|
1
|
Vaghari-Tabari M, Ferns GA, Qujeq D,
Andevari AN, Sabahi Z and Moein ZS: Signaling, metabolism, and
cancer: An important relationship for therapeutic intervention. J
Cell Physiol. 236:5512–5532. 2021. View Article : Google Scholar
|
|
2
|
Giunta S: Decoding human cancer with whole
genome sequencing: A review of PCAWG Project studies published in
February 2020. Cancer Metastasis Rev. 40:909–924. 2021. View Article : Google Scholar
|
|
3
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar
|
|
4
|
Ramezankhani R, Solhi R, Es HA, Vosough M
and Hassan M: Novel molecular targets in gastric adenocarcinoma.
Pharmacol Ther. 220:1077142021. View Article : Google Scholar
|
|
5
|
Guo N, Liu JB, Li W, Ma YS and Fu D: The
power and the promise of CRISPR/Cas9 genome editing for clinical
application with gene therapy. J Adv Res. 40:135–152. 2022.
View Article : Google Scholar
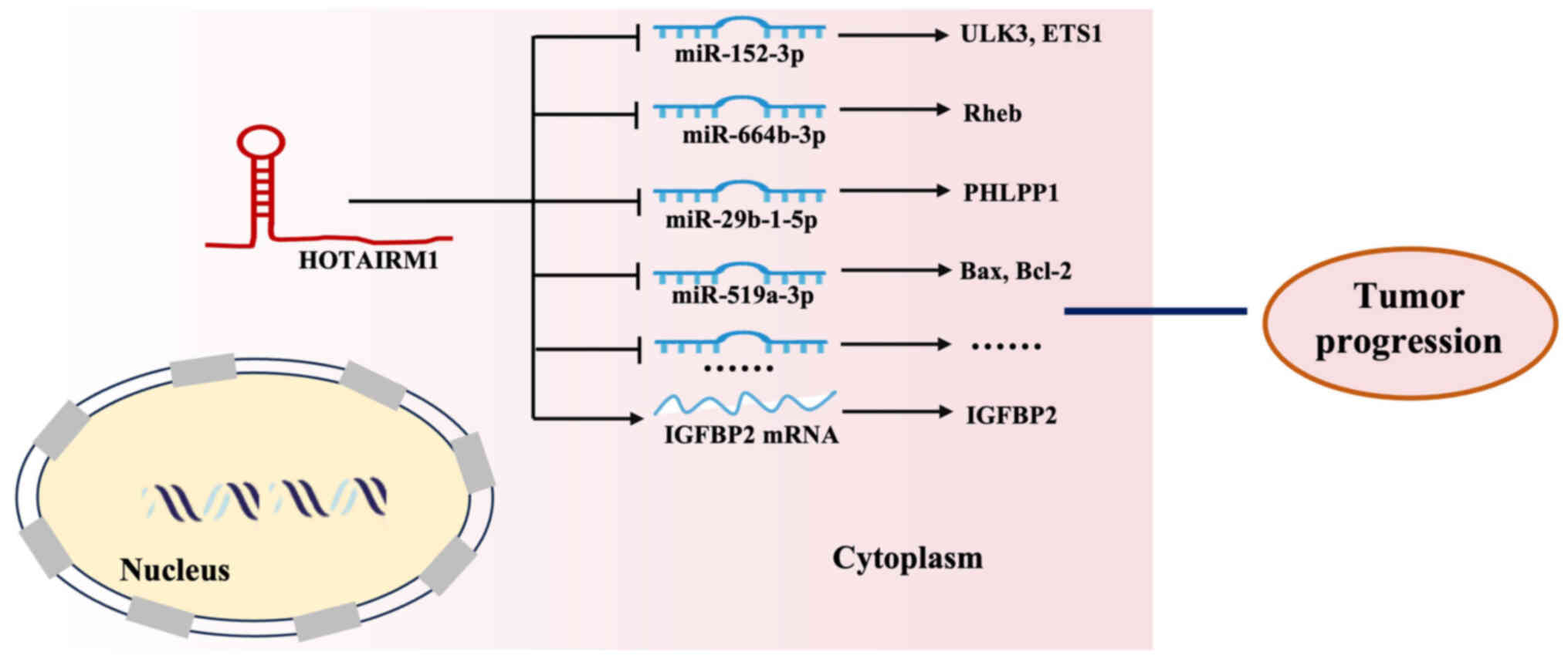
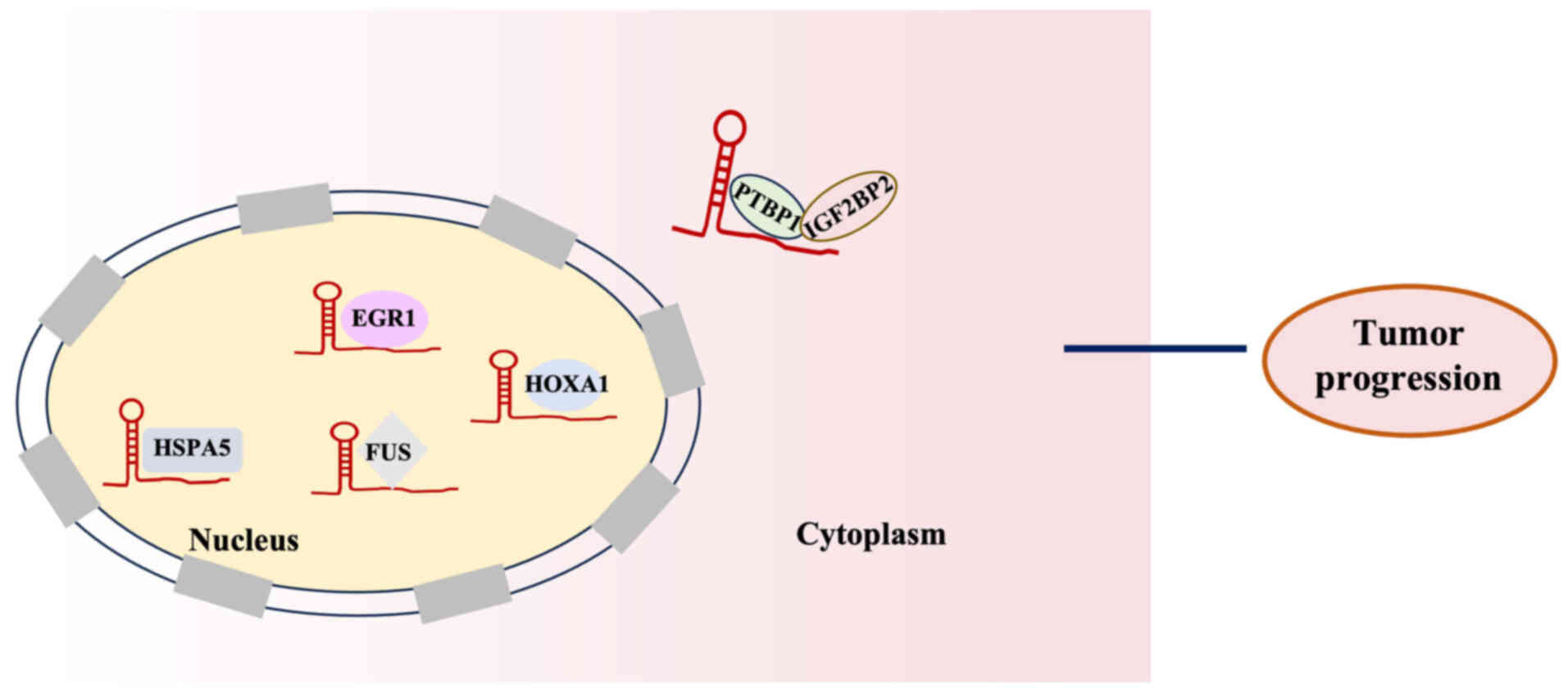
|
|
6
|
Traba J, Sack MN, Waldmann TA and Anton
OM: Immunometabolism at the nexus of cancer therapeutic efficacy
and resistance. Front Immunol. 12:6572932021. View Article : Google Scholar
|
|
7
|
Ghafouri-Fard S, Khoshbakht T, Taheri M
and Hajiesmaeili M: Long intergenic non-protein coding RNA 460:
Review of its role in carcinogenesis. Pathol Res Pract.
225:1535562021. View Article : Google Scholar
|
|
8
|
Hovhannisyan H and Gabaldón T: The long
non-coding RNA landscape of Candida yeast pathogens. Nat Commun.
12:73172021. View Article : Google Scholar
|
|
9
|
Kovalenko TF, Yadav B, Anufrieva KS,
Rubtsov YP, Zatsepin TS, Shcherbinina EY, Solyus EM, Staroverov DB,
Larionova TD, Latyshev YA, et al: Functions of long non-coding RNA
ROR in patient-derived glioblastoma cells. Biochimie. 200:131–139.
2022. View Article : Google Scholar
|
|
10
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar
|
|
11
|
Herman AB, Tsitsipatis D and Gorospe M:
Integrated lncRNA function upon genomic and epigenomic regulation.
Mol Cell. 82:2252–2266. 2022. View Article : Google Scholar
|
|
12
|
Sun KK, Zu C, Wu XY, Wang QH, Hua P, Zhang
YF, Shen XJ and Wu YY: Identification of lncRNA and mRNA regulatory
networks associated with gastric cancer progression. Front Oncol.
13:11404602023. View Article : Google Scholar
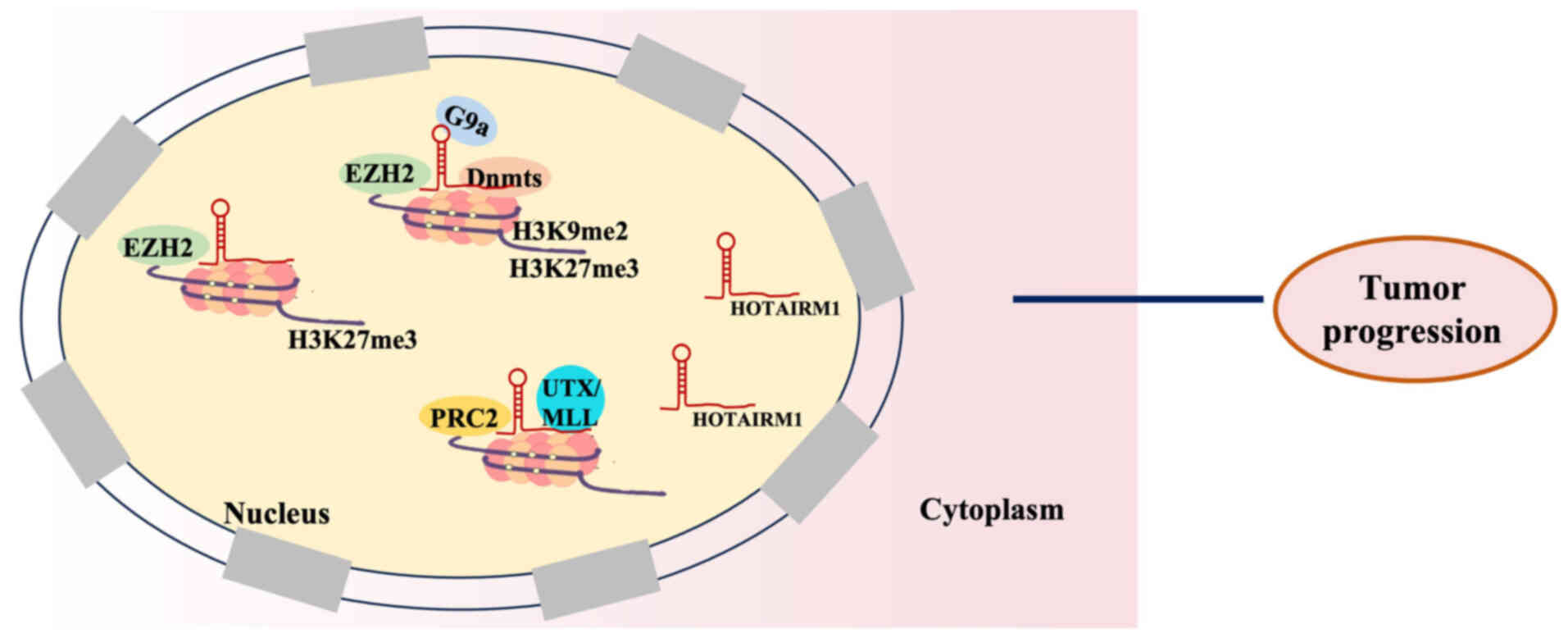
|
|
13
|
Pierce JB, Zhou H, Simion V and Feinberg
MW: Long noncoding RNAs as therapeutic targets. Adv Exp Med Biol.
1363:161–175. 2022. View Article : Google Scholar
|
|
14
|
Sun W, Xu J, Wang L, Jiang Y, Cui J, Su X,
Yang F, Tian L, Si Z and Xing Y: Non-coding RNAs in cancer
therapy-induced cardiotoxicity: Mechanisms, biomarkers, and
treatments. Front Cardiovasc Med. 9:9461372022. View Article : Google Scholar
|
|
15
|
Zhu Y, Ren J, Wu X, Zhang Y, Wang Y, Xu J,
Tan Q, Jiang Y and Li Y: lncRNA ENST00000422059 promotes cell
proliferation and inhibits cell apoptosis in breast cancer by
regulating the miR-145-5p/KLF5 axis. Acta Biochim Biophys Sin
(Shanghai). 55:1892–1901. 2023.
|
|
16
|
Bah I, Youssef D, Yao ZQ, McCall CE and El
Gazzar M: Hotairm1 controls S100A9 protein phosphorylation in
myeloid-derived suppressor cells during sepsis. J Clin Cell
Immunol. 14:10006912023.
|
|
17
|
Zhang X, Weissman SM and Newburger PE:
Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle
progression during myeloid maturation in NB4 human promyelocytic
leukemia cells. RNA Biol. 11:777–787. 2014. View Article : Google Scholar
|
|
18
|
Basyegit H, Tatar BG, Kose S, Gunduz C,
Ozmen Yelken B and Yilmaz Susluer S: Exploring the role of long
non-coding RNAs in predicting outcomes for hepatitis B patients.
Asian Pac J Cancer Prev. 25:4313–4321. 2024. View Article : Google Scholar
|
|
19
|
Bagheri-Mohammadi S, Karamivandishi A,
Mahdavi SA and Siahposht-Khachaki A: New sights on long non-coding
RNAs in glioblastoma: A review of molecular mechanism. Heliyon.
10:e397442024. View Article : Google Scholar
|
|
20
|
Nekoeian S, Rostami T, Norouzy A, Hussein
S, Tavoosidana G, Chahardouli B, Rostami S, Asgari Y and Azizi Z:
Identification of lncRNAs associated with the progression of acute
lymphoblastic leukemia using a competing endogenous RNAs network.
Oncol Res. 30:259–268. 2023. View Article : Google Scholar
|
|
21
|
Chen R, Zhou D, Chen Y, Chen M and Shuai
Z: Understanding the role of exosomal lncRNAs in rheumatic
diseases: A review. PeerJ. 11:e164342023. View Article : Google Scholar
|
|
22
|
Ramos TAR, Urquiza-Zurich S, Kim SY,
Gillette TG, Hill JA, Lavandero S, Rêgo TG and Maracaja-Coutinho V:
Single-cell transcriptional landscape of long non-coding RNAs
orchestrating mouse heart development. Cell Death Dis. 14:8412023.
View Article : Google Scholar
|
|
23
|
Hu C, Dai Q, Zhang R, Yang H, Wang M, Gu
K, Yang J, Meng W, Chen P and Xu M: Case report: Identification of
a novel LYN::LINC01900 transcript with promyelocytic phenotype and
TP53 mutation in acute myeloid leukemia. Front Oncol.
13:13224032023. View Article : Google Scholar
|
|
24
|
Wang D, Zhao X, Li S, Guo H, Li S and Yu
D: The impact of LncRNA-SOX2-OT/let-7c-3p/SKP2 Axis on head and
neck squamous cell carcinoma progression: Insights from
bioinformatics analysis and experimental validation. Cell Signal.
115:1110182024. View Article : Google Scholar
|
|
25
|
Yu M, He X, Liu T and Li J: lncRNA
GPRC5D-AS1 as a ceRNA inhibits skeletal muscle aging by regulating
miR-520d-5p. Aging (Albany NY). 15:13980–13997. 2023. View Article : Google Scholar
|
|
26
|
Zhang X, Zhong Y, Liu L, Jia C, Cai H,
Yang J, Wu B and Lv Z: Fasting regulates mitochondrial function
through lncRNA PRKCQ-AS1-mediated IGF2BPs in papillary thyroid
carcinoma. Cell Death Dis. 14:8272023. View Article : Google Scholar
|
|
27
|
Wu Z, Lin Y and Wei N:
N6-methyladenosine-modified HOTAIRM1 promotes vasculogenic mimicry
formation in glioma. Cancer Sci. 114:129–141. 2023. View Article : Google Scholar
|
|
28
|
Liu L, Zhou Y, Dong X, Li S, Cheng S, Li
H, Li Y, Yuan J, Wang L and Dong J: HOTAIRM1 maintained the
malignant phenotype of tMSCs transformed by GSCs via E2F7 by
binding to FUS. J Oncol. 2022:77344132022.
|
|
29
|
Shi T, Guo D, Xu H, Su G, Chen J, Zhao Z,
Shi J, Wedemeyer M, Attenello F, Zhang L and Lu W: HOTAIRM1, an
enhancer lncRNA, promotes glioma proliferation by regulating
long-range chromatin interactions within HOXA cluster genes. Mol
Biol Rep. 47:2723–2733. 2020. View Article : Google Scholar
|
|
30
|
Wang H, Li H, Jiang Q, Dong X, Li S, Cheng
S, Shi J, Liu L, Qian Z and Dong J: HOTAIRM1 promotes malignant
progression of transformed fibroblasts in glioma stem-like cells
remodeled microenvironment via regulating miR-133b-3p/TGFβ Axis.
Front Oncol. 11:6031282021. View Article : Google Scholar
|
|
31
|
Xia H, Liu Y, Wang Z, Zhang W, Qi M, Qi B
and Jiang X: Long noncoding RNA HOTAIRM1 maintains tumorigenicity
of glioblastoma stem-like cells through regulation of HOX gene
expression. Neurotherapeutics. 17:754–764. 2020. View Article : Google Scholar
|
|
32
|
Hao Y, Li X, Chen H, Huo H, Liu Z and Chai
E: Over-expression of long noncoding RNA HOTAIRM1 promotes cell
proliferation and invasion in human glioblastoma by up-regulating
SP1 via sponging miR-137. Neuroreport. 31:109–117. 2020. View Article : Google Scholar
|
|
33
|
Wu Z and Wei N: METTL3-mediated HOTAIRM1
promotes vasculogenic mimicry icontributionsn glioma via regulating
IGFBP2 expression. J Transl Med. 21:8552023. View Article : Google Scholar
|
|
34
|
Xie P, Li X, Chen R, Liu Y, Liu DC, Liu W,
Cui G and Xu J: Upregulation of HOTAIRM1 increases migration and
invasion by glioblastoma cells. Aging (Albany NY). 13:2348–2364.
2020. View Article : Google Scholar
|
|
35
|
Ahmadov U, Picard D, Bartl J, Silginer M,
Trajkovic-Arsic M, Qin N, Blümel L, Wolter M, Lim JKM, Pauck D, et
al: The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness
and radiotherapy resistance in glioblastoma. Cell Death Dis.
12:8852021. View Article : Google Scholar
|
|
36
|
Jing Y, Jiang X, Lei L, Peng M, Ren J,
Xiao Q, Tao Y, Tao Y, Huang J, Wang L, et al: Mutant NPM1-regulated
lncRNA HOTAIRM1 promotes leukemia cell autophagy and proliferation
by targeting EGR1 and ULK3. J Exp Clin Cancer Res. 40:3122021.
View Article : Google Scholar
|
|
37
|
Wei S, Zhao M, Wang X, Li Y and Wang K:
PU.1 controls the expression of long noncoding RNA HOTAIRM1 during
granulocytic differentiation. J Hematol Oncol. 9:442016. View Article : Google Scholar
|
|
38
|
Hu N, Chen L, Li Q and Zhao H: LncRNA
HOTAIRM1 is involved in the progression of acute myeloid leukemia
through targeting miR-148b. RSC Adv. 9:10352–10359. 2019.
View Article : Google Scholar
|
|
39
|
Yu X, Duan W, Wu F, Yang D, Wang X, Wu J,
Zhou D and Shen Y: LncRNA-HOTAIRM1 promotes aerobic glycolysis and
proliferation in osteosarcoma via the miR-664b-3p/Rheb/mTOR
pathway. Cancer Sci. 114:3537–3552. 2023. View Article : Google Scholar
|
|
40
|
Li X, Pang L, Yang Z, Liu J, Li W and Wang
D: LncRNA HOTAIRM1/HOXA1 axis promotes cell proliferation,
migration and invasion in endometrial cancer. Onco Targets Ther.
12:10997–11015. 2019. View Article : Google Scholar
|
|
41
|
Zhang L, Zhang J, Li S, Zhang Y, Liu Y,
Dong J, Zhao W, Yu B, Wang H and Liu J: Genomic amplification of
long noncoding RNA HOTAIRM1 drives anaplastic thyroid cancer
progression via repressing miR-144 biogenesis. RNA Biol.
18:547–562. 2021. View Article : Google Scholar
|
|
42
|
Li C, Chen X, Liu T and Chen G: lncRNA
HOTAIRM1 regulates cell proliferation and the metastasis of thyroid
cancer by targeting Wnt10b. Oncol Rep. 45:1083–1093. 2021.
View Article : Google Scholar
|
|
43
|
Chen D, Li Y, Wang Y and Xu J: LncRNA
HOTAIRM1 knockdown inhibits cell glycolysis metabolism and tumor
progression by miR-498/ABCE1 axis in non-small cell lung cancer.
Genes Genomics. 43:183–194. 2021. View Article : Google Scholar
|
|
44
|
Xiong F, Yin H, Zhang H, Zhu C, Zhang B,
Chen S, Ling C and Chen X: Clinicopathologic features and the
prognostic implications of long noncoding RNA HOTAIRM1 in non-small
cell lung cancer. Genet Test Mol Biomarkers. 24:47–53. 2020.
View Article : Google Scholar
|
|
45
|
Yu Y, Niu J, Zhang X, Wang X, Song H, Liu
Y, Jiao X and Chen F: Identification and validation of HOTAIRM1 as
a novel biomarker for oral squamous cell carcinoma. Front Bioeng
Biotechnol. 9:7985842022. View Article : Google Scholar
|
|
46
|
Wang L, Wang L, Wang Q, Yosefi B, Wei S,
Wang X and Shen D: The function of long noncoding RNA HOTAIRM1 in
the progression of prostate cancer cells. Andrologia.
53:e138972021. View Article : Google Scholar
|
|
47
|
Luo Y, He Y, Ye X, Song J, Wang Q, Li Y
and Xie X: High expression of long noncoding RNA HOTAIRM1 is
associated with the proliferation and migration in pancreatic
ductal adenocarcinoma. Pathol Oncol Res. 25:1567–1577. 2019.
View Article : Google Scholar
|
|
48
|
Zhou Y, Gong B, Jiang ZL, Zhong S, Liu XC,
Dong K, Wu HS, Yang HJ and Zhu SK: Microarray expression profile
analysis of long non-coding RNAs in pancreatic ductal
adenocarcinoma. Int J Oncol. 48:670–680. 2016. View Article : Google Scholar
|
|
49
|
Ye L, Meng X, Xiang R, Li W and Wang J:
Investigating function of long noncoding RNA of HOTAIRM1 in
progression of SKOV3 ovarian cancer cells. Drug Dev Res.
82:1162–1168. 2021. View Article : Google Scholar
|
|
50
|
Chao H, Zhang M, Hou H, Zhang Z and Li N:
HOTAIRM1 suppresses cell proliferation and invasion in ovarian
cancer through facilitating ARHGAP24 expression by sponging
miR-106a-5p. Life Sci. 243:1172962020. View Article : Google Scholar
|
|
51
|
Li D, Chai L, Yu X, Song Y, Zhu X, Fan S,
Jiang W, Qiao T, Tong J, Liu S, et al: The HOTAIRM1/miR-107/TDG
axis regulates papillary thyroid cancer cell proliferation and
invasion. Cell Death Dis. 11:2272020. View Article : Google Scholar
|
|
52
|
Zheng M, Liu X, Zhou Q and Liu G: HOTAIRM1
competed endogenously with miR-148a to regulate DLGAP1 in head and
neck tumor cells. Cancer Med. 7:3143–3156. 2018. View Article : Google Scholar
|
|
53
|
Zhang Y, Mi L, Xuan Y, Gao C, Wang YH,
Ming HX and Liu J: LncRNA HOTAIRM1 inhibits the progression of
hepatocellular carcinoma by inhibiting the Wnt signaling pathway.
Eur Rev Med Pharmacol Sci. 22:4861–4868. 2018.
|
|
54
|
Chen TJ, Gao F, Yang T, Li H, Li Y, Ren H
and Chen MW: LncRNA HOTAIRM1 inhibits the proliferation and
invasion of lung adenocarcinoma cells via the miR-498/WWOX Axis.
Cancer Manag Res. 12:4379–4390. 2020. View Article : Google Scholar
|
|
55
|
Xu F, Chen M, Chen H, Wu N, Qi Q, Jiang X,
Fang D, Feng Q, Jin R and Jiang L: The curcumin analog Da0324
inhibits the proliferation of gastric cancer cells via
HOTAIRM1/miR-29b-1-5p/PHLPP1 Axis. J Cancer. 13:2644–2655. 2022.
View Article : Google Scholar
|
|
56
|
Lu R, Zhao G, Yang Y, Jiang Z, Cai J,
Zhang Z and Hu H: Long noncoding RNA HOTAIRM1 inhibits cell
progression by regulating miR-17-5p/PTEN axis in gastric cancer. J
Cell Biochem. 120:4952–4965. 2019. View Article : Google Scholar
|
|
57
|
Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M
and Zhang H: HOTAIRM1 as a potential biomarker for diagnosis of
colorectal cancer functions the role in the tumour suppressor. J
Cell Mol Med. 20:2036–2044. 2016. View Article : Google Scholar
|
|
58
|
Ren T, Hou J, Liu C, Shan F, Xiong X, Qin
A, Chen J and Ren W: The long non-coding RNA HOTAIRM1 suppresses
cell progression via sponging endogenous miR-17-5p/B-cell
translocation gene 3 (BTG3) axis in 5-fluorouracil resistant
colorectal cancer cells. Biomed Pharmacother. 117:1091712019.
View Article : Google Scholar
|
|
59
|
Zhang W, Dang R, Liu H, Dai L, Liu H,
Adegboro AA, Zhang Y, Li W, Peng K, Hong J and Li X: Machine
learning-based investigation of regulated cell death for predicting
prognosis and immunotherapy response in glioma patients. Sci Rep.
14:41732024. View Article : Google Scholar
|
|
60
|
Zafar N, Ghias K and Fadoo Z: Genetic
aberrations involved in relapse of pediatric acute myeloid
leukemia: A literature review. Asia Pac J Clin Oncol. 17:e135–e141.
2021. View Article : Google Scholar
|
|
61
|
Hayatigolkhatmi K, Valzelli R, El Menna O
and Minucci S: Epigenetic alterations in AML: Deregulated functions
leading to new therapeutic options. Int Rev Cell Mol Biol.
387:27–75. 2024. View Article : Google Scholar
|
|
62
|
Ding Y and Chen Q: Wnt/β-catenin signaling
pathway: An attractive potential therapeutic target in
osteosarcoma. Front Oncol. 14:14569592025. View Article : Google Scholar
|
|
63
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The Eighth Edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
“personalized” approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017.
|
|
64
|
Ding S, Hao Y, Qi Y, Wei H, Zhang J and Li
H: Molecular mechanism of tumor-infiltrating immune cells
regulating endometrial carcinoma. Genes Dis. 12:1014422024.
View Article : Google Scholar
|
|
65
|
Menendez-Santos M, Gonzalez-Baerga C,
Taher D, Waters R, Virarkar M and Bhosale P: Endometrial cancer:
2023 Revised FIGO staging system and the role of imaging. Cancers
(Basel). 16:18692024. View Article : Google Scholar
|
|
66
|
Chen DW, Lang BHH, McLeod DSA, Newbold K
and Haymart MR: Thyroid cancer. Lancet. 401:1531–1544. 2023.
View Article : Google Scholar
|
|
67
|
Boucai L, Zafereo M and Cabanillas ME:
Thyroid cancer: A review. JAMA. 331:425–435. 2024. View Article : Google Scholar
|
|
68
|
Meyer ML, Fitzgerald BG, Paz-Ares L,
Cappuzzo F, Jänne PA, Peters S and Hirsch FR: New promises and
challenges in the treatment of advanced non-small-cell lung cancer.
Lancet. 404:803–822. 2024. View Article : Google Scholar
|
|
69
|
Badwelan M, Muaddi H, Ahmed A, Lee KT and
Tran SD: Oral squamous cell carcinoma and concomitant primary
tumors, what do we know? A review of the literature. Curr Oncol.
30:3721–3734. 2023. View Article : Google Scholar
|
|
70
|
Jagadeesan D, Sathasivam KV, Fuloria NK,
Balakrishnan V, Khor GH, Ravichandran M, Solyappan M, Fuloria S,
Gupta G, Ahlawat A, et al: Comprehensive insights into oral
squamous cell carcinoma: Diagnosis, pathogenesis, and therapeutic
advances. Pathol Res Pract. 261:1554892024. View Article : Google Scholar
|
|
71
|
Almeeri MNE, Awies M and Constantinou C:
Prostate cancer, pathophysiology and recent developments in
management: A narrative review. Curr Oncol Rep. 26:1511–1519. 2024.
View Article : Google Scholar
|
|
72
|
Wilson TK and Zishiri OT: Prostate cancer:
A review of genetics, current biomarkers and personalised
treatments. Cancer Rep (Hoboken). 7:e700162024.
|
|
73
|
Dhillon J and Betancourt M: Pancreatic
ductal adenocarcinoma. Monogr Clin Cytol. 26:74–91. 2020.
View Article : Google Scholar
|
|
74
|
Konstantinopoulos PA and Matulonis UA:
Clinical and translational advances in ovarian cancer therapy. Nat
Cancer. 4:1239–1257. 2023. View Article : Google Scholar
|
|
75
|
Chow LQM: Head and neck cancer. N Engl J
Med. 382:60–72. 2020. View Article : Google Scholar
|
|
76
|
Kudrimoti A and Kudrimoti MR: Head and
neck cancers. Prim Care. 52:139–155. 2025. View Article : Google Scholar
|
|
77
|
Da BL, Suchman KI, Lau L, Rabiee A, He AR,
Shetty K, Yu H, Wong LL, Amdur RL, Crawford JM, et al: Pathogenesis
to management of hepatocellular carcinoma. Genes Cancer. 13:72–87.
2022. View Article : Google Scholar
|
|
78
|
Akhras A, Beran A, Guardiola J,
Bhavsar-Burke I, Reyes B and Ur Rahman A: S2808 direct oral
anticoagulants versus warfarin in elderly patients with child-pugh
C cirrhosis and atrial fibrillation: A real-world perspective. Am J
Gastroenterol. 120((10S2)): pS6032025.
|
|
79
|
Tawfiq RK, de Camargo Correia GS, Li S,
Zhao Y, Lou Y and Manochakian R: Targeting lung cancer with
precision: The ADC therapeutic revolution. Curr Oncol Rep.
27:669–686. 2025. View Article : Google Scholar
|
|
80
|
Merle G, Friedlaender A, Desai A and Addeo
A: Antibody drug conjugates in lung cancer. Cancer J. 28:429–435.
2022. View Article : Google Scholar
|
|
81
|
López MJ, Carbajal J, Alfaro AL, Saravia
LG, Zanabria D, Araujo JM, Quispe L, Zevallos A, Buleje JL, Cho CE,
et al: Characteristics of gastric cancer around the world. Crit Rev
Oncol Hematol. 181:1038412023. View Article : Google Scholar
|
|
82
|
Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh
Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M and Saad A:
Colorectal cancer epidemiology: Recent trends and impact on
outcomes. Curr Drug Targets. 22:998–1009. 2021. View Article : Google Scholar
|
|
83
|
Mahmoud NN: Colorectal cancer:
Preoperative evaluation and staging. Surg Oncol Clin N Am.
31:127–141. 2022. View Article : Google Scholar
|
|
84
|
Salido-Guadarrama I, Romero-Cordoba SL and
Rueda-Zarazua B: Multi-Omics Mining of lncRNAs with biological and
clinical relevance in cancer. Int J Mol Sci. 24:166002023.
View Article : Google Scholar
|
|
85
|
Mahato RK, Bhattacharya S, Khullar N,
Sidhu IS, Reddy PH, Bhatti GK and Bhatti JS: Targeting long
non-coding RNAs in cancer therapy using CRISPR-Cas9 technology: A
novel paradigm for precision oncology. J Biotechnol. 379:98–119.
2024. View Article : Google Scholar
|
|
86
|
Yang Q, Fu Y, Wang J, Yang H and Zhang X:
Roles of lncRNA in the diagnosis and prognosis of triple-negative
breast cancer. J Zhejiang Univ Sci B. 24:1123–1140. 2023.(In
English, Chinese). View Article : Google Scholar
|
|
87
|
Alharthi NS, Al-Zahrani MH, Hazazi A,
Alhuthali HM, Gharib AF, Alzahrani S, Altalhi W, Almalki WH and
Khan FR: Exploring the lncRNA-VEGF axis: Implications for cancer
detection and therapy. Pathol Res Pract. 253:1549982023. View Article : Google Scholar
|
|
88
|
Mehmandar-Oskuie A, Jahankhani K,
Rostamlou A, Mardafkan N, Karamali N, Razavi ZS and Mardi A:
Molecular mechanism of lncRNAs in pathogenesis and diagnosis of
auto-immune diseases, with a special focus on lncRNA-based
therapeutic approaches. Life Sci. 336:1223222024. View Article : Google Scholar
|
|
89
|
Li Q, Dong C, Cui J, Wang Y and Hong X:
Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion
through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away
from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer
Res. 37:2652018. View Article : Google Scholar
|
|
90
|
Kim CY, Oh JH, Lee JY and Kim MH: The
LncRNA HOTAIRM1 promotes tamoxifen resistance by mediating HOXA1
expression in ER+ breast cancer cells. J Cancer. 11:3416–3423.
2020. View Article : Google Scholar
|
|
91
|
Wang XQ and Dostie J: Reciprocal
regulation of chromatin state and architecture by HOTAIRM1
contributes to temporal collinear HOXA gene activation. Nucleic
Acids Res. 45:1091–1104. 2017.
|
|
92
|
Usman M, Li A, Wu D, Qinyan Y, Yi LX, He G
and Lu H: The functional role of lncRNAs as ceRNAs in both ovarian
processes and associated diseases. Noncoding RNA Res. 9:165–177.
2023. View Article : Google Scholar
|
|
93
|
Kim S: LncRNA-miRNA-mRNA regulatory
networks in skin aging and therapeutic potentials. Front Physiol.
14:13031512023. View Article : Google Scholar
|
|
94
|
Lin Y, Wen H, Yang B, Wang C and Liang R:
Integrated bioinformatics and validation to construct
lncRNA-miRNA-mRNA ceRNA network in status epilepticus. Heliyon.
9:e222052023. View Article : Google Scholar
|
|
95
|
Jiang M, Wang Z, Lu T, Li X, Yang K, Zhao
L, Zhang D, Li J and Wang L: Integrative analysis of long noncoding
RNAs dysregulation and synapse-associated ceRNA regulatory axes in
autism. Transl Psychiatry. 13:3752023. View Article : Google Scholar
|
|
96
|
Wang X, Liu Y and Lei P: LncRNA HOTAIRM1
promotes osteogenic differentiation of human bone marrow-derived
mesenchymal stem cells by targeting miR-152-3p/ETS1 axis. Mol Biol
Rep. 50:5597–5608. 2023. View Article : Google Scholar
|
|
97
|
Wang G, Yu Y and Wang Y: Effects of
propofol on neuroblastoma cells via the HOTAIRM1/miR-519a-3p axis.
Transl Neurosci. 13:57–69. 2022. View Article : Google Scholar
|
|
98
|
Han W, Wang S, Qi Y, Wu F, Tian N, Qiang B
and Peng X: Targeting HOTAIRM1 ameliorates glioblastoma by
disrupting mitochondrial oxidative phosphorylation and serine
metabolism. iScience. 25:1048232022. View Article : Google Scholar
|
|
99
|
Chen W, Liu J, Ge F, Chen Z, Qu M, Nan K,
Gu J, Jiang Y, Gao S and Liao Y: Long noncoding RNA HOTAIRM1
promotes immunosuppression in sepsis by inducing T cell exhaustion.
J Immunol. 208:618–632. 2022. View Article : Google Scholar
|
|
100
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar
|
|
101
|
Ge T, Gu X, Jia R, Ge S, Chai P, Zhuang A
and Fan X: Crosstalk between metabolic reprogramming and
epigenetics in cancer: Updates on mechanisms and therapeutic
opportunities. Cancer Commun (Lond). 42:1049–1082. 2022. View Article : Google Scholar
|
|
102
|
Zhou Y, Wu Q, Long X, He Y and Huang J:
lncRNA HOTAIRM1 Activated by hoxa4 drives huvec proliferation
through direct interaction with protein partner HSPA5.
Inflammation. 47:421–437. 2024. View Article : Google Scholar
|
|
103
|
Guo H, Li T and Sun X: LncRNA HOTAIRM1,
miR-433-5p and PIK3CD function as a ceRNA network to exacerbate the
development of PCOS. J Ovarian Res. 14:192021. View Article : Google Scholar
|
|
104
|
Dahariya S, Raghuwanshi S, Thamodaran V,
Velayudhan SR and Gutti RK: Role of long non-coding RNAs in
human-induced pluripotent stem cells derived megakaryocytes: A p53,
HOX antisense intergenic RNA Myeloid 1, and miR-125b interaction
study. J Pharmacol Exp Ther. 384:92–101. 2023. View Article : Google Scholar
|
|
105
|
Li YR, Yan WJ, Cai LL and Deng XL: Effect
of down-regulation of LncRNA-HOTAIRM1 to proliferation, apoptosis
and KIT/AKT pathway of jurkat cells. Zhongguo Shi Yan Xue Ye Xue Za
Zhi. 29:1123–1128. 2021.(In Chinese).
|
|
106
|
Liu WB, Li GS, Shen P, Li YN and Zhang FJ:
Long non-coding RNA HOTAIRM1-1 silencing in cartilage tissue
induces osteoarthritis through microRNA-125b. Exp Ther Med.
22:9332021. View Article : Google Scholar
|
|
107
|
Tollis P, Vitiello E, Migliaccio F,
D'Ambra E, Rocchegiani A, Garone MG, Bozzoni I, Rosa A, Carissimo
A, Laneve P and Caffarelli E: The long noncoding RNA nHOTAIRM1 is
necessary for differentiation and activity of iPSC-derived spinal
motor neurons. Cell Death Dis. 14:7412023. View Article : Google Scholar
|
|
108
|
Segal D, Coulombe S, Sim J and Josée
Dostie J: A conserved HOTAIRM1-HOXA1 regulatory axis contributes
early to neuronal differentiation. RNA Biol. 20:1523–1539. 2023.
View Article : Google Scholar
|
|
109
|
Zamame Ramirez JA, Romagnoli GG and Kaneno
R: Inhibiting autophagy to prevent drug resistance and improve
anti-tumor therapy. Life Sci. 265:1187452021. View Article : Google Scholar
|
|
110
|
Liang L, Gu W, Li M, Gao R, Zhang X, Guo C
and Mi S: The long noncoding RNA HOTAIRM1 controlled by AML1
enhances glucocorticoid resistance by activating RHOA/ROCK1 pathway
through suppressing ARHGAP18. Cell Death Dis. 12:7022021.
View Article : Google Scholar
|
|
111
|
Gu D, Tong M, Wang J, Zhang B, Liu J, Song
G and Zhu B: Overexpression of the lncRNA HOTAIRM1 promotes
lenvatinib resistance by downregulating miR-34a and activating
autophagy in hepatocellular carcinoma. Discov Oncol. 14:662023.
View Article : Google Scholar
|
|
112
|
Chen L, Hu N, Wang C and Zhao H: HOTAIRM1
knockdown enhances cytarabine-induced cytotoxicity by suppression
of glycolysis through the Wnt/β-catenin/PFKP pathway in acute
myeloid leukemia cells. Arch Biochem Biophys. 680:1082442020.
View Article : Google Scholar
|
|
113
|
Gustafson MP, Ligon JA, Bersenev A, McCann
CD, Shah NN and Hanley PJ: Emerging frontiers in immuno- and gene
therapy for cancer. Cytotherapy. 25:20–32. 2023. View Article : Google Scholar
|
|
114
|
Kaur R, Bhardwaj A and Gupta S: Cancer
treatment therapies: Traditional to modern approaches to combat
cancers. Mol Biol Rep. 50:9663–9676. 2023. View Article : Google Scholar
|
|
115
|
Hashemi M, Moosavi MS, Abed HM, Dehghani
M, Aalipour M, Heydari EA, Behroozaghdam M, Entezari M,
Salimimoghadam S, Gunduz ES, et al: Long non-coding RNA (lncRNA)
H19 in human cancer: From proliferation and metastasis to therapy.
Pharmacol Res. 184:1064182022. View Article : Google Scholar
|
|
116
|
McCabe EM and Rasmussen TP: lncRNA
involvement in cancer stem cell function and epithelial-mesenchymal
transitions. Semin Cancer Biol. 75:38–48. 2021. View Article : Google Scholar
|