Introduction
The incidence rate and mortality of prostate cancer
(PCa) pose a huge threat to global health (1). The androgen receptor (AR) signaling
pathway, as the most important growth signaling axis, plays an
important role in PCa progression (2,3). This
is mainly closely related to the numerous downstream target genes
regulated by AR, which are widely involved in multiple tumor
characteristics of PCa (2).
Therefore, identification of novel AR downstream target genes and
clarification of their functions are expected to bring new insights
into PCa diagnosis and treatment. In the present study, it was
found that SENP5 acts as a downstream target of AR and
contributions to PCa growth.
SUMO-specific proteases (SENPs) mainly consist of 7
genes, namely SENP1, SENP2, SENP3, SENP5, SENP6, SENP7 and SENP8,
and their abnormal expression are closely related to the
development of multiple tumors (4).
Currently, the main research hotspots are SENP1 and SENP2 of this
family, and a large number of studies have shown that SENP1/2 can
act as oncogenes involving PCa progression (4,5).
Further research reveals that SENP1 and SENP2 are downstream target
genes of AR (6). However, the roles
of other genes in this family have not yet been clarified in
PCa.
Materials and methods
Ethics
The present study and all related procedures were
approved (approval no. 2021-1142) by the Ethics Committee of
Zhejiang University School of Medicine Second Affiliated Hospital
(Hangzhou, China). Prior to conducting the study, informed consent
forms were obtained from all patients with PCa participating in the
study.
Bioinformatics
JASPARA database (https://jaspar.elixir.no/) was used to predicted
sequences of transcription factor AR binding to the promoter of
SENPs genes.
Survival analysis
The survival analysis of SENPs was obtained from the
GEPIA 2.0 database (http://gepia.cancer-pku.cn/), including overall
survival (OS) and disease-free survival (DFS) in patients with
cancer. Patients were divided into two groups according to the
tumor types and the levels of SENPs from The Cancer Genome Atlas
(TCGA) database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga),
either lower or higher than the mean level. P<0.05 was
considered to indicate a significantly significant difference. The
95% confidence interval (CI) is presented as dotted line, hazards
ratio (HR).
Cell lines and cell culture
LNCaP cells (cat. no. CRL-1740) were purchased from
the American Type Culture Collection, cells were cultured in
RPMI-1640 media (cat. no. PYG0006; Wuhan Boster Biological
Technology, Ltd.) supplemented with 10% fetal bovine serum (FBS) in
37°C and 5% CO2 atmosphere. To mimic patients undergoing
ADT treatment, androgen-independent cell lines, had already been
generated: LNCaP-AI cells derived from androgen-sensitive LNCaP
cells cultured under androgen-depleted conditions (7). LNCaP-AI cells were cultured in
RPMI-1640 medium with 10% dextran-charcoal stripped FBS (CS-FBS;
Gibco; Thermo Fisher Scientific, Inc.).
Reagents and antibodies
Enzalutamide (cat. no. MDV3100) was purchased from
Selleck Chemicals and dihydrotestosterone (DHT; cat. no. A8380)
from MilliporeSigma. Antibody against GAPDH (cat. no. 5174) was
obtained from Cell Signaling Technology, Inc. Anti-human SENP5 (cat
no. 19529-1-AP) antibody was purchased from Proteintech Group, Inc.
Secondary antibodies [HRP-conjugated sheep anti-rabbit antibodies
(cat. no. AP510P) or HRP-conjugated sheep anti-mouse antibodies
(cat. no. AC111P)] for western blotting were obtained from
MilliporeSigma. DyLight 488 AffiniPure Goat Anti-Rabbit IgG (H+L)
for immunofluorescence were obtained from Abbkine Scientific Co.,
Ltd.
Immunofluorescence staining of
tissues
Immunofluorescence staining with SENP5 was performed
as previously described (8).
Transfection
LNCaP and LNCaP-AI cells were seeded in six-well
plates (5×105/well) and transfected with the siRNAs (100
nM) using Lipofectamine 2000® according to the
manufacturer's instructions (Thermo Fisher Scientific, Inc.). The
siRNAs used in the study were as follows: siSENP5 #1:
5′-CCAACACTTGTGCATTCTGAA-3′; siSENP5 #2:
5′-CCTTACCAGAACATCGTTCTA-3′; siAR: 5′-GGAACTCGATCGTATCATTGC-3′;
negative control: 5′-UUCUCCGAACGUGUCACGUTT-3′. Cell transfection
was performed for 6 h at 37°C, after which the medium was replaced.
After 48 h, the cells were collected for subsequent
experiments.
Western blot analysis
Total protein of cells was extracted with RIPA
buffer (Beyotime Institute of Biotechnology), and western blot
analysis was performed as previously described (9).
Cell Counting Kit-8 assay
LNCaP and LNCaP-AI cells were transfected with siScr
or siSENP5 for 48 h. Cell proliferation assay was performed as
previously described (9).
Reverse transcription-quantitative
PCR
RNA isolation and qPCR were performed as previously
described (8). Primers used were as
follows: SENP5 forward, 5′-CTTTAGGTCAGGCCAATGGTC-3′ and reverse,
5′-CAGCAGCCGTAACAAAAGCC-3; and GAPDH forward,
5′-AACAGCCTCAAGATCATCAGCA-3′ and reverse,
5′-CATGAGTCCTTCCACGATACCA-3′.
Chromatin
immunoprecipitation-quantitative PCR assays (ChIP-qPCR)
ChIP-qPCR was performed as previously described
(10). Cells (5×107)
were collected, followed by ChIP assays with anti-AR (Cell
Signaling Technology, Inc.; cat. no. 5153) or IgG (Proteintech
Group, Inc.; cat no. 30000-0-AP). qPCR analysis was carried out on
ChIPed and input DNA. Data are presented as percent of input DNA,
and error bars represent S.D. for technical duplicates of the qPCR
analysis. Primer sequences to detect the AR binding site along the
SENP5 promoter were as follows: SENP5-ChIP-F1:
TGGGGCGGGTAAGACATAGA; SENP5-ChIP-R1: CCATTCCAGCTCTGGACGTT.
Statistical analysis
Data were analyzed with GraphPad Prism 8
(Dotmatics). The values were shown as the mean ± S.D. for
triplicate experiments and the statistical differences were
calculated by one-way ANOVA analysis of variance with Dunnett's
test or Newman-Keuls test and Student's two-tailed t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
SENPs are potential downstream target
genes of transcription factor AR
In order to clarify the expression and potential
clinical values of SENPs in PCa, firstly, the mRNA expression level
of SENPs were analyzed in PCa and normal tissues using the TCGA
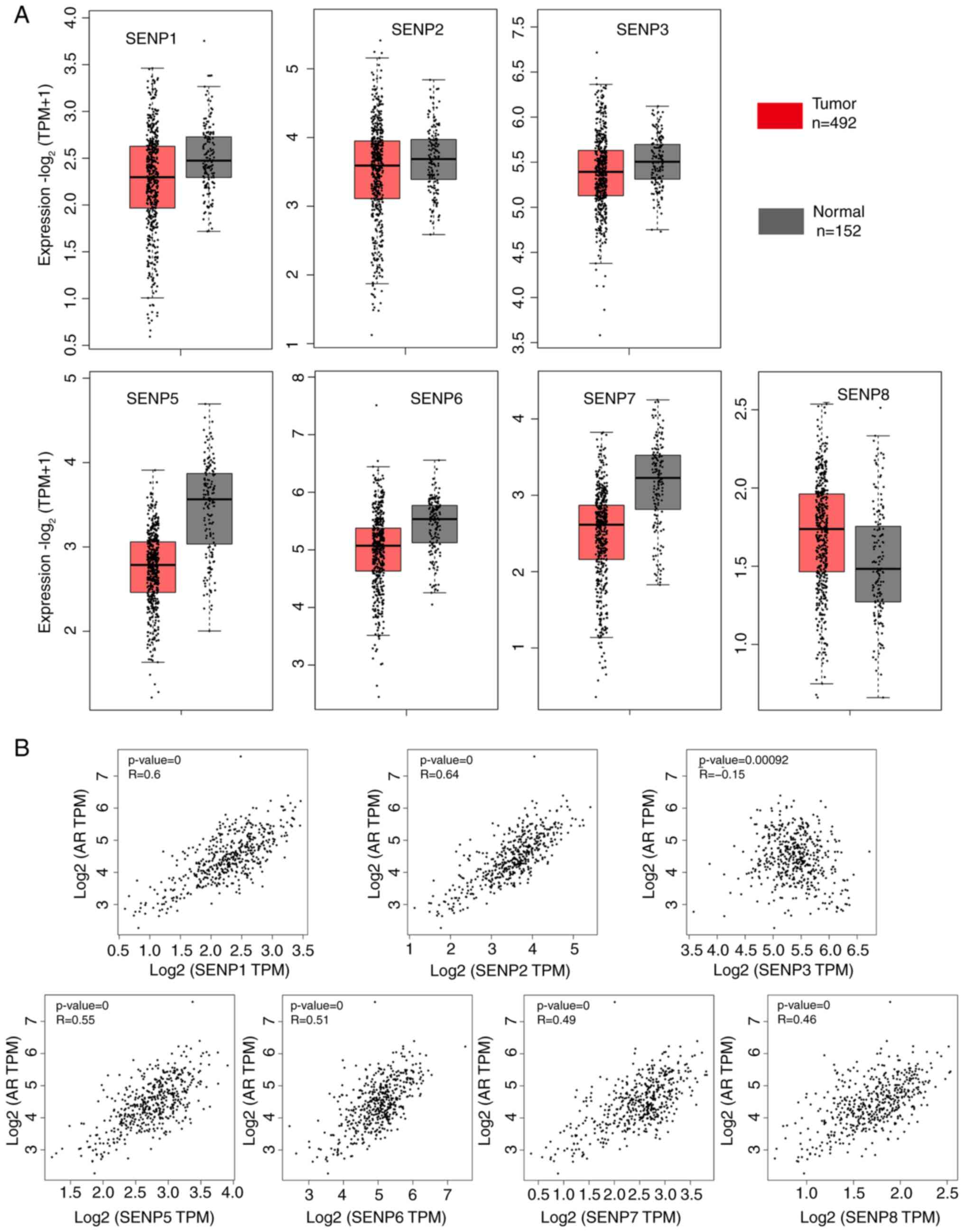
database. As revealed in Fig. 1A,
the expression level of SENPs were almost no difference between PCa
and normal tissues. However, interestingly, significant
co-expression between SENPs and AR was found (Fig. 1B), except for SENP3, strongly
indicating a potential regulatory relationship between SENPs and
AR. It has been previously reported that AR can transcriptionally
regulate SENP1 and SENP2 (6).
Therefore, it was hypothesized that AR may also be able to
transcriptionally regulate other genes in the SENP family. Using
the public online tool JASPARA, an interaction was found between
the promoter regions of SENPs genes and the transcription factor AR
(Table SI, Table SII, Table SIII, Table SIV, Table SV, Table SVI, Table SVII). These data indicated that
SENPs are potential downstream target genes of transcription factor
AR.
SENP5 acts as a downstream target of
AR and contributes to PCa growth
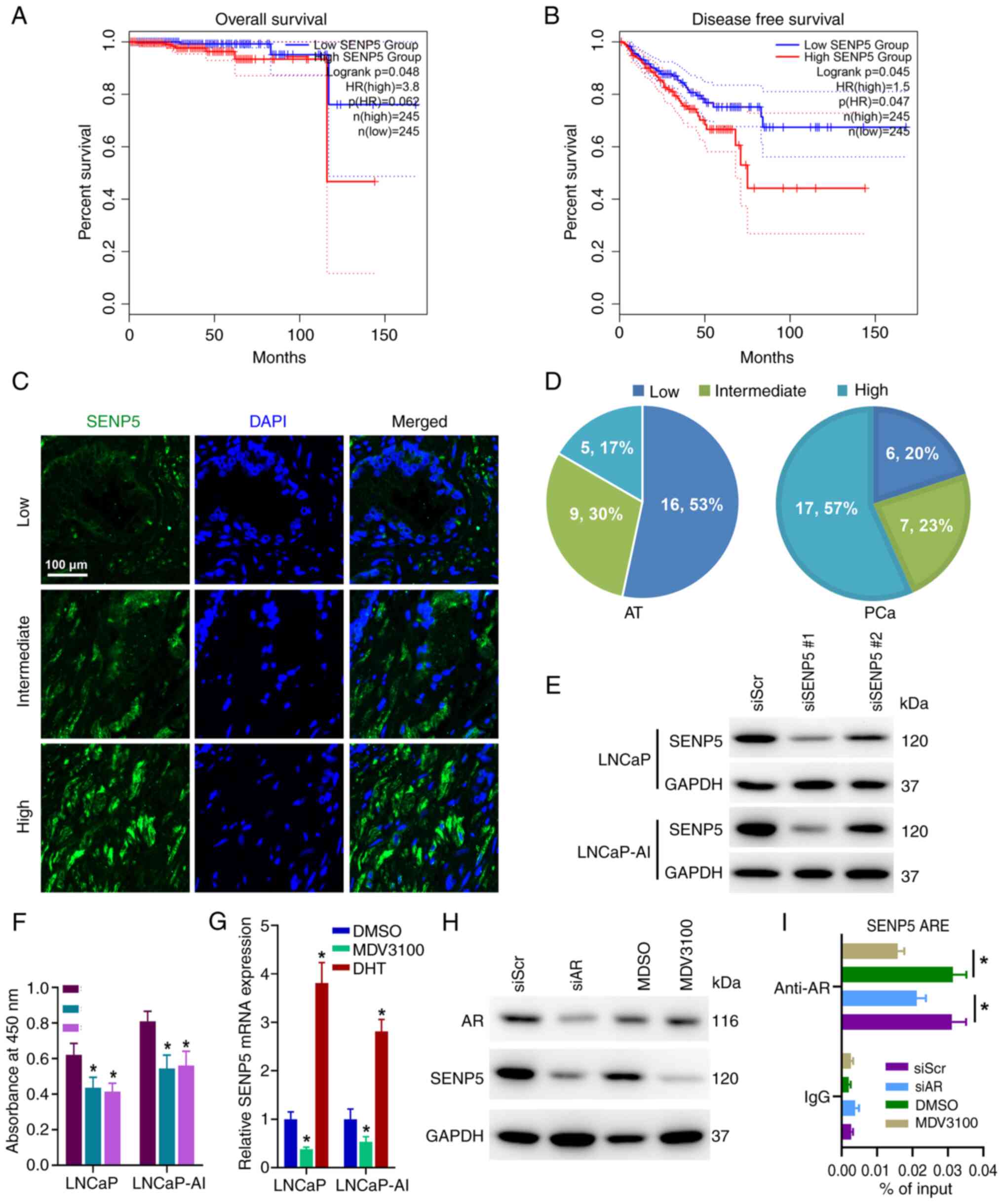
Despite there was almost no difference in the
expression level of SENPs between PCa and normal tissues, it was
attempted to further clarify whether the expression of SENPs is
related to the prognosis of PCa. Using the TCGA database, it was
found that the mRNA expression of SENP5 was significantly
negatively associated with OS (Fig.
2A) and DFS (Fig. 2B) in PCa,
while other genes in the SENP family were not markedly associated
with OS and DFS (Fig. S1),
indicating that high expression of SENP5 is associated with poor
prognosis in PCa. Furthermore, it was found that the protein level
of SENP5 was markedly higher in PCa tissues compared with normal
adjacent tissues through immunofluorescence analysis (Fig. 2C and D). In addition, SENP5
silencing significantly inhibited the proliferation activity of
LNCaP and LNCaP AI cells (Fig. 2E and
F). These results suggested that SENP5 may act as an oncogene
in PCa.
To further illuminate the regulatory mechanism
between AR and SENP5, PCa cell lines (LNCaP and LNCaP-AI) were
treated with MDV3100 and DHT respectively. DHT, the most potent
androgen, is usually synthesized in the prostate from testosterone
secreted by the testis, which acts as the activating ligand of AR
(11). Furthermore, MDV3100 is an
antagonist of AR that is widely used in the treatment of patients
with PCa (12). Compared with the
control group, qPCR results showed that the mRNA level of SENP5 was
significantly reduced after MDV3100 treatment (Fig. 2G). On the contrary, the mRNA level
of SENP5 significantly increased after DHT treatment (Fig. 2G). Consistent with the mRNA results,
similar results were also obtained at the protein level (Fig. 2H). In addition, it was further
confirmed that AR could transcriptionally activate SENP5 expression
using CHIP-qPCR (Fig. 2I). These
results further confirmed that SENP5 is indeed a direct downstream
target gene of AR.
Discussion
SUMO-mediated signaling is a proteome regulatory
pathway that plays a key role in maintaining cellular physiological
functions (13,14). Aberrant SENPs expression promotes
the development and progression of multiple cancers, including
colon cancer (15), ovarian cancer
(16), and breast cancer (17). In the present study, it was
demonstrated that SENP5 protein level was markedly elevated in PCa
tissues compared to normal adjacent tissues. Moreover, knocking
down SENP5 significantly inhibited the growth activity of PCa
cells, indicating that SENP5 may act as an oncogene in PCa.
High-level expression of SENP5 is significantly associated with
poor prognosis, further suggests that SENP5 is an oncogene in PCa
(Fig. 2B). Our finding is
consistent with its role in promoting drug resistance in colon
cancer (18).
Furthermore, it was confirmed that SENP5 is a direct
downstream target gene of AR. Firstly, TCGA dataset showed
significant co-expression between SENP5 and AR (Fig. 1B). Using JASPARA database, we
further found an interaction between the promoter regions of SENP5
and AR, strongly indicating that SENP5 is a potential downstream
target gene of AR. Thus, we further verified the result using
CHIP-qPCR, as expected, SENP5 is indeed a direct downstream target
gene of AR (Fig. 2I). Our finding
further expands the types of SENPs targeted by AR, in addition to
SENP1 and SENP2 (6), but also
SENP5. To some extent, this also implies that inhibition of SENP5
may induce the expression of SENP1 and SENP2 as a negative feedback
mechanism to maintain the growth of PCa cells, and the combined
blockade of AR and SENP5 may be an improved therapeutic
strategy.
Taken together, our results suggest that SENP5 is a
novel downstream target gene of AR, and its mRNA level is
significantly associated with poor prognosis in PCa, indicating
that SENP5 can participate in the occurrence and development of PCa
as an oncogene, the underlying molecular mechanism of which needs
further investigation. Thus, SENP5 has great potential to become a
new target for the diagnosis and treatment of PCa.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank the Clinical
Research Center, Zhejiang University School of Medicine Second
Affiliated Hospital for essential technical support and the use of
the laboratory to perform experiments.
Funding
The present study was supported by the National Natural Science
Foundation of China Youth Science Foundation Project (grant no.
81802571) and Zhejiang Medical and Health Science and Technology
Project (grant no. 2019RC039).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
WL and JZ designed the study. XC, XG and WL
performed the experiments. XC, XG and WL supervised specific
experiments and revised the manuscript. WL wrote the manuscript.
All authors have read and approved the final version of the
manuscript. WL and JZ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
All samples were collected from patients with
informed consent, and all related procedures were performed with
the approval of the internal review and ethics boards of Zhejiang
University School of Medicine Second Affiliated Hospital (approval
no. 2021-1142; Hangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
|
|
2
|
Mills IG: Maintaining and reprogramming
genomic androgen receptor activity in prostate cancer. Nat Rev
Cancer. 14:187–198. 2014. View
Article : Google Scholar
|
|
3
|
Dai C, Dehm SM and Sharifi N: Targeting
the androgen signaling axis in prostate cancer. J Clin Oncol.
41:4267–4278. 2023. View Article : Google Scholar
|
|
4
|
Claessens LA and Vertegaal ACO: SUMO
proteases: From cellular functions to disease. Trends Cell Biol.
34:901–912. 2024. View Article : Google Scholar
|
|
5
|
Chang HM and Yeh ETH: SUMO: From bench to
bedside. Physiol Rev. 100:1599–1619. 2020. View Article : Google Scholar
|
|
6
|
Kaikkonen S, Jääskeläinen T, Karvonen U,
Rytinki MM, Makkonen H, Gioeli D, Paschal BM and Palvimo JJ:
SUMO-specific protease 1 (SENP1) reverses the hormone-augmented
SUMOylation of androgen receptor and modulates gene responses in
prostate cancer cells. Mol Endocrinol. 23:292–307. 2009. View Article : Google Scholar
|
|
7
|
Xu G, Wu J, Zhou L, Chen B, Sun Z, Zhao F
and Tao Z: Characterization of the small RNA transcriptomes of
androgen dependent and independent prostate cancer cell line by
deep sequencing. PLoS One. 5:e155192010. View Article : Google Scholar
|
|
8
|
Liu W, Wang X, Wang Y, Dai Y, Xie Y, Ping
Y, Yin B, Yu P, Liu Z, Duan X, et al: SGK1 inhibition-induced
autophagy impairs prostate cancer metastasis by reversing EMT. J
Exp Clin Cancer Res. 37:732018. View Article : Google Scholar
|
|
9
|
Liu W, Wang X, Wang Y, Dai Y, Xie Y, Ping
Y, Yin B, Yu P, Liu Z, Duan X, et al: SGK1 inhibition induces
autophagy-dependent apoptosis via the mTOR-Foxo3a pathway. Br J
Cancer. 117:1139–1153. 2017. View Article : Google Scholar
|
|
10
|
Liang D, Feng Y, Zandkarimi F, Wang H,
Zhang Z, Kim J, Cai Y, Gu W, Stockwell BR and Jiang X: Ferroptosis
surveillance independent of GPX4 and differentially regulated by
sex hormones. Cell. 186:2748–2764.e2722. 2023. View Article : Google Scholar
|
|
11
|
Chang KH, Li R, Kuri B, Lotan Y, Roehrborn
CG, Liu J, Vessella R, Nelson PS, Kapur P, Guo X, et al: A
gain-of-function mutation in DHT synthesis in castration-resistant
prostate cancer. Cell. 154:1074–1084. 2013. View Article : Google Scholar
|
|
12
|
Higano CS, Beer TM, Taplin ME, Efstathiou
E, Hirmand M, Forer D and Scher HI: Long-term safety and antitumor
activity in the phase 1–2 study of enzalutamide in pre- and
post-docetaxel castration-resistant prostate cancer. Eur Urol.
68:795–801. 2015. View Article : Google Scholar
|
|
13
|
Claessens LA, Verlaan-de Vries M, de Graaf
IJ and Vertegaal ACO: SENP6 regulates localization and nuclear
condensation of DNA damage response proteins by group
deSUMOylation. Nat Commun. 14:58932023. View Article : Google Scholar
|
|
14
|
Lin H, Suzuki K, Smith N, Li X, Nalbach L,
Fuentes S, Spigelman AF, Dai XQ, Bautista A, Ferdaoussi M, et al: A
role and mechanism for redox sensing by SENP1 in β-cell responses
to high fat feeding. Nat Commun. 15:3342024. View Article : Google Scholar
|
|
15
|
Wei M, Huang X, Liao L, Tian Y and Zheng
X: SENP1 decreases RNF168 phase separation to promote DNA damage
repair and drug resistance in colon cancer. Cancer Res.
83:2908–2923. 2023. View Article : Google Scholar
|
|
16
|
Zhang Y, Wei H, Zhou Y, Li Z, Gou W, Meng
Y, Zheng W, Li J, Li Y and Zhu W: Identification of potent SENP1
inhibitors that inactivate SENP1/JAK2/STAT signaling pathway and
overcome platinum drug resistance in ovarian cancer. Clin Transl
Med. 11:e6492021. View
Article : Google Scholar
|
|
17
|
Xiao M, Bian Q, Lao Y, Yi J, Sun X, Sun X
and Yang J: SENP3 loss promotes M2 macrophage polarization and
breast cancer progression. Mol Oncol. 16:1026–1044. 2021.
View Article : Google Scholar
|
|
18
|
Liu T, Wang H, Chen Y, Wan Z, Du Z, Shen
H, Yu Y, Ma S, Xu Y, Li Z, et al: SENP5 promotes homologous
recombination-mediated DNA damage repair in colorectal cancer cells
through H2AZ deSUMOylation. J Exp Clin Cancer Res. 42:2342023.
View Article : Google Scholar
|