Introduction
Hypopharyngeal cancer (HPC) is a highly aggressive
subtype of head and neck squamous cell carcinoma (HNSCC) developing
from the epithelial lining of the hypopharynx. In the United
States, HPC accounts for 3–5% of all head and neck malignancies and
is often diagnosed at an advanced stage due to its asymptomatic
early progression and the anatomical complexity of the hypopharynx
(1,2). Risk factors, including tobacco use,
excessive alcohol consumption and human papillomavirus infection,
contribute markedly to its pathogenesis (3,4). HPC
is clinically classified according to the TNM staging system;
early-stage disease (stage I and II) is often localized, while in
its advanced stages (stage III and IV), regional lymph node
involvement and distant metastasis are observed, leading to poor
prognosis (5). Due to the
aggressive nature of late-stage HPC, treatment strategies involve a
combination of surgery, radiotherapy and chemotherapy; however,
five-year survival rates have been reported to be 15–45%, due to
high recurrence and resistance to therapy (6).
The progression of HPC, at the molecular level, is
driven by a complex network of signaling pathways that regulate
tumor growth, angiogenesis and metastasis. A hallmark of HPC is
aberrant activation of the EGFR pathway, promoting uncontrolled
cell proliferation through downstream effectors such as
Ras-Raf-MEK-extracellular signal-regulated kinase and
phosphoinositide 3 kinase-mammalian target of rapamycin, which
promote tumor cell survival, proliferation and resistance to
apoptosis (7). The
epithelial-mesenchymal transition (EMT) process, mediated by
transforming growth factor-β, Wnt/β-catenin and Notch signaling, is
pivotal in enhancing tumor cell motility and invasiveness (8). Therefore, angiogenesis is a key factor
supporting metastasis, driven by hypoxia-inducible factor-1α
(HIF-1α) and vascular endothelial growth factor for
neovascularization, ensuring an adequate supply of oxygen and
nutrients to proliferating tumor cells (9). Increased angiogenic activity in
advanced HPC is associated with increased metastatic potential and
worse clinical outcomes (10).
Recent studies have highlighted the role of HIF-1α in promoting
metabolic reprogramming and angiogenesis, contributing to tumor
progression and resistance to therapy (11,12).
Suppression of interferon (IFN) signaling has been
identified as a key mechanism of immune evasion in HPC, reducing
the susceptibility of the tumor to immune surveillance.
IFN-stimulated genes (ISGs) are required for the enhancement of
immune recognition and antiviral responses, while they have been
reported to be frequently downregulated, impairing antitumor
immunity (13,14). The loss of IFN signaling not only
reduces immune-mediated tumor clearance but also promotes
resistance to immunotherapy.
The present study aimed to identify the
differentially expressed genes (DEGs) between parental FaDu HPC
cells and advanced FaDu (FaDuex) cells, which were isolated from a
late-stage xenograft tumor, were analyzed using the RNA-sequencing
(RNA-seq) technique.
Materials and methods
Cell lines
The human HPC FaDu cells (cat no. HTB-43TM; American
Type Culture Collection) and FaDuex cells were cultured in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) containing 10%
FBS (Thermo Fisher Scientific, Inc.), 1% penicillin/streptomycin
and 1% L-glutamine (both MilliporeSigma; Merck KGaA). The FaDu
cells were originally derived from a punch biopsy of a
hypopharyngeal tumor obtained from a 56-year-old White male patient
(15). FaDuex cells were previously
established and considered as late-stage or advanced HPC cells,
isolated from FaDu cells-derived xenograft tumors that reached
2,000 mm3 at 40 days (10). In brief, when the tumor size reached
600 mm3 exhibiting a necrotic region, the tumor was
excised and cultured in RPMI-1640 medium supplemented with
high-dose antibiotics, allowing viable cells to migrate from the
tumor mass. These cells have been demonstrated to be more
tumorigenic, exhibiting an increased rate of proliferation,
angiogenesis and invasive capacity (10). The biological characteristics of
FaDuex cells closely reflect the clinical features of human HPC
diagnosed at a late stage (16).
RNA extraction and RNA-seq
analysis
RNA was extracted from cells using the GENEZol™
TriRNA Pure Kit (cat. no. GZX100; Geneaid Biotech), according to
the manufacturer's instructions. Up to 5×106 cells were
harvested by centrifugation at 300 × g for 5 min at room
temperature and the culture medium was completely removed. The cell
pellet was thoroughly mixed in 700 µl of GENEzol™ reagent by
pipetting and incubated for 5 min at room temperature. Ethanol was
added in a 1:1 ratio to the lysate and the mixture was processed
through a spin column-based protocol. RNA was eluted using
RNase-free water and stored at −80°C until further use. For each
condition (FaDu and FaDuex), there were two biological replicates
and RNA was independently extracted from separate cultures, and
reproducibility was ascertained. RNA library preparation and
next-generation sequencing were performed by Biotools Co., Ltd.
using their RNA-Seq (Q)20M package (service code: B-IRQTNT-E20P).
Libraries were constructed using poly(A) mRNA enrichment and
sequenced with 150 bp paired-end reads on the Illumina platform.
All samples were processed and sequenced in a single batch.
The raw sequencing reads were processed for quality
control using Trimmomatic (version 0.38) by Biotools Co., Ltd. and
assessed using FastQC and MultiQC (17,18).
Clean reads were aligned using HISAT2 to the human reference genome
(GRCh38) and read counts for each gene were quantified using
‘featureCounts’. DEGs were analyzed by comparing the transcripts
per million (TPM) values of genes between FaDuex and FaDu
samples using ‘DESeq2’ to calculate fold-changes and statistical
significance of the outcomes (19).
Gene set enrichment analysis (GSEA) in ‘clusterProfiler’ (version
4.6.2, Bioconductor release 3.16) (20,21)
was performed to identify significant pathways in the Hallmark and
C2 gene sets from the Human Molecular Signatures Database (MSigDB;
h.all.v2023.2.Hs and c2.all.v2023.2.Hs) (22–24),
as well as other pathways of interest, including 23 downstream NFE2
like BZIP transcription factor 2 gene sets
[Transcription_Factor_PPIs (25),
Rummagene_transcription_factors (26), TRRUST_Transcription_Factors
(27) and TRANSFAC_and_JASPAR_PWMs
(28)] (26,28,29)
and 20 cancer stem cell-related gene sets (30) collected from Enrichr (25,31,32).
Only pathways with an adjusted P<0.05, as determined by
Benjamini-Hochberg correction, were considered.
Complementary DNA (cDNA) synthesis and
quantitative polymerase chain reaction (qPCR)
The cDNA was synthesized using the ToolsQuant II
Fast RT kit (cat. no. KRT-BA-06; TOOLS Biotech) following the
manufacturer's instructions. Briefly, after quantifying total RNA
using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher
Scientific, Inc.), 300 ng of RNA was used as the template for
reverse transcription. Genomic DNA contamination was removed by
incubating with gDNA Eraser at 42°C for 3 min, followed by reverse
transcription at 42°C for 15 min. The resulting cDNA was diluted
with 80 µl nuclease-free water and utilized directly for subsequent
qPCR analysis employing the StepOnePlus™ Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Primers used
for qPCR are summarized in Table I.
Each reaction was carried out in a final volume of 20 µl,
comprising 6 µl nuclease-free water, 2 µl primer mix (6 µM forward
primer and 6 µM reverse primer), 2 µl cDNA template and 10 µl,
EnTurbo™ SYBR Green PCR SuperMix (cat. no. EQ013; High ROX
Premixed; ELK Biotechnology). The thermal cycling program was as
follows: Initial denaturation at 95°C for 20 sec, followed by 40
cycles of denaturation at 95°C for 3 sec and annealing/extension at
60°C for 30 sec, during which fluorescence signals were recorded.
At the end of the amplification reaction, the melt curve was
analyzed to confirm product specificity. The expression of each
target gene, along with the internal control gene (S26), was
analyzed in four technical replicates. Each datum represented a
mean of four repeats and was compared using unpaired t-tests.
Relative expression levels were estimated based on amplification
curves (33).
 | Table I.Sequences of primers for each gene
analyzed by quantitative PCR. |
Table I.
Sequences of primers for each gene
analyzed by quantitative PCR.
| Gene | Primer sequence
(5′-3′) |
|---|
| KRT13 | F:
GATGCTGAGGAATGGTTCCACG |
|
| R:
AGCTCCGTGATCTCTGTCTTGC |
| IFI44L | F:
TGCACTGAGGCAGATGCTGCG |
|
| R:
TCATTGCGGCACACCAGTACAG |
| IFI6 | F:
TGATGAGCTGGTCTGCGATCCT |
|
| R:
GTAGCCCATCAGGGCACCAATA |
| STC1 | F:
GCAGGAAGAGTGCTACAGCAAG |
|
| R:
CATTCCAGCAGGCTTCGGACAA |
| MUC16 | F:
GATGTCAAGCCAGGCAGCACAA |
|
| R:
GAGAGTGGTAGACATTTCTGGGC |
| S26 | F:
CCGTGCCTCCAAGATGACAAAG |
|
| R:
ACTCAGCTCCTTACATGGGCTT |
Results and Discussion
Analysis of DEGs between parental and
advanced HPC cells isolated from late-stage xenograft tumors
The present study first analyzed the DEGs between
FaDu and FaDuex cells (advanced FaDu cells) to identify
transcriptional differences associated with their phenotypic
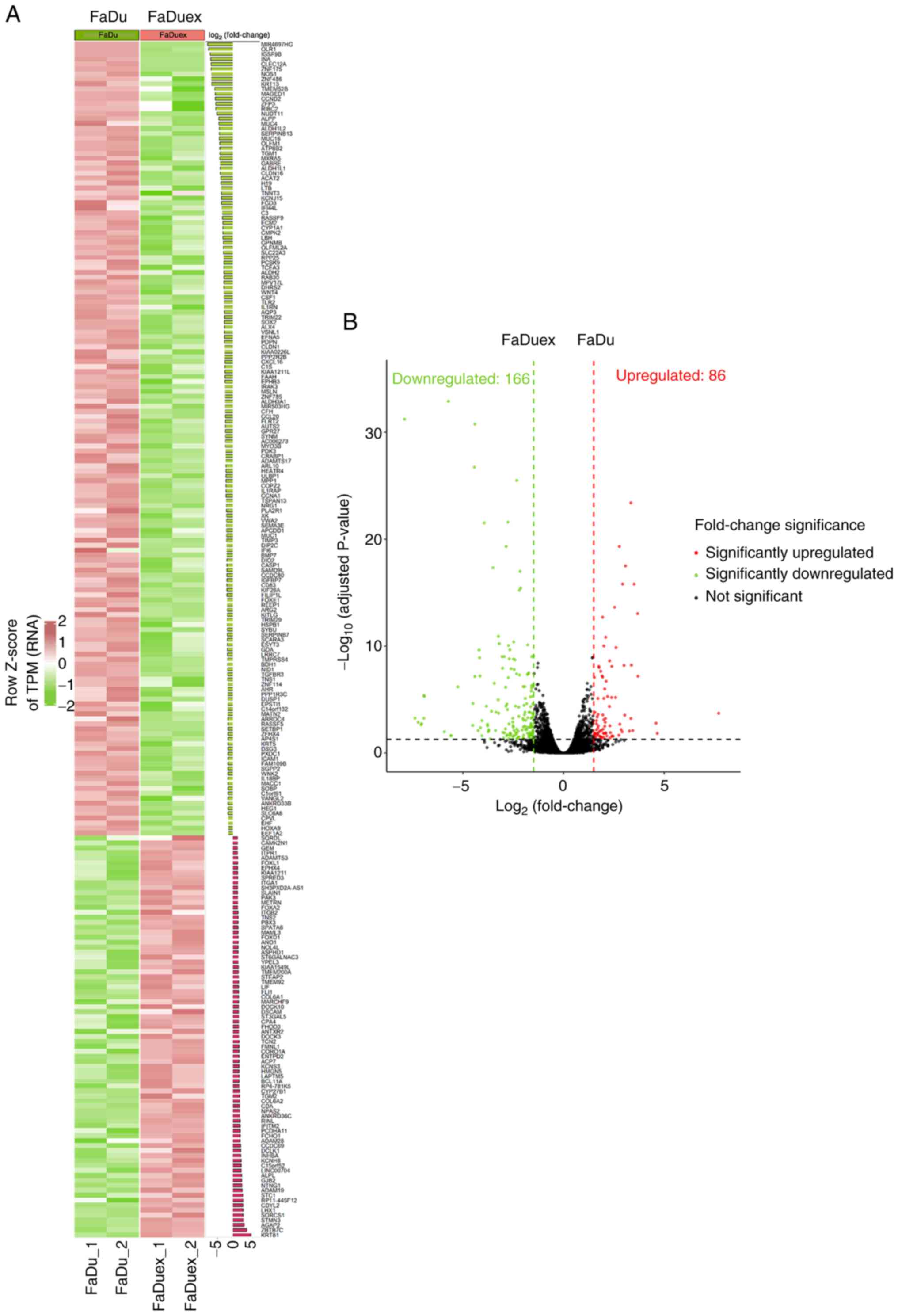
variation. The heatmap in Fig. 1A
presents distinct clustering patterns; genes upregulated in FaDuex
cells are displayed in red and the downregulated genes are
presented in green, indicating significant transcriptional
reprogramming (Fig. 1A). The
differences are further illustrated in the volcano plot (Fig. 1B). A total of 86 genes were
significantly upregulated, while 166 genes were significantly
downregulated in FaDuex cells relative to their expression in FaDu
cells (adjusted P<0.05). These findings suggest that the
expression levels of genes in FaDuex cells undergo significant
changes, potentially explaining their distinct biological
properties.
Comparison of specific signaling
pathways activated and suppressed in advanced HPC cells
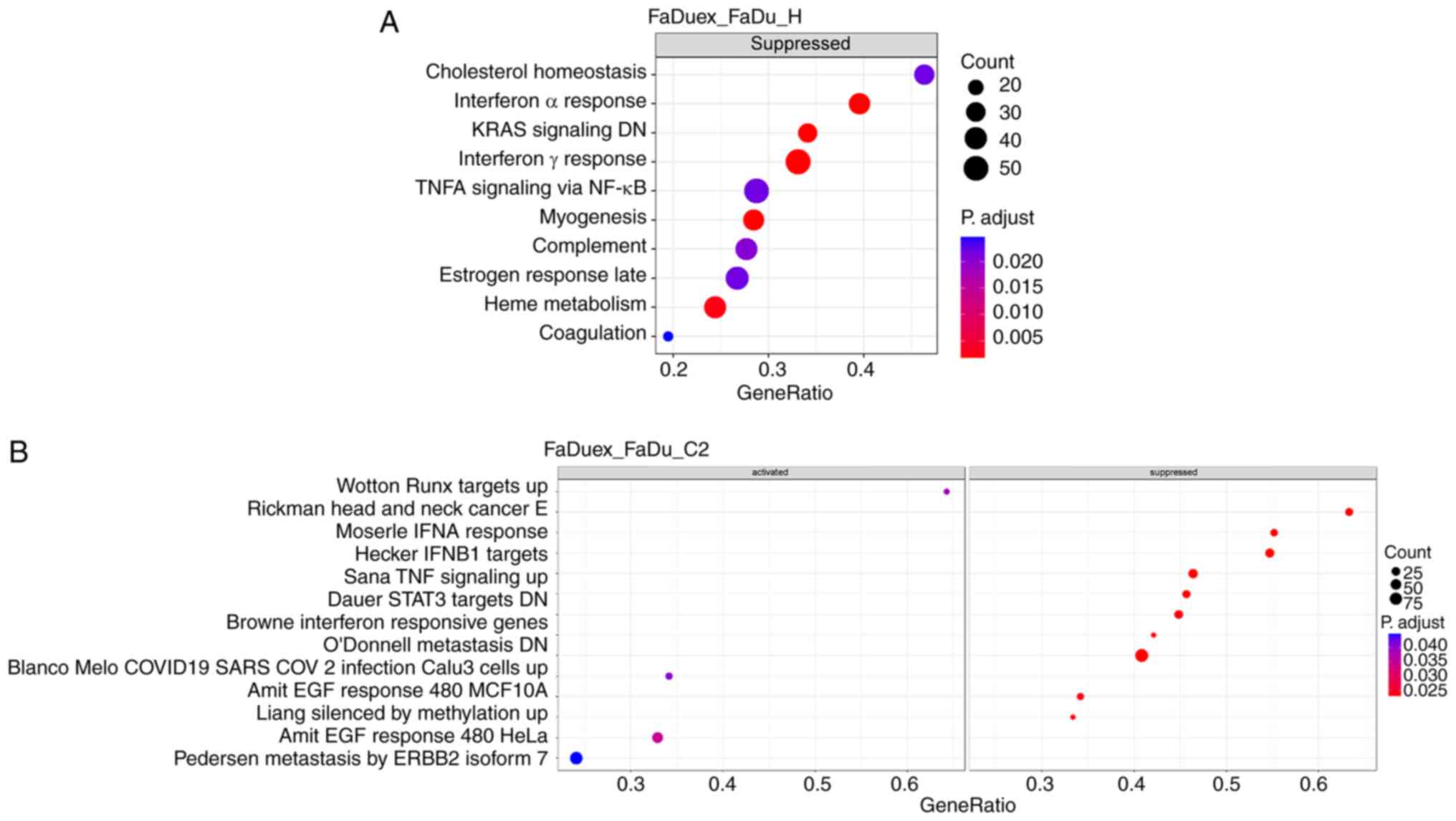
GSEA was performed next using the Hallmark and C2
gene sets to investigate the biological processes associated with
the DEGs between FaDuex and FaDu cells. Significantly enriched
pathways were subsequently categorized into activated and
suppressed groups. In the Hallmark gene set, only suppressed
enriched pathways were significant and the most suppressed pathways
included those involved in ‘IFNα response’, downregulated ‘KRAS
signaling’, ‘IFNγ response’, ‘myogenesis’ and ‘heme metabolism’
(Fig. 2A). In the C2 gene set with
significant changes, four enriched pathways (upregulated ‘Wotton
RUNX targets’, ‘AMIT EGF response’ in MCF10A and HeLa cells and
‘Pedersen metastasis by ERBB2 isoform’) were activated, while 10
pathways were suppressed, including those involved in ‘BROWNE IFN
response’ and ‘Pedersen IFN-α response’ (Fig. 2B). Since the IFN pathways are key
for antiviral defense and tumor immune surveillance (34,35),
their suppression in FaDuex cells possibly indicates an immune
evasion phenomenon, potentially facilitating tumor progression and
resistance to immune-mediated clearance. These findings highlighted
a significant suppression of IFN signaling in FaDuex cells,
potentially contributing to their altered immune landscape and
tumorigenic potential.
Gene regulations of advanced HPC cells
involved in different cancerous associated signaling pathways
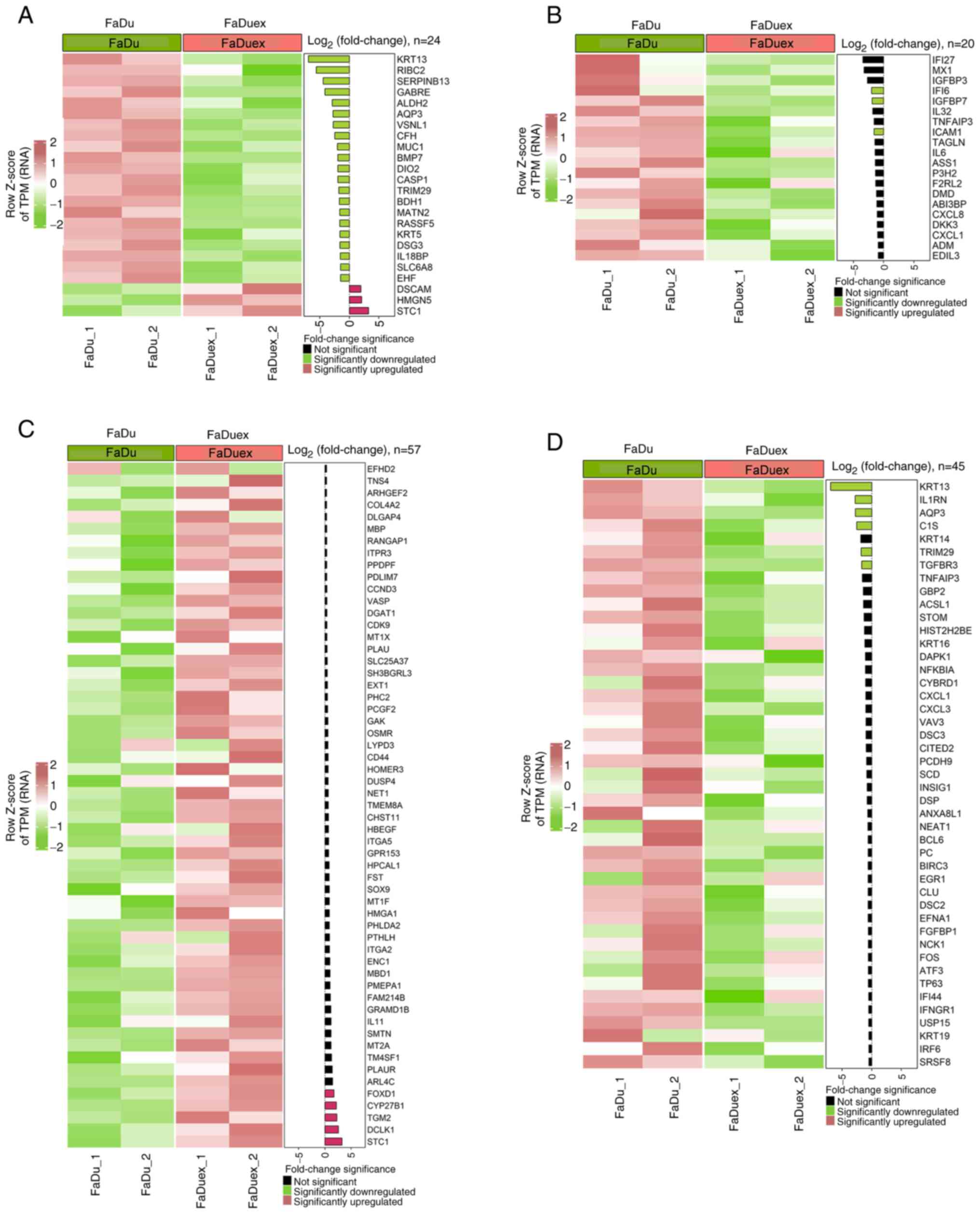
The changes in the expression levels of genes
associated with several pathways involved in metastasis,
angiogenesis, EGF signaling and EMT in FaDuex compared with FaDu
cells were further examined. Assessment of differential expression
of genes revealed that several metastasis-related genes were
altered, with keratin 13 (KRT13), RIBC2 and serpin
family B member 13 being the most significantly downregulated
metastasis-inhibiting genes, and stanniocalcin-1 (STC1),
high mobility group nucleosome binding domain 5 and down syndrome
cell adhesion molecule being the most significantly upregulated
metastasis-promoting genes, suggesting an increase in metastatic
potential in FaDuex cells (Fig.
3A). Regarding the angiogenesis pathway, only interferon
α-inducible protein 6 (IFI6), insulin-like growth factor
binding protein 7 and intercellular adhesion molecule 1
(ICAM1) were significantly downregulated
angiogenesis-inhibiting genes in FaDuex cells, indicating a
possible promotion of neovascularization (Fig. 3B). Furthermore, a significant
upregulation of STC1, doublecortin-like kinase 1,
transglutaminase 2, cytochrome P450 family 27 subfamily B member 1
and forkhead box D1 genes was noted, indicating the activation of
the EGF-related pathway, which may contribute to enhanced
proliferative and survival signaling in FaDuex cells (Fig. 3C). Furthermore, the genes involved
in the inhibition of the EMT pathway, including KRT13,
interleukin 1 receptor antagonist, aquaporin-3, complement
component 1s, tripartite motif-containing protein 29 and
transforming growth factor-β receptor 3, were downregulated in
FaDuex cells, suggesting a shift toward EMT phenotypes (Fig. 3D). These findings indicated that
FaDuex cells exhibit enhanced metastatic, angiogenic, proliferative
and EMT-related potential, reflecting a more aggressive
phenotype.
Downregulation of interferon
signaling-associated genes in advanced HPC cells
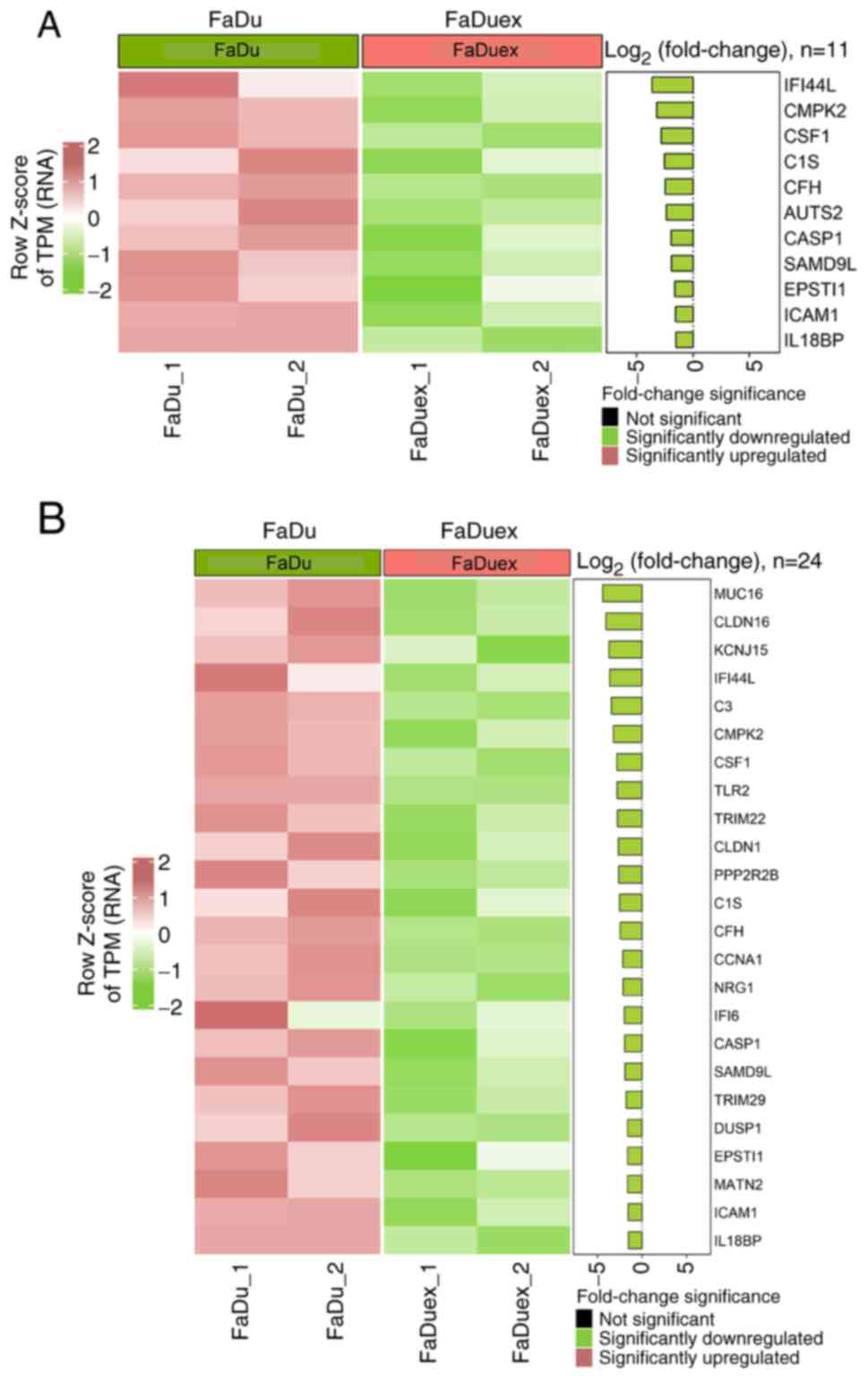
The present study analyzed the expression of
IFN-responsive genes using the C2 and Hallmark gene sets to
evaluate the differences in IFN signaling between FaDu and FaDuex
cells. Both analyses revealed a broad suppression of ISGs in FaDuex
cells, indicating a significant suppression of IFN signaling. In
the C2 gene set (Fig. 4A), key ISGs
such as interferon-induced protein 44-like (IFI44L),
IFI6 and tripartite motif-containing protein 22, all of
which are key mediators of the IFN response and antiviral defense
mechanisms, were markedly downregulated. The Hallmark gene set
(Fig. 4B) further confirmed the
suppression of IFN signaling, demonstrating a decrease in the
expression of genes such as IFI44L, caspase-1 and
ICAM1, which are involved in immune activation and
inflammatory responses. In the Hallmark gene set, mucin-16
(MUC16) was the most significantly downregulated; however,
its expression was only stimulated by the combined action of tumor
necrosis factor-α and IFN-γ (36).
The downregulation of these ISGs suggested that FaDuex cells may
have reduced sensitivity to IFN-mediated immune responses,
potentially altering their interaction with the immune
microenvironment and affecting their ability to respond to external
immune challenges.
Validation of gene expression using
qPCR
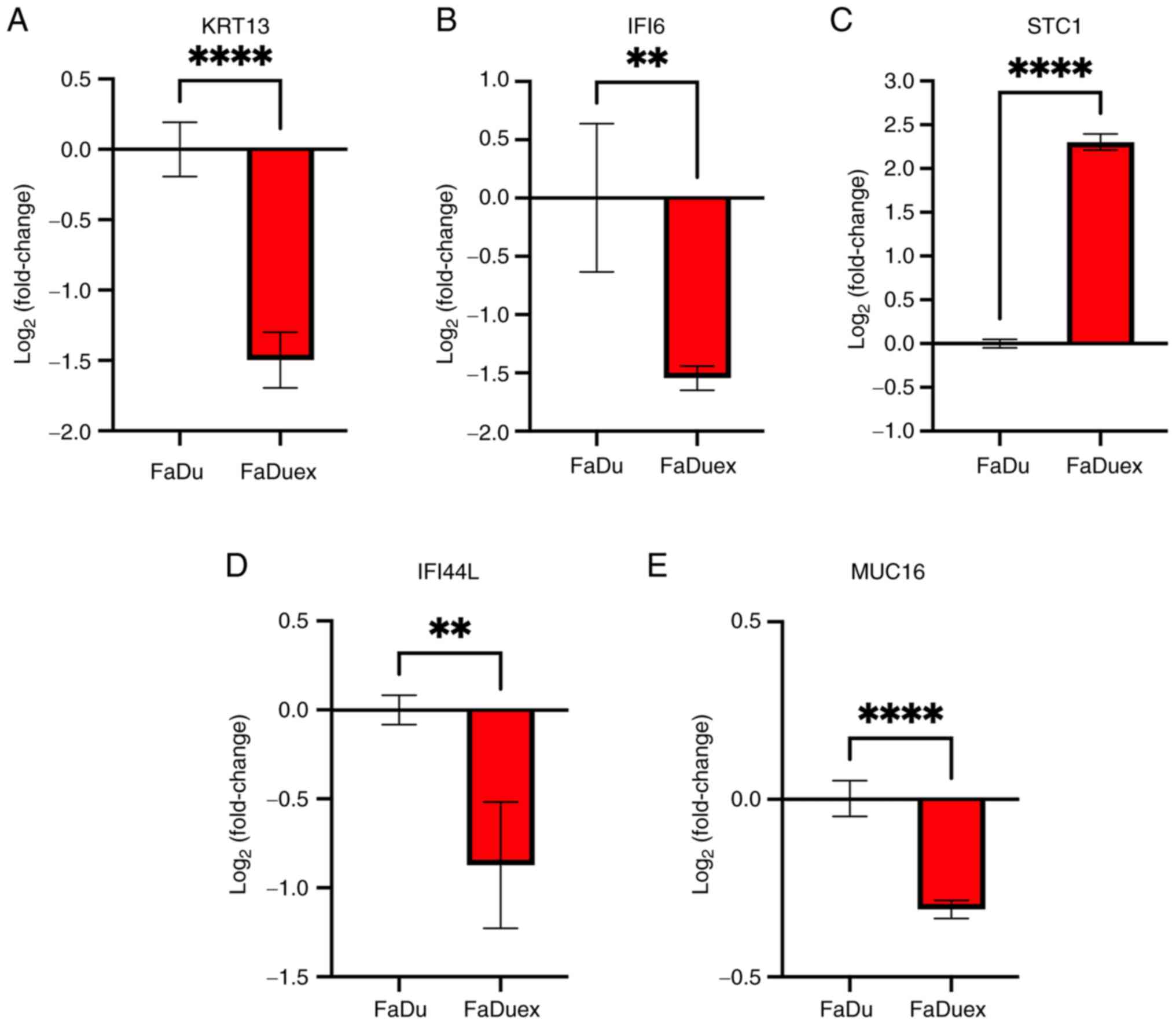
The results of DEGs analysis were validated by
performing qPCR by selecting genes, in FaDu cells and FaDuex cells,
whose expression levels exhibited the most significant changes.
According to the DEGs analysis, KRT13 and IFI6 were
most significantly downregulated in metastasis/EMT and
angiogenesis, respectively, whereas STC1 was the most
significantly upregulated in proliferative and survival-related
gene of FaDuex cells. In addition, IFI44L and MUC16
were most downregulated in the IFN-responsive genes of FaDuex cells
according to the C2 and Hallmark gene sets. The qPCR analysis
further demonstrated the downregulation of KRT13, IFI44L,
IFI6 and MUC16 transcripts (Fig. 5A, B, D and E) and STC1 was
upregulated in FaDuex cells (Fig.
5C), consistent with DEG analysis. These results suggested that
RNA-seq determined gene expression is useful for the prediction of
cancer-related signaling pathways.
The limitations of the present study include a small
sample size; in addition, the results were analyzed from human HPC
cell lines rather than clinical samples. As mentioned, HPC is a
rare subtype of HNSCC (6). In
Taiwan, the age-standardized incidence rate for HPC was increased
from 1980 and reached 6.46 per 100,000 in 2019 (37). A pilot study may be initiated in the
future to determine notable changes of gene expression using a
widely used HPC cell line and its derivatives from xenograft tumor.
FaDu cells are isolated from the primary tumor of a White male
patient with HPC in 1968 (15).
Recently, another HPC cell type (CZH1) isolated from a Chinese
patient with HPC has been reported, exhibiting a greater capacity
for invasion and increased radiosensitivity compared with FaDu
cells (38). The present study
would be key in exploring the gene expression differences of
patients with HPC from different ethnicities. FaDuex cells are
isolated from xenograft tumors that have reached 2,000
mm3 and the present study defined them as late-stage
tumors because of increased metastatic, tumorigenic, angiogenic and
chemoradiotherapy-resistant capacity (10). However, it is difficult to explain
the TNM stage of clinical HPC through late-stage FaDuex cells, as
they exhibit similar characteristics as advanced human HPC
diagnosed in clinics. In addition, the translational limitation may
still exist when using FaDuex cells to represent late-stage HPC in
clinical settings. Another limitation of the present study was the
use of parental FaDu cells for comparison with tumor-derived FaDuex
cells rather than the early stage of FaDu tumors (for example,
tumor size at 100 mm3). It may be key to compare potent
DEGs in FaDu-derived xenograft tumors at the early (small size)
stage and the late (large size) stage in the future.
To conclude, the present study demonstrated that
FaDuex cells, the arbitrarily defined late-stage HPC cells isolated
from xenograft tumors, exhibited several genes - including
KRT13, IFI6, STC1, MUC16 and IFI44L - that were
differentially expressed and were associated with malignant
characteristics, including increased metastasis, angiogenesis,
proliferation and decreased inflammatory responses. The present
study also observed that RNA-seq data-based DEGs analysis could be
validated using qPCR. Although the use of human cell lines may
encounter several of the aforementioned limitations, the variation
in samples would be low and easy to interpret. The present study
results suggest that several genes are associated with the advanced
malignancy of HPC cells, which makes this information potentially
key for diagnostic and therapeutic considerations in the
future.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant of Taipei City
Hospital, RenAi Branch (grant no. TPCH-112-12) and that of National
Science and Technology Council (grant no. NSTC
114-2314-B-A49-065-MY3).
Availability of data and materials
The data generated in the present study may be found
in the Sequence Read Archive (SRA) of National Library of Medicine
under accession no. PRJNA1282043 or at the following URL:
https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA1282043.
Authors' contributions
CWK conducted the cell culture experiments, RNA
preparation, RNA-sequencing arrangement and data analysis. YPY
conducted the differential gene expression and gene set enrichment
analyses. TWC, JDL and YJL confirmed the authenticity of all the
raw data. TWC interpreted the RNA sequencing data, and resolved
questions related to the accuracy of this work. MYL isolated and
identified different types of FaDu cells. JDL and YJL contributed
to the conception and the design of the present study. YJL drafted
the manuscript. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, AI tools were
used to improve the readability and language of the manuscript or
to generate images, and subsequently, the authors revised and
edited the content produced by the AI tools as necessary, taking
full responsibility for the ultimate content of the present
manuscript.
References
|
1
|
Petersen JF, Timmermans AJ, van Dijk BAC,
Overbeek LIH, Smit LA, Hilgers FJM, Stuiver MM and van den Brekel
MWM: Trends in treatment, incidence and survival of hypopharynx
cancer: A 20-year population-based study in the Netherlands. Eur
Arch Otorhinolaryngol. 275:181–189. 2018. View Article : Google Scholar
|
|
2
|
Cordunianu AV, Ganea G, Cordunianu MA,
Cochior D, Moldovan CA and Adam R: Hypopharyngeal cancer trends in
a high-incidence region: A retrospective tertiary single center
study. World J Clin Cases. 11:5666–5677. 2023. View Article : Google Scholar
|
|
3
|
Sanders O and Pathak S: Hypopharyngeal
Cancer. StatPearls. StatPearls Publishing; Treasure Island, FL:
2025
|
|
4
|
Patel EJ, Oliver JR, Jacobson AS, Li Z, Hu
KS, Tam M, Vaezi A, Morris LGT and Givi B: Human papillomavirus in
patients with hypopharyngeal squamous cell carcinoma. Otolaryngol
Head Neck Surg. 166:109–117. 2022. View Article : Google Scholar
|
|
5
|
Rami-Porta R, Nishimura KK, Giroux DJ,
Detterbeck F, Cardillo G, Edwards JG, Fong KM, Giuliani M, Huang J,
Kernstine KH Sr, et al: The International association for the study
of lung cancer lung cancer staging project: Proposals for revision
of the TNM stage groups in the forthcoming (Ninth) Edition of the
TNM classification for lung cancer. J Thorac Oncol. 19:1007–1027.
2024. View Article : Google Scholar
|
|
6
|
Bozec A, Benezery K, Chamorey E, Ettaiche
M, Vandersteen C, Dassonville O, Poissonnet G, Riss JC,
Hannoun-Lévi JM, Chand ME, et al: Nutritional status and
feeding-tube placement in patients with locally advanced
hypopharyngeal cancer included in an induction chemotherapy-based
larynx preservation program. Eur Arch Otorhinolaryngol.
273:2681–2687. 2016. View Article : Google Scholar
|
|
7
|
Malecki K, Glinski B, Mucha-Malecka A, Rys
J, Kruczak A, Roszkowski K, Urbańska-Gąsiorowska M and Hetnał M:
Prognostic and predictive significance of p53, EGFr, Ki-67 in
larynx preservation treatment. Rep Pract Oncol Radiother. 15:87–92.
2010. View Article : Google Scholar
|
|
8
|
Zhang J, Tian XJ and Xing J: Signal
transduction pathways of EMT Induced by TGF-beta, SHH, and WNT and
their crosstalks. J Clin Med. 5:412016. View Article : Google Scholar
|
|
9
|
Ahluwalia A and Tarnawski AS: Critical
role of hypoxia sensor-HIF-1α in VEGF gene activation. Implications
for angiogenesis and tissue injury healing. Curr Med Chem.
19:90–97. 2012. View Article : Google Scholar
|
|
10
|
Lin MY, Wang CY, Chan YH, Su SP, Chiang
HK, Yang MH and Lee YJ: The emergence of tumor-initiating cells in
an advanced hypopharyngeal tumor model exhibits enhanced
angiogenesis and nuclear factor erythroid 2-related factor
2-associated antioxidant effects. Antioxid Redox Signal.
41:505–521. 2024. View Article : Google Scholar
|
|
11
|
Taneja N, Chauhan A, Kulshreshtha R and
Singh S: HIF-1 mediated metabolic reprogramming in cancer:
Mechanisms and therapeutic implications. Life Sci. 352:1228902024.
View Article : Google Scholar
|
|
12
|
Zhong X, Wang Y, He X, He X, Hu Z, Huang
H, Chen J, Chen K, Wei P, Zhao S, et al: HIF1A-AS2 promotes the
metabolic reprogramming and progression of colorectal cancer via
miR-141-3p/FOXC1 axis. Cell Death Dis. 15:6452024. View Article : Google Scholar
|
|
13
|
Meyer SP, Bauer R, Brune B and Schmid T:
The role of type I interferon signaling in myeloid anti-tumor
immunity. Front Immunol. 16:15474662025. View Article : Google Scholar
|
|
14
|
Critchley-Thorne RJ, Yan N, Nacu S, Weber
J, Holmes SP and Lee PP: Down-regulation of the interferon
signaling pathway in T lymphocytes from patients with metastatic
melanoma. PLoS Med. 4:e1762007. View Article : Google Scholar
|
|
15
|
Rangan SR: A new human cell line (FaDu)
from a hypopharyngeal carcinoma. Cancer. 29:117–121. 1972.
View Article : Google Scholar
|
|
16
|
Keski-Santti H, Luukkaa M, Carpen T,
Jouppila-Matto A, Lehtio K, Maenpaa H, Vuolukka K, Vahlberg T and
Mäkitie A: Hypopharyngeal carcinoma in Finland from 2005 to 2014:
Outcome remains poor after major changes in treatment. Eur Arch
Otorhinolaryngol. 280:1361–1367. 2023. View Article : Google Scholar
|
|
17
|
Ewels P, Magnusson M, Lundin S and Kaller
M: MultiQC: Summarize analysis results for multiple tools and
samples in a single report. Bioinformatics. 32:3047–3048. 2016.
View Article : Google Scholar
|
|
18
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar
|
|
19
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar
|
|
20
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2:1001412021.
|
|
21
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar
|
|
22
|
Liberzon A, Birger C, Thorvaldsdottir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar
|
|
23
|
Liberzon A, Subramanian A, Pinchback R,
Thorvaldsdottir H, Tamayo P and Mesirov JP: Molecular signatures
database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 2011.
View Article : Google Scholar
|
|
24
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar
|
|
25
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016. View Article : Google Scholar
|
|
26
|
Clarke DJB, Marino GB, Deng EZ, Xie Z,
Evangelista JE and Ma'ayan A: Rummagene: Massive mining of gene
sets from supporting materials of biomedical research publications.
Commun Biol. 7:4822024. View Article : Google Scholar
|
|
27
|
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D,
Yang S, Kim CY, Lee M, Kim E, et al: TRRUST v2: An expanded
reference database of human and mouse transcriptional regulatory
interactions. Nucleic Acids Res. 46:D380–D386. 2018. View Article : Google Scholar
|
|
28
|
Rauluseviciute I, Riudavets-Puig R,
Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB,
Lucas J, Chèneby J, Baranasic D, et al: JASPAR 2024: 20th
anniversary of the open-access database of transcription factor
binding profiles. Nucleic Acids Res. 52:D174–D182. 2024. View Article : Google Scholar
|
|
29
|
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D,
et al: TRRUST v2: an expanded reference database of human and mouse
transcriptional regulatory interactions. Nucleic Acids Res.
46:D380–D386. 2018. View Article : Google Scholar
|
|
30
|
Hu C, Li T, Xu Y, Zhang X, Li F, Bai J,
Chen J, Jiang W, Yang K, Ou Q, et al: CellMarker 2.0: An updated
database of manually curated cell markers in human/mouse and web
tools based on scRNA-seq data. Nucleic Acids Res. 51:D870–D876.
2023. View Article : Google Scholar
|
|
31
|
Xie Z, Bailey A, Kuleshov MV, Clarke DJB,
Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML,
Kropiwnicki E, Jagodnik KM, et al: Gene set knowledge discovery
with enrichr. Curr Protoc. 1:e902021. View
Article : Google Scholar
|
|
32
|
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z,
Meirelles GV, Clark NR and Ma'ayan A: Enrichr: Interactive and
collaborative HTML5 gene list enrichment analysis tool. BMC
Bioinformatics. 14:1282013. View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Bonjardim CA, Ferreira PC and Kroon EG:
Interferons: Signaling, antiviral and viral evasion. Immunol Lett.
122:1–11. 2009. View Article : Google Scholar
|
|
35
|
Kaplan DH, Shankaran V, Dighe AS, Stockert
E, Aguet M, Old LJ and Schreiber RD: Demonstration of an interferon
gamma-dependent tumor surveillance system in immunocompetent mice.
Proc Natl Acad Sci USA. 95:7556–7561. 1998. View Article : Google Scholar
|
|
36
|
Morgado M, Sutton MN, Simmons M, Warren
CR, Lu Z, Constantinou PE, Liu J, Francis LL, Conlan RS, Bast RC Jr
and Carson DD: Tumor necrosis factor-α and interferon-ү stimulate
MUC16 (CA125) expression in breast, endometrial and ovarian cancers
through NFĸB. Oncotarget. 7:14871–14884. 2016. View Article : Google Scholar
|
|
37
|
Tsai YS, Chen YC, Chen TI, Lee YK, Chiang
CJ, You SL, Hsu WL and Liao LJ: Incidence trends of oral cavity,
oropharyngeal, hypopharyngeal and laryngeal cancers among males in
Taiwan, 1980–2019: A population-based cancer registry study. BMC
Cancer. 23:2132023. View Article : Google Scholar
|
|
38
|
Ma J, Zhu X, Heng Y, Ding X, Tao L and Lu
L: Establishment and characterization of a novel hypopharyngeal
squamous cell carcinoma cell line CZH1 with genetic abnormalities.
Hum Cell. 37:546–559. 2024. View Article : Google Scholar
|