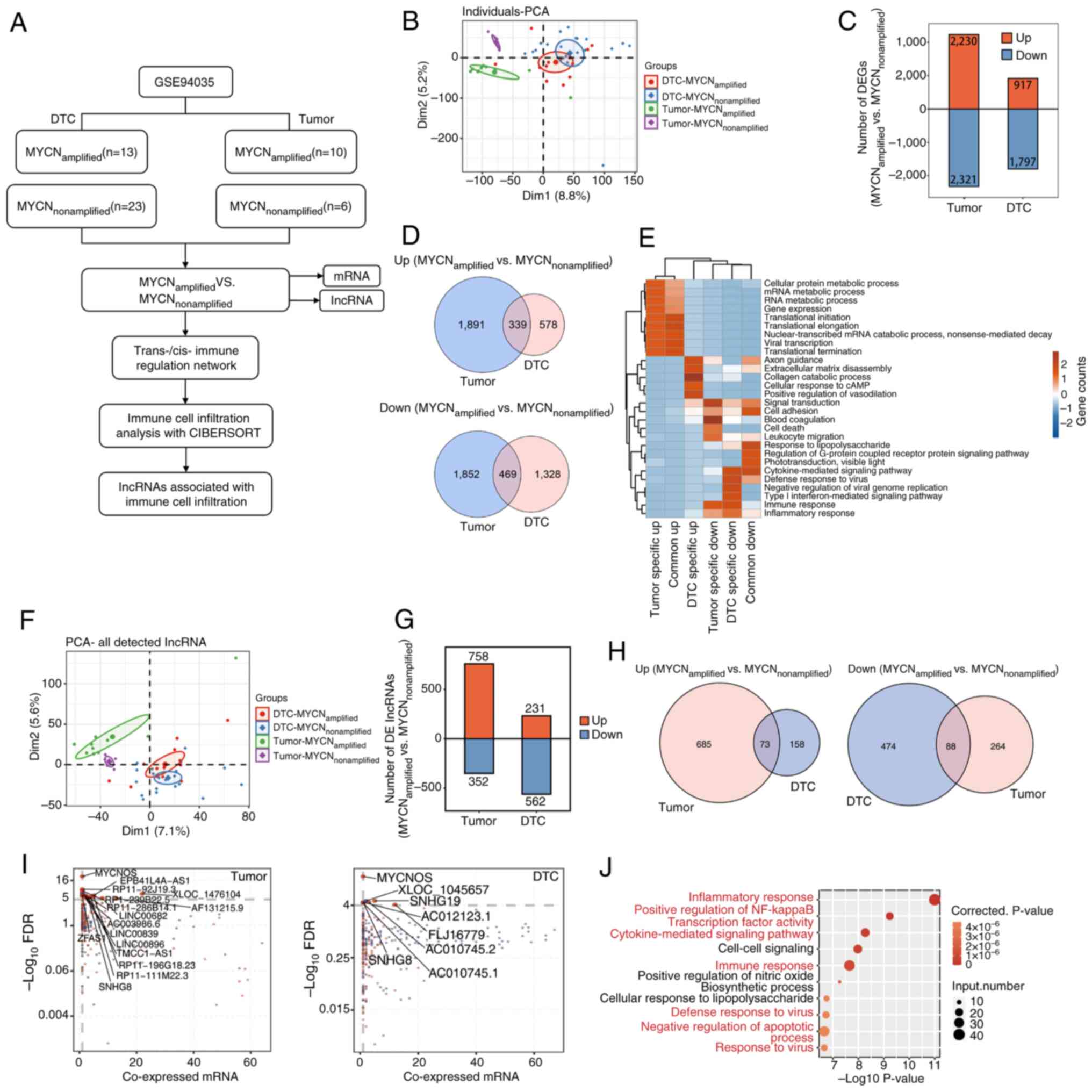
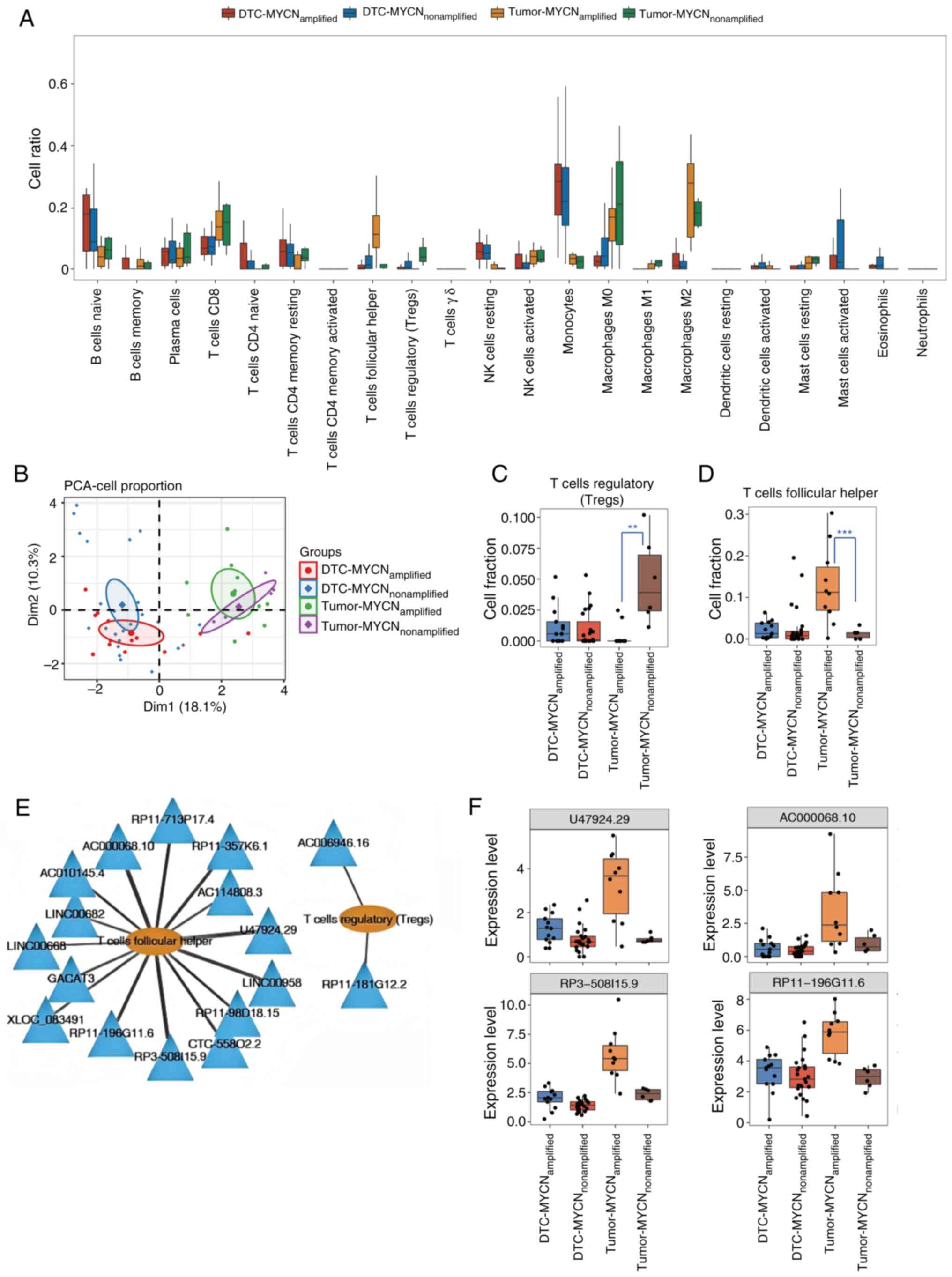
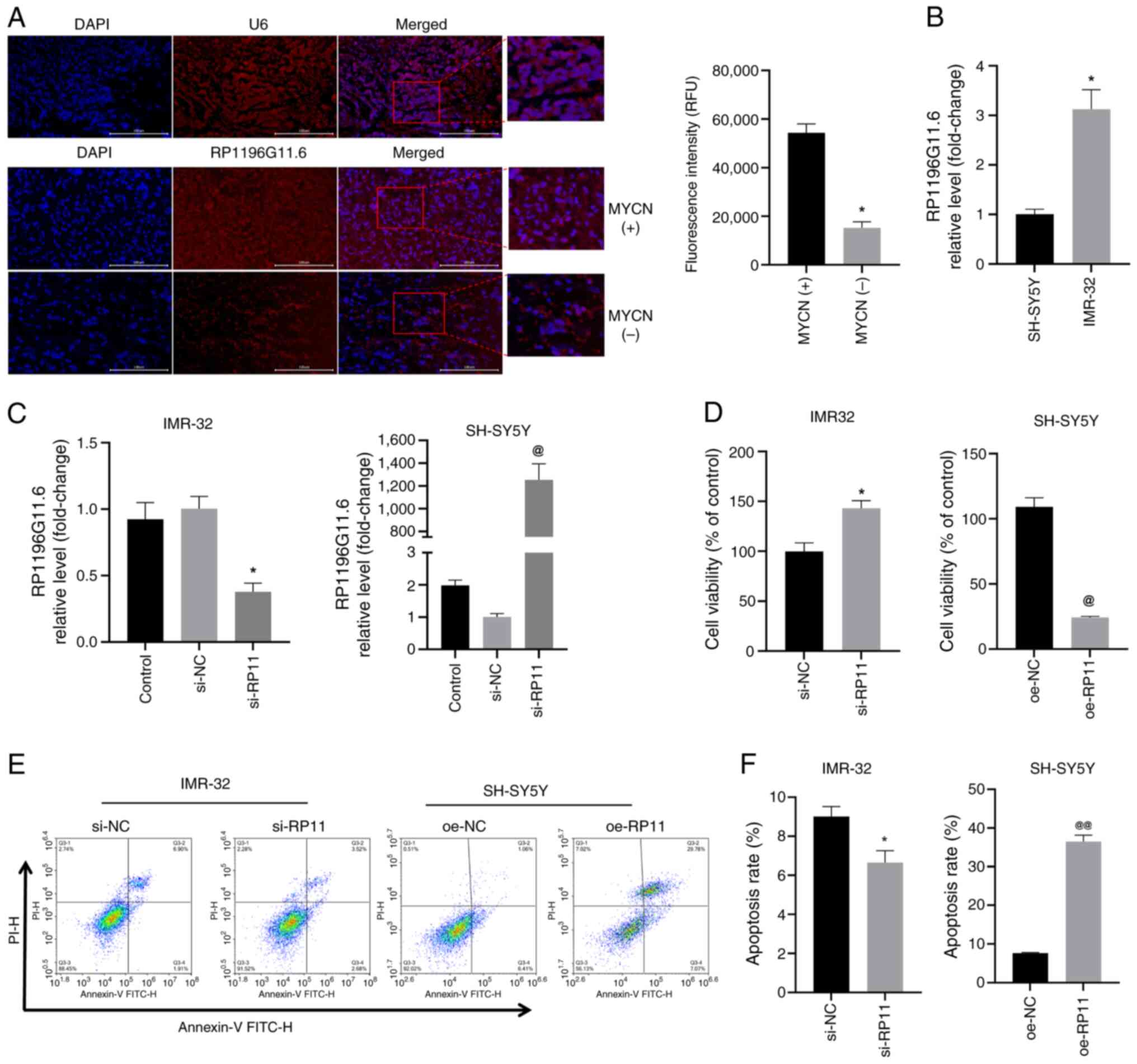
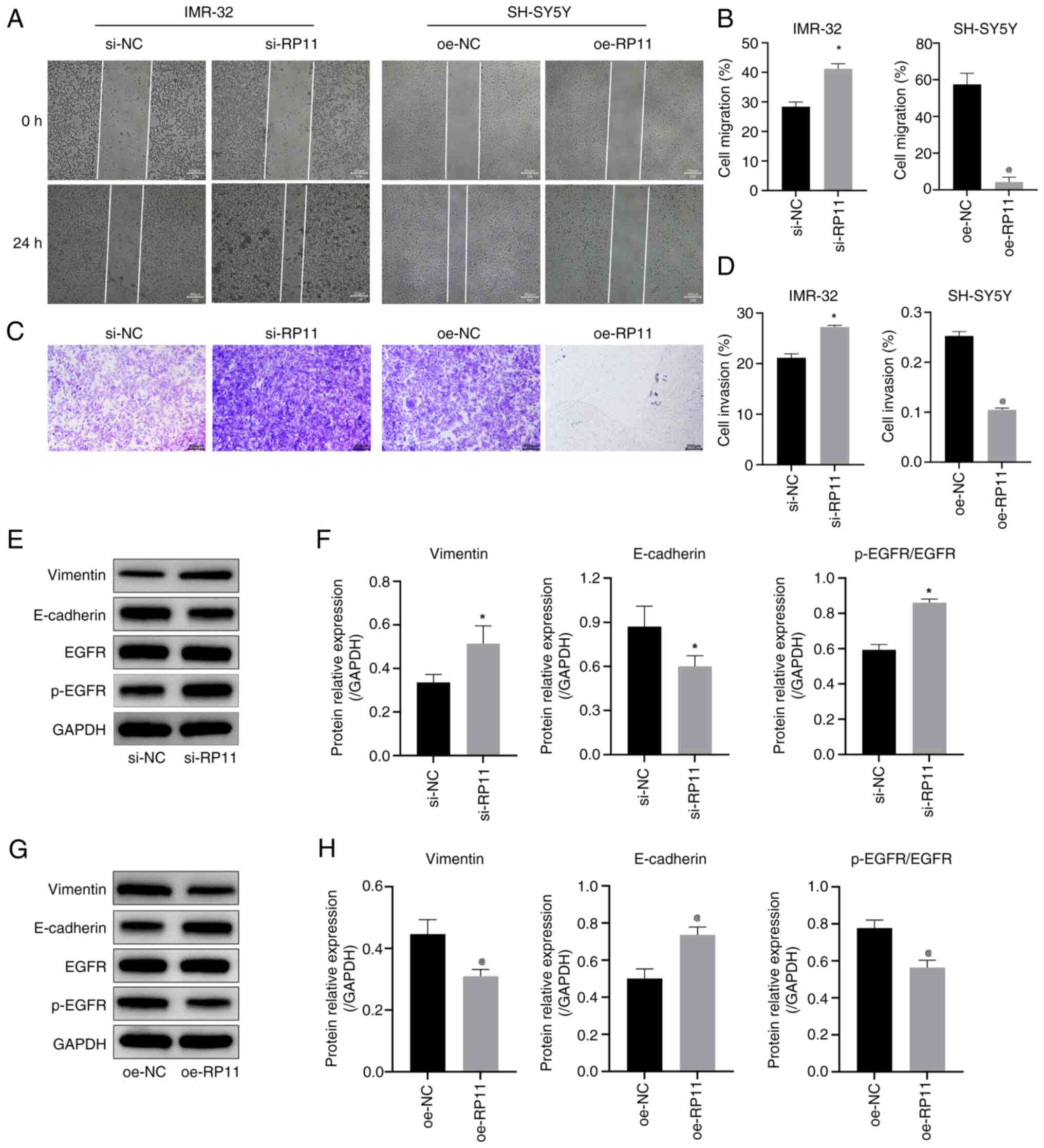
|
1
|
Irwin MS and Park JR: Neuroblastoma:
Paradigm for precision medicine. Pediatr Clin North Am. 62:225–256.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suh E, Stratton KL, Leisenring WM, Nathan
PC, Ford JS, Freyer DR, McNeer JL, Stock W, Stovall M, Krull KR, et
al: Late mortality chronic health conditions in long-term survivors
of early-adolescent, young adult cancers: A retrospective cohort
analysis from the childhood cancer survivor study. Lancet Oncol.
21:421–435. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ponzoni M, Bachetti T, Corrias MV,
Brignole C, Pastorino F, Calarco E, Bensa V, Giusto E, Ceccherini I
and Perri P: Recent advances in the developmental origin of
neuroblastoma: An overview. J Exp Clin Cancer Res. 41:922022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Körber V, Stainczyk SA, Kurilov R, Henrich
KO, Hero B, Brors B, Westermann F and Höfer T: Neuroblastoma arises
in early fetal development and its evolutionary duration predicts
outcome. Nat Genet. 55:619–630. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Monclair T, Brodeur GM, Ambros PF, Brisse
HJ, Cecchetto G, Holmes K, Kaneko M, London WB, Matthay KK,
Nuchtern JG, et al: The international neuroblastoma risk group
(INRG) staging system: An INRG task force report. J Clin Oncol.
27:298–303. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Castel V, García-Miguel P, Cañete A,
Melero C, Navajas A, Ruíz-Jiménez JI, Navarro S and Badal MD:
Prospective evaluation of the international neuroblastoma staging
system (INSS) and the international neuroblastoma response criteria
(INRC) in a multicentre setting. Eur J Cancer. 35:606–611. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shimada H, Ambros IM, Dehner LP, Hata J,
Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK and
Castleberry RP: The international neuroblastoma pathology
classification (the Shimada system). Cancer. 86:364–372. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pinto NR, Applebaum MA, Volchenboum SL,
Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F,
Schleiermacher G, Park JR, et al: Advances in risk classification
and treatment strategies for neuroblastoma. J Clin Oncol.
33:3008–3017. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Steliarova-Foucher E, Colombet M, Ries
LAG, Moreno F, Dolya A, Bray F, Hesseling P, Shin HY and Stiller
CA: IICC-3 contributors: International incidence of childhood
cancer, 2001-10: A population-based registry study. Lancet Oncol.
18:719–731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Diede SJ: Spontaneous regression of
metastatic cancer: Learning from neuroblastoma. Nat Rev Cancer.
14:71–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brodeur GM, Seeger RC, Schwab M, Varmus HE
and Bishop JM: Amplification of N-myc in untreated human
neuroblastomas correlates with advanced disease stage. Science.
224:1121–1124. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ruiz-Pérez MV, Henley AB and
Arsenian-Henriksson M: The MYCN protein in health and disease.
Genes (Basel). 8:1132017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang M and Weiss WA: Neuroblastoma and
MYCN. Cold Spring Harb Perspect Med. 3:a0144152013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taniue K and Akimitsu N: The functions and
unique features of LncRNAs in cancer development and tumorigenesis.
Int J Mol Sci. 22:6322021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang R, Liu N, Li T, Liu F, Zhang J, Zhao
H, Zou L and He X: LncRNA AC142119.1 facilitates the progression of
neuroblastoma by epigenetically initiating the transcription of
MYCN. J Transl Med. 21:6592023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu CL, Yin CF, Chang YW, Fan YC, Lin SH,
Wu YC, Huang HC and Juan HF: LncRNA SNHG1 regulates neuroblastoma
cell fate via interactions with HDAC1/2. Cell Death Dis.
13:8092022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu Z, Xu W, Wang H, Li M, Wang J, Sun C
and Yang X: CARM1-induced lncRNA NEAT1 synchronously activates MYCN
and GalNAcT-I to accelerate the progression of neuroblastoma. Gene.
938:1491642025. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu S, Wang Z, Chen D, Zhang B, Tian RR,
Wu J, Zhang Y, Xu K, Yang LM, Cheng C, et al: Annotation and
cluster analysis of spatiotemporal- and sex-related lncRNA
expression in rhesus macaque brain. Genome Res. 27:1608–1620. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sturm G, Finotello F and List M:
Immunedeconv: An R package for unified access to computational
methods for estimating immune cell fractions from bulk
RNA-sequencing data. Methods Mol Biol. 2120:223–232. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39((Web Server Issue)): W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan K, Zhang X, Cong X, Huang B, Chen H
and Chen D: Tumor suppressor RYBP harbors three nuclear
localization signals and its cytoplasm-located mutant exerts more
potent anti-cancer activities than corresponding wild type. Cell
Signal. 29:127–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu X, Yan M, Luo W, Liu W, Ren Y, Bei C,
Tang G, Chen R and Tan S: Expression and clinical significance of
PcG-associated protein RYBP in hepatocellular carcinoma. Oncol
Lett. 13:141–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morinaka T, Sakai N, Takayashiki T, Kuboki
S, Takano S, Ohira G, Matsubara H and Ohtsuka M: RYBP contributes
to improved prognosis in colorectal cancer via regulation of cell
cycle, apoptosis and oxaliplatin sensitivity. Int J Oncol.
63:1202023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zafar A, Wang W, Liu G, Wang X, Xian W,
McKeon F, Foster J, Zhou J and Zhang R: Molecular targeting
therapies for neuroblastoma: Progress and challenges. Med Res Rev.
41:961–1021. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qiu B and Matthay KK: Advancing therapy
for neuroblastoma. Nat Rev Clin Oncol. 19:515–533. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Floros KV, Cai J, Jacob S, Kurupi R,
Fairchild CK, Shende M, Coon CM, Powell KM, Belvin BR, Hu B, et al:
MYCN-amplified neuroblastoma is addicted to iron and vulnerable to
inhibition of the system Xc-/glutathione axis. Cancer Res.
81:1896–1908. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vaid R, Thombare K, Mendez A,
Burgos-Panadero R, Djos A, Jachimowicz D, Lundberg KI, Bartenhagen
C, Kumar N, Tümmler C, et al: METTL3 drives telomere targeting of
TERRA lncRNA through m6A-dependent R-loop formation: A therapeutic
target for ALT-positive neuroblastoma. Nucleic Acids Res.
52:2648–2671. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Heward JA and Lindsay MA: Long non-coding
RNAs in the regulation of the immune response. Trends Immunol.
35:408–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Otte J, Dyberg C, Pepich A and Johnsen JI:
MYCN function in neuroblastoma development. Front Oncol.
10:6240792021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Braoudaki M, Hatziagapiou K, Zaravinos A
and Lambrou GI: MYCN in neuroblastoma: ‘Old wine into new
wineskins’. Diseases. 78:782021. View Article : Google Scholar
|
|
36
|
Chen L and Tweddle DA: p53, SKP2, and DKK3
as MYCN target genes and their potential therapeutic significance.
Front Oncol. 28:1732012.PubMed/NCBI
|
|
37
|
Kobayashi K, Jakt LM and Nishikawa SI:
Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn.
Oncogene. 32:2640–2648. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Weiss WA, Aldape K, Mohapatra G,
Feuerstein BG and Bishop JM: Targeted expression of MYCN causes
neuroblastoma in transgenic mice. EMBO J. 16:2985–1995. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Winkle M, van den Berg A, Tayari M,
Sietzema J, Terpstra M, Kortman G, de Jong D, Visser L, Diepstra A,
Kok K and Kluiver J: Long noncoding RNAs as a novel component of
the Myc transcriptional network. FASEB J. 29:2338–2346. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Q, Wei J, Li N and Liu B: LINC00839
promotes neuroblastoma progression by sponging miR-454-3p to
Up-regulate NEUROD1. Neurochem Res. 47:2278–2293. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xin X, Xu Z, Wei J and Zhang Y:
MiR-376a-3p increases cell apoptosis in acute myeloid leukemia by
targeting MT1X. Cancer Biol Ther. 23:234–242. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Jiang F, Xiong Y, Cheng X, Qiu Z
and Song R: LncRNA TTN-AS1 sponges miR-376a-3p to promote
colorectal cancer progression via upregulating KLF15. Life Sci.
244:1169362020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xie F, Li L, Luo Y, Chen R and Mei J: Long
non-coding RNA LINC00488 facilitates thyroid cancer cell
progression through miR-376a-3p/PON2. Biosci Rep.
41:BSR202016032021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shan F, Somasundaram A, Bruno TC, Workman
CJ and Vignali DAA: Therapeutic targeting of regulatory T cells in
cancer. Trends Cancer. 8:944–961. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yan S, Zhang Y and Sun B: The function and
potential drug targets of tumour-associated Tregs for cancer
immunotherapy. Sci China Life Sci. 62:179:1862019. View Article : Google Scholar
|
|
46
|
Overacre-Delgoffe AE, Bumgarner HJ, Cillo
AR, Burr AHP, Tometich JT, Bhattacharjee A, Bruno TC, Vignali DAA
and Hand TW: Microbiota-specific T follicular helper cells drive
tertiary lymphoid structures and anti-tumor immunity against
colorectal cancer. Immunity. 54:2812–2824.e4. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou H, Li J, Zhang Z, Ye R, Shao N,
Cheang T and Wang S: RING1 and YY1 binding protein suppresses
breast cancer growth and metastasis. Int J Oncol. 49:2442–2452.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xian Y, Wang L, Yao B, Yang W, Mo H, Zhang
L and Tu K: MicroRNA-769-5p contributes to the proliferation,
migration and invasion of hepatocellular carcinoma cells by
attenuating RYBP. Biomed Pharmacother. 118:1093432019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ganesan K, Xu C, Wu S, Sui Y, Du B, Zhang
J, Gao F, Chen J and Tang H: Ononin inhibits tumor bone metastasis
and osteoclastogenesis by targeting mitogen-activated protein
kinase pathway in breast cancer. Research (Wash D C).
7:05532024.PubMed/NCBI
|
|
50
|
Zeng Y, Du W, Huang Z, Wu S, Ou X, Zhang
J, Peng C, Sun X and Tang H: Hsa_circ_0060467 promotes breast
cancer liver metastasis by complexing with eIF4A3 and sponging
miR-1205. Cell Death Discov. 9:1532023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Han M, Yang J, Chen P, Li S, Tang H, Fan
H, Wang Y, Li X, Pan W, Koutouratsas V, et al: Isocucurbitacin B
inhibits gliomas through the promotion of anoikis by targeting
caveolin 1. Cancer Lett. 629:2178732025. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dinglin X, Ding L, Li Q, Liu Y, Zhang J
and Yao H: RYBP inhibits progression and metastasis of lung cancer
by suppressing EGFR signaling and epithelial-mesenchymal
transition. Transl Oncol. 10:280–287. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tong AH, Tan J, Zhang JH, Xu FJ, Li FY and
Cao CY: Overexpression of RYBP inhibits proliferation, invasion,
and chemoresistance to cisplatin in anaplastic thyroid cancer cells
via the EGFR pathway. J Biochem Mol Toxicol. 33:e222412019.
View Article : Google Scholar : PubMed/NCBI
|