribution. J Cell Biochem.
36:185–198. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schumacher C, Clark-Lewis I, Baggiolini M
and Moser B: High- and low-affinity binding of GRO alpha and
neutrophil-activating peptide 2 to interleukin 8 receptors on human
neutrophils. Proc Natl Acad Sci USA. 89:10542–10546. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhuo C, Ruan Q, Zhao X, Shen Y and Lin R:
CXCL1 promotes colon cancer progression through activation of
NF-κB/P300 signaling pathway. Biol Direct. 17:342022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Man X, Yang X, Wei Z, Tan Y, Li W, Jin H
and Wang B: High expression level of CXCL1/GROα is linked to
advanced stage and worse survival in uterine cervical cancer and
facilitates tumor cell malignant processes. BMC Cancer. 22:7122022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Purdue MP, Rhee J, Moore L, Gao X, Sun X,
Kirk E, Bencko V, Janout V, Mates D, Zaridze D, et al: Differences
in risk factors for molecular subtypes of clear cell renal cell
carcinoma. Int J Cancer. 149:1448–1454. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Do HTT, Lee CH and Cho J: Chemokines and
their receptors: Multifaceted roles in cancer progression and
potential value as cancer prognostic markers. Cancers (Basel).
12:2872020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qi YL, Li Y, Man XX, Sui HY, Zhao XL,
Zhang PX, Qu XS, Zhang H, Wang BX, Li J, et al: CXCL3
overexpression promotes the tumorigenic potential of uterine
cervical cancer cells via the MAPK/ERK pathway. J Cell Physiol.
235:4756–4765. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chow MT and Luster AD: Chemokines in
cancer. Cancer Immunol Res. 2:1125–1131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ha H, Debnath B and Neamati N: Role of the
CXCL8-CXCR1/2 axis in cancer and inflammatory diseases.
Theranostics. 7:1543–1588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang W, Wang H, Sun M, Deng X, Wu X, Ma
Y, Li M, Shuoa SM, You Q and Miao L: CXCL5/CXCR2 axis in tumor
microenvironment as potential diagnostic biomarker and therapeutic
target. Cancer Commun (Lond). 40:69–80. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gui SL, Teng LC, Wang SQ, Liu S, Lin YL,
Zhao XL, Liu L, Sui HY, Yang Y, Liang L, et al: Overexpression of
CXCL3 can enhance the oncogenic potential of prostate cancer. Int
Urol Nephrol. 48:701–709. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xin H, Cao Y, Shao ML, Zhang W, Zhang CB,
Wang JT, Liang LC, Shao WW, Qi YL, Li Y, et al: Chemokine CXCL3
mediates prostate cancer cells proliferation, migration and gene
expression in an autocrine/paracrine fashion. Int Urol Nephrol.
50:861–868. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Largeot A, Pagano G, Gonder S, Moussay E
and Paggetti J: The B-side of cancer immunity: The underrated tune.
Cells. 8:4492019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vilgelm AE and Richmond A: Chemokines
modulate immune surveillance in tumorigenesis, metastasis, and
response to immunotherapy. Front Immunol. 10:3332019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sionov RV, Fridlender ZG and Granot Z: The
multifaceted roles neutrophils play in the tumor microenvironment.
Cancer Microenviron. 8:125–158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuan M, Zhu H, Xu J, Zheng Y, Cao X and
Liu Q: Tumor-Derived CXCL1 promotes lung cancer growth via
recruitment of tumor-associated neutrophils. J Immunol Res.
2016:65304102016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han KQ, He XQ, Ma MY, Guo XD, Zhang XM,
Chen J, Han H, Zhang WW, Zhu QG and Zhao WZ: Targeted silencing of
CXCL1 by siRNA inhibits tumor growth and apoptosis in
hepatocellular carcinoma. Int J Oncol. 47:2131–2140. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miyake M, Hori S, Morizawa Y, Tatsumi Y,
Nakai Y, Anai S, Torimoto K, Aoki K, Tanaka N, Shimada K, et al:
CXCL1-Mediated interaction of cancer cells with tumor-associated
macrophages and cancer-associated fibroblasts promotes tumor
progression in human bladder cancer. Neoplasia. 18:636–646. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Noorolyai S, Shajari N, Baghbani E,
Sadreddini S and Baradaran B: The relation between PI3K/AKT
signalling pathway and cancer. Gene. 698:120–128. 2019. View Article : Google Scholar : PubMed/NCBI
|