Introduction
Cancer is a multifactorial disease due to various
causes and impacts, such as environmental factors, infectious
agents, genetic alterations and epigenetic shifts (1,2).
Advances in genetic research have not only elucidated the
pathogenesis of several cancer types, such as acute myeloid
leukemia (AML), non-small cell lung cancer (NSCLC) and prostate
cancer (PCa), but have also directly contributed to the development
of treatments (3–6). For example, imatinib is an effective
treatment for leukemia caused by mutations in the breakpoint
cluster region-Abelson, offering the possibility of long-term
disease remission. Furthermore, <2% of the entire genome encode
proteins, while the rest of the genetic material is composed of
non-coding genes, which are key contributors to tumorigenesis among
multiple cancer types, such as PCa and colorectal cancer (CRC)
(7–9).
Transcripts >200 nucleotides in length and which
have little or no ability to code for proteins are termed long
non-coding RNAs (lncRNAs) (10,11).
Previous studies identified that several dysregulated lncRNAs
contribute to the development and progression of cancer types,
working as oncogenes or tumor suppressors (12–15).
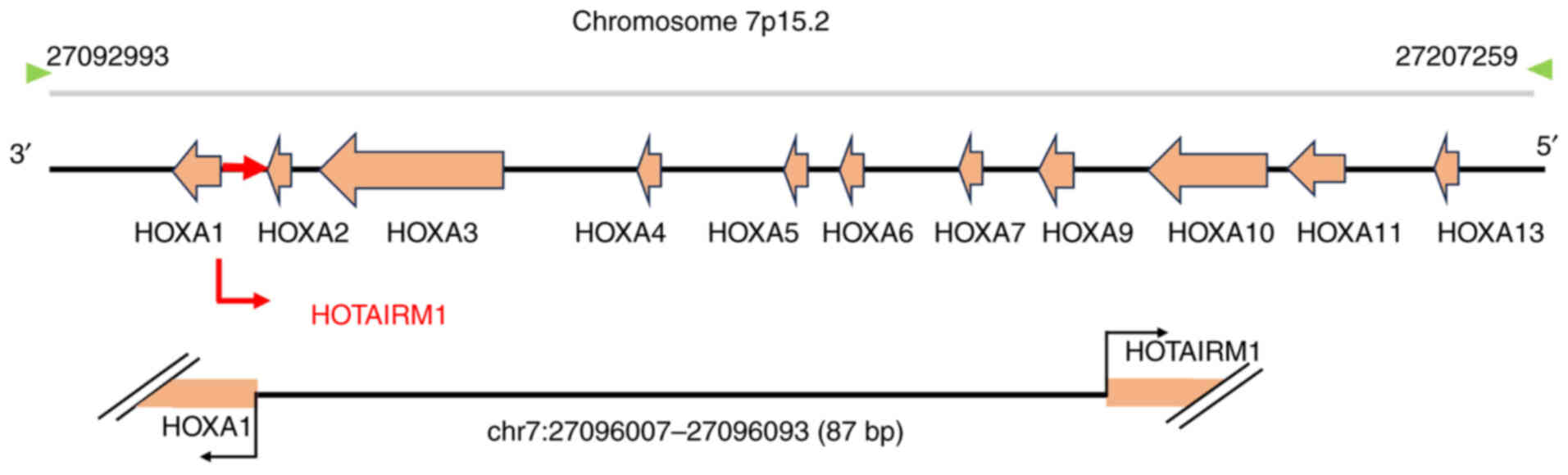
HOTAIRM1 is a recently identified lncRNA located in the HOXA gene
cluster on the human chromosome 7p15.2 (Fig. 1) (16). HOTAIRM1 was first identified as
involved in the differentiation of granulocytes in the NB4
promyelocytic leukemia model (17).
Since its identification, HOTAIRM1 has gained notable attention in
cancer research (18–20).
The relevant studies in the present review were
collected using PubMed (https://pubmed.ncbi.nlm.nih.gov) with a combination of
the following terms: ‘HOTAIRM1’, ‘cancer’, ‘tumor’ and ‘disease’.
English-language publications from the past 10 years were selected.
Two reviewers (YJ and XL) independently performed an initial
screening of titles and abstracts. The reference lists of
potentially eligible articles were cross-checked to ensure
extensive coverage of the literature. A total of 103 articles
meeting the predefined literature searching criteria were
identified by May 2025.
Therefore, the present review provides a
comprehensive overview of the current understanding of the
expression, roles and molecular mechanisms of HOTAIRM1 to regulate
cancer.
HOTAIRM1 expression in cancer
Several previous studies have documented the
abnormal expression of lncRNAs in different diseases, particularly
in cancer (21–23). The dysregulation of lncRNAs
contributes to the development of tumors through the promotion,
proliferation, invasion and metastasis of cancer cells (24–26).
The lncRNA HOTAIRM1 is upregulated in different types of cancer
such as glioma (27–35), AML (36–38),
osteosarcoma (OS) (39),
endometrial cancer (EC) (40),
thyroid cancer (TC) (41,42), NSCLC (43,44),
oral squamous cell carcinoma (OSCC) (45), PCa (46), pancreatic ductal adenocarcinoma
(PDAC) (47,48) and ovarian cancer (OC) (49,50).
The involvement of HOTAIRM1 in governing tumor characteristics,
including proliferation, invasion and metastasis, has been
demonstrated. However, HOTAIRM1 expression is downregulated in
papillary TC (PTC) (51), head and
neck tumor (HNT) (52),
hepatocellular carcinoma (HCC) (53), lung adenocarcinoma (ADC) (54), gastric cancer (GC) (55,56)
and CRC (Table I) (57,58).
These collective findings position HOTAIRM1 as a key tumor-related
lncRNA, whose dysregulation could drive oncogenic pathways and
offers a promising target for future cancer interventions.
 | Table I.HOTAIRM1 expression in several cancer
types. |
Table I.
HOTAIRM1 expression in several cancer
types.
| Cancer type | Expression | Functions | Associated
genes | Role | (Refs.) |
|---|
| Glioma | Up | Migration,
invasion, VM formation, proliferation, stemness and
radiosensitivity | IGFBP2, FUS, HOXAs,
miR-133b-3p, miR-137, miR-153-5p and TGM2 | Oncogene | (27–35) |
| AML | Up | Autophagy,
proliferation, apoptosis, cell cycle and differentiation | EGR1, miR-152-3p
and miR-148b | Oncogene | (36–38) |
| OS | Up | Proliferation,
apoptosis | miR-664b-3p | Oncogene | (39) |
| EC | Up | Proliferation,
migration, invasion and EMT | HOXA1 | Oncogene | (40) |
| TC | Up | Proliferation,
apoptosis, migration and invasion | ILF3, pri-miR-144
and miR148a | Oncogene | (41,42) |
| NSCLC | Up | Proliferation,
apoptosis, migration, invasion and glycolysis metabolism | miR-498 | Oncogene | (43,44) |
| OSCC | Up | Proliferation and
cell cycle | PCNA, cyclin D1,
p53, p21, CDK4 and CDK6 | Oncogene | (45) |
| PCa | Up | Proliferation and
apoptosis | β-catenin | Oncogene | (46) |
| PDAC | Up | Proliferation,
apoptosis, cell cycle and migration | CDK1, cyclin D1,
p21, Bax, Bad and Bcl-2 | Oncogene | (47,48) |
| OC | Up | Proliferation and
apoptosis | MMP2 | Oncogene | (49) |
| PTC | Down | Proliferation,
migration and invasion | miR-107 | Anticancer | (51) |
| OC | Down | Proliferation
invasion and apoptosis | miR-106a-5p | Anticancer | (50) |
| HNT | Down | Proliferation,
apoptosis, migration and invasion | miR-148a | Anticancer | (52) |
| HCC | Down | Proliferation and
apoptosis | β-catenin | Anticancer | (53) |
| ADC | Down | Cell cycle,
proliferation and invasion | miR-498 | Anticancer | (54) |
| GC | Down | Proliferation,
migration and apoptosis | miR-29b-1-5p and
miR-17-5p | Anticancer | (55,56) |
| CRC | Down | Invasion, migration
and multi-drug resistance | miR-17-5p | Anticancer | (57,58) |
HOTAIRM1 expression in glioma
Glioma is the most common primary central nervous
system tumor (59). Several
previous studies have reported a notable increase in HOTAIRM1
expression in glioblastoma tissues and cell lines when compared
with their normal counterparts (27–32).
Glioblastoma tissues and cells exhibit an abnormal HOTAIRM1
upregulation, which promotes glioma malignancy by enhancing cell
proliferation, migration, invasion and VM formation. The high
HOTAIRM1 expression is mediated by METTL3-dependent m6A
modification (33). Furthermore,
Snail family transcriptional repressor 2 transcriptionally
activated HOTAIRM1 and a strong association was observed between
increased HOTAIRM1 expression and worse prognosis in glioma
(34). HOTAIRM1 is also strongly
associated with decreased survival rates of patients diagnosed with
glioblastoma (35).
HOTAIRM1 expression in AML
AML is a heterogeneous disease characterized by
genetic irregularities (including mutations in genes like FLT3,
NPM1 and RUNX1) and epigenetic alterations (such as mutations in
regulators of DNA methylation like DNMT3A, TET2 and IDH1/2)
(60,61). Jing et al (36) observed that HOTAIRM1 expression was
increased in 14 AML samples with nucleophosmin 1 (NPM1) mutations
when compared with AML samples without NPM1 mutations. This finding
suggested a potential functional link between HOTAIRM1 and this
specific genetic irregularity, implying that HOTAIRM1 upregulation
may be part of the oncogenic machinery in NPM1-mutated AML.
Furthermore, Kaplan-Meier survival analysis on patients with AML
demonstrated a notably reduced survival time in the group with high
HOTAIRM1 expression. A different investigation demonstrated that
the treatment of acute promyelocytic leukemia cells with all-trans
retinoic acid led to an increase in HOTAIRM1 expression, which is
key to myeloid differentiation. Notably, the transcription factor
PU.1 binds to a specific DNA site (+1,100) in the promoter of the
HOTAIRM1 gene, therefore activating it and increasing its
expression (37). Furthermore, Hu
et al (38) demonstrated
that HOTAIRM1 expression is increased in patients with AML compared
with individuals without the disease. The functional analysis
indicated that HOTAIRM1 downregulation inhibits the growth and
triggers cell death in AML cells, indicating its tumorigenic
function in AML.
HOTAIRM1 expression in OS
OS is a prevalent and aggressive form of primary
bone cancer that mostly affects children and teenagers (62). HOTAIRM1 expression is markedly
increased in both OS samples and cell lines compared with their
non-cancerous counterparts. The association between HOTAIRM1
expression and the clinicopathological features of patients with OS
revealed that individuals with high HOTAIRM1 expression are prone
to having an advanced TNM stage (as defined by the American Joint
Committee on Cancer, 8th edition) (63). HOTAIRM1 upregulation stimulates the
growth and movement of cells while inhibiting cell death. These
findings suggest that HOTAIRM1, a cancer-causing gene in OS,
potentially holds promise in the identification and management of
this disease (39).
HOTAIRM1 expression in EC
In the female reproductive system, EC ranks as one
of the top three prevalent malignancies (64). Li et al (40) identified markedly higher HOTAIRM1
expression in type I EC tissues compared with normal endometrium
tissues and showed it is associated with the clinicopathological
features of affected patients. The increased expression of HOTAIRM1
is also strongly associated with advanced International Federation
of Gynecology and Obstetrics (FIGO) stage (according to the FIGO
staging system, 2023 edition) (65)
and the presence of lymph node metastasis. The suppression of
HOTAIRM1 inhibits the growth, movement, infiltration of type I EC
cells, epithelial-mesenchymal transition (EMT) and tumor growth
in vivo (40).
HOTAIRM1 expression in TC
TC is divided into three main histological groups:
i) Differentiated TC, which includes papillary, follicular and
oncocytic carcinomas; ii) medullary TC, often associated with
multiple endocrine neoplasia type 2 syndrome; and iii) anaplastic
TC, an aggressive malignancy that often arises from pre-existing
differentiated lesions and is characterized by a high mortality
rate (66,67). Zhang et al (41) reported that HOTAIRM1 gene is
amplified and its expression is increased in anaplastic TC compared
with PTC and normal thyroid tissue. Furthermore, patients with
anaplastic TC indicating higher HOTAIRM1 copy number and expression
have worse survival outcomes. The role of HOTAIRM1 in TC appears to
be complex and potentially context-dependent. While one study by Li
et al (42) reported that
elevated HOTAIRM1 expression in TC cells and tissues was associated
with advanced TNM stage and lymph node metastasis, another
investigation found contrary evidence, demonstrating that HOTAIRM1
expression was notably reduced in PTC tissues and that lower levels
correlated with lymph node metastasis and more advanced disease
(51). This discrepancy highlights
the need for further research to clarify the precise function of
HOTAIRM1 in TC progression.
HOTAIRM1 expression in NSCLC
NSCLC is responsible for ~85% of cancer-related
mortality globally, making it the primary contributor to lung
cancer mortalities worldwide (68).
Chen et al (43) observed
that HOTAIRM1 expression is markedly increased in NSCLC tissues
compared with tissues of a control group. A different study
demonstrated that HOTAIRM1 expression is associated with tumor
histological differentiation, tumor size, TNM stage and Ki-67
expression in patients with NSCLC. Furthermore, HOTAIRM1 expression
is increased in NSCLC compared with that in adjacent non-cancerous
tissues. Patients with NSCLC with low HOTAIRM1 expression have a
markedly longer overall survival compared with those with high
expression (44).
HOTAIRM1 expression in OSCC
OSCC is a malignant tumor with the highest
occurrence rate among tumors affecting mouth and face. OSCC is well
known for its tendency to recur and spread to other parts of the
body (69,70). Yu et al (45) reported that HOTAIRM1 expression is
increased in OSCC and it is closely associated with poor prognosis.
Systematic bioinformatics analyses revealed that HOTAIRM1 is
associated with tumor stage, overall survival, genomic instability,
tumor cell stemness, tumor microenvironment activity and immunocyte
infiltration.
HOTAIRM1 expression in PCa
PCa is a major contributor to cancer-associated
mortality among men, particularly in Western countries, with Africa
and Asia having the lowest incidence rates (71,72).
Wang et al (46) reported
that HOTAIRM1 expression is increased in PCa cells. The inhibition
of HOTAIRM1 expression prevents tumor cell proliferation while
inducing programmed cell death through the modulation of proteins
associated with apoptosis. The inhibition of HOTAIRM1 expression
limits the activity of the Wnt pathway in PCa cells, thereby
suppressing the malignant characteristics of tumor cells.
HOTAIRM1 expression in PDAC
PDAC is the most common form of PCa arising from the
epithelial lining of the pancreatic duct (73). Samples of 47 PDAC tissues and 5 cell
lines exhibit an abnormal increase of HOTAIRM1 expression compared
with its expression in a control group (47). Similarly, Zhou et al
(48) reported that HOTAIRM1
expression is increased in 12 PDAC tissue samples when compared
with the corresponding non-tumor samples.
HOTAIRM1 expression in OC
OC is the eighth most common type of cancer among
women worldwide and is the third most frequent gynecological cancer
after cervical cancer and EC (74).
Ye et al (49) reported an
overexpression of HOTAIRM1 in the human ovarian cancer cell line
SKOV3. Inhibition of HOTAIRM1 expression reduces cell proliferation
and increases cell death. Chao et al (50) observed that HOTAIRM1 expression is
reduced in both ovarian cancer tissues and cells and advanced FIGO
stage and lymphatic metastasis are associated with reduced HOTAIRM1
expression. HOTAIRM1 overexpression inhibits the growth and
invasion of OC cells, while enhancing apoptosis. Furthermore,
HOTAIRM1 slows OC tumor growth in vivo.
HOTAIRM1 expression in head and neck
cancer
Head and neck cancer is a frequently diagnosed form
of cancer globally, with an annual incidence of >600,000 new
cases (75,76). Zheng et al (52) reported that HOTAIRM1 expression is
decreased in 43 head carcinoma tissues and 41 neck carcinoma
tissues when compared with the corresponding adjacent normal
tissues. Furthermore, no association was identified between
HOTAIRM1 expression and age, sex or tumor location. However,
patients with increased HOTAIRM1 expression have a higher
probability to develop an advanced TNM stage, suggesting that
although HOTAIRM1 is generally downregulated in head and neck
cancer, tumors that maintain a relatively higher expression
(although still lower compared with normal tissues) are associated
with increased malignancy and progression.
HOTAIRM1 expression in HCC
HCC is classified as the sixth most prevalent tumor
and the third main reason of cancer fatality (77). Zhang et al (53) reported that HOTAIRM1 expression is
lower in HCC tissues compared with that in the adjacent
non-cancerous tissues. Furthermore, the receiver operating
characteristic curve demonstrated that HOTAIRM1 expression so a
notable level of sensitivity and specificity in detecting HCC. The
absence of disease progression in patients with HCC is associated
with tumor size and HOTAIRM1 expression. However, no association
was observed with age, sex, γ-glutamyl transferase levels,
α-fetoprotein levels, Child-Pugh grade (as defined by the American
Association for the Study of Liver Diseases practice guidelines)
(78), hepatitis B surface antigen
status, presence of cirrhosis, number of tumors, micro-vessel
metastasis, tumor differentiation and TNM stage of HCC.
HOTAIRM1 expression in ADC
Among NSCLCs, ADC is one of the key histological
subtypes with high incidence and mortality (79,80).
Thus, current studies on HOTAIRM1 primarily focus on ADC rather
than other NSCLC subtypes, such as squamous cell carcinoma or large
cell carcinoma. Chen et al (54) reported a notable decrease in
HOTAIRM1 expression in ADC tissues compared with that in normal
lung tissues. A clear association exists between the decrease of
HOTAIRM1 expression and clinical stage, metastasis to lymph nodes
and tumor size. Furthermore, HOTAIRM1 inhibition is associated with
poor overall survival in patients with ADC, as demonstrated by the
Kaplan-Meier analysis.
HOTAIRM1 expression in GC
GC is a prevalent malignancy worldwide. GC ranks as
the second most prevalent form of cancer in China and ranks as the
third leading cause of cancer-associated mortality (81). Xu et al (55) identified a notable decrease of
HOTAIRM1 expression in GC tissues is associated with a low survival
rate among patients with GC. These findings are consistent with
those reported by Lu et al (56) suggesting a marked decrease in
HOTAIRM1 expression in 20 gastric cancer tissues and cell lines.
Furthermore, HOTAIRM1 expression is associated with the
clinicopathological features of patients with GC and a strong
association exists between decreased HOTAIRM1 expression and
advanced TNM stage as well as lymph node metastasis.
HOTAIRM1 expression in CRC
CRC is one of the most prevalent malignancies
worldwide, with particularly high incidence in Western countries
(82,83). Wan et al (57) reported that HOTAIRM1 expression is
reduced in CRC tissues compared with that in normal tissues.
Furthermore, in the matched cohort, plasma HOTAIRM1 levels were
markedly lower in patients with CRC compared with those in healthy
controls. Ren et al (58)
identified a similar scenario where HOTAIRM1 expression is
downregulated in both CRC tissues and cell lines and is even lower
in 5-fluorouracil (FU)-resistant CRC tissues and cell lines. This
progressive downregulation in resistant tissues and cell lines
strongly implied that the loss of HOTAIRM1 is a key event in the
acquisition of chemoresistance, consistent with its established
role in suppressing cancer progression through mechanisms such as
the miR-17-5p/BTG3 axis (58).
Mechanisms involved in the regulation of
HOTAIRM1
LncRNAs interact with DNA, RNA or proteins as
molecular absorbers, frameworks and stimulators, exerting
regulatory functions in different biological processes, including
gene regulation, cellular differentiation and human diseases,
particularly cancer (84–88). HOTAIRM1 is involved in normal and
abnormal biological processes. Several molecular functions have
been identified after years of research and they are categorized
into three primary pathways: i) Interaction with DNA; ii)
interaction with RNA; and iii) interaction with proteins.
Interaction with DNA
HOTAIRM1 is involved in the methylation alteration
of several genes that are associated with tumors and in the
modification of histones (Fig. 2).
HOTAIRM1 controls gene expression through its interaction with
polycomb repressive complex 2 (PRC2), which consists of enhancer of
zeste homolog 2 (EZH2), suppressor of zeste 12 homolog and
embryonic ectoderm development protein. HOTAIRM1 catalyzes the
dimethylation and trimethylation of the histone 3 lysine residue 27
(H3K27me3), thus controlling the expression of its gene. Li et
al (89) identified that
HOTAIRM1 triggers the transcription of the homeobox A1 (HOXA1) gene
by reducing the levels of histone H3 lysine 9 (H3K9) dimethylation,
H3K27me3 and DNA methylation, which are markers associated with the
suppression of gene expression. HOTAIRM1 prevents the recruitment
of histone H3K9 methyltransferase, EZH2 and DNA methyltransferases
to the HOXA1 promoter during its interaction with them, thereby
reducing their local abundance at this site (89). Kim et al (90) reported that HOTAIRM1 directly
interacts with EZH2, the histone methyltransferases responsible for
H3K27me3 trimethylation, thereby preventing the deposition of
H3K27me3 marks at the putative HOXA1 promoter. Therefore, HOXA1
expression is increased in ER+ breast cancer cells.
Furthermore, HOTAIRM1 interacts with the histone demethylases PRC2
and ubiquitously transcribed tetratricopeptide repeat on chromosome
X/mixed lineage leukemia to control chromatin structure and
subsequently impacts the transcriptional activity of the HOXA gene
cluster (91).
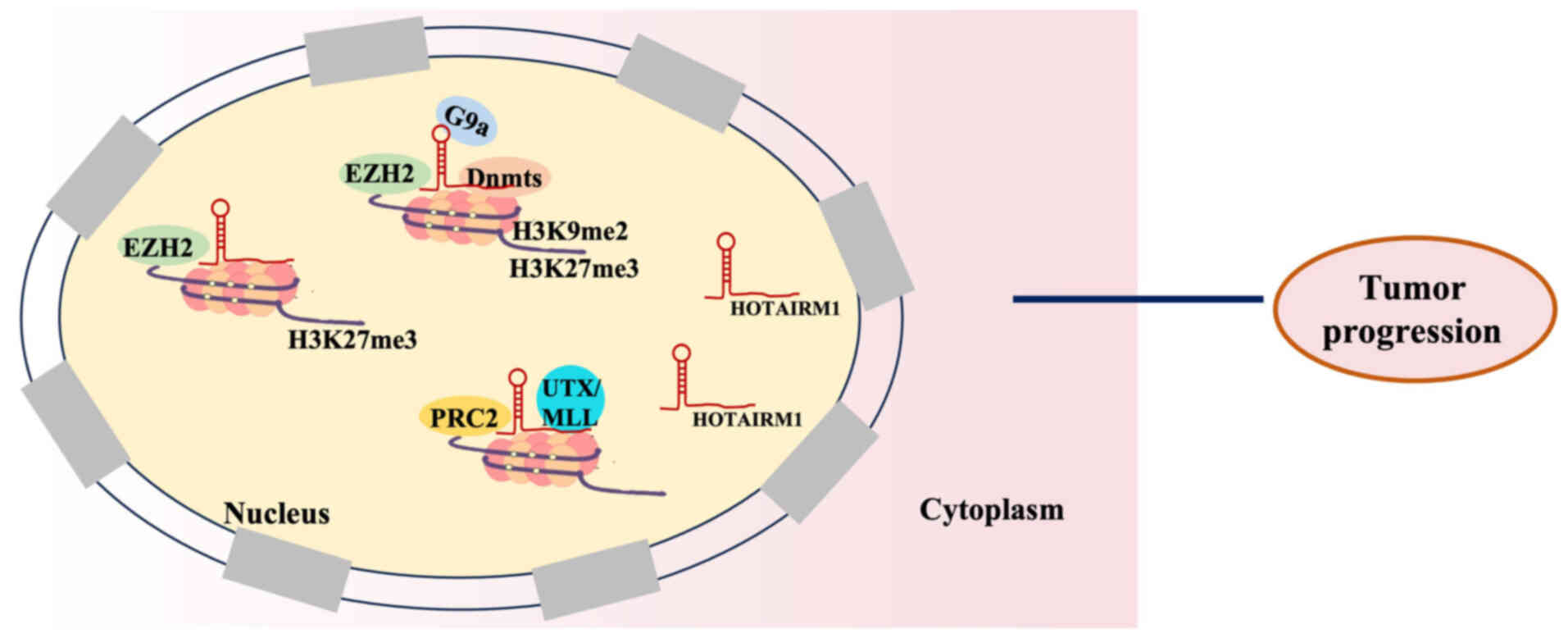 | Figure 2.HOTAIRM1 interaction with DNA to
exert a regulatory role in tumor progression. Interaction with
PRC2/EZH2: HOTAIRM1 directly interacts with EZH2 of the PRC2
complex, preventing the deposition of the repressive H3K27me3 mark
at target gene promoters; Interaction with H3K9 methyltransferases
and DNMTs: HOTAIRM1 binds to and prevents the recruitment of H3K9
methyltransferases and DNA methyltransferases to promoters, thereby
reducing repressive H3K9me2 and DNA methylation. Interaction with
histone demethylases: HOTAIRM1 interacts with histone demethylases
to help control chromatin structure and activate transcription of
target gene cluster. HOTAIRM1, HOXA transcript antisense RNA
myeloid-specific 1; EZH2, enhancer of zeste homolog 2; H3K27me3,
lysine residue 27 of histone 3; H3K9me2, histone H3 lysine 9
dimethylation; UTX, ubiquitously transcribed tetratricopeptide
repeat on chromosome X; MLL, mixed lineage leukemia; PRC2, polycomb
repressive complex 2; G9a, euchromatic histone-lysine
N-methyltransferase 2. |
Interaction with RNA
HOTAIRM1 also interacts with RNA to mediate several
molecular functions, including the promotion of cell proliferation
(33,36), invasion and metastasis (40,58).
The relevance of competitive endogenous RNA (ceRNA) networks in
cancer initiation and progression has become apparent in recent
years (92,93). MicroRNAs (miR/miRNA) are involved in
the ceRNA network, exerting a negative influence on mRNA
expression. The canonical mechanism involves miRNAs binding to the
3′-untranslated regions of target mRNAs, which leads to their
demethylation and subsequent destabilization (94,95).
HOTAIRM1 competitively binds to specific miRNAs through the ceRNA
mechanism, thereby relieving the repression of their target mRNAs
and modulating tumor progression (Fig.
3). HOTAIRM1 competitively binds to miR-152-3p, functioning as
a ceRNA that inhibits miR-152-3p activity and therefore upregulates
its target Unc-51 like kinase 3 (ULK3), thereby modulating various
leukemic cell processes, including autophagy, proliferation and
apoptosis (36). Yu et al
(39) identified that HOTAIRM1
functions as a molecular sponge for miR-664b-3p, thereby
upregulating Ras homolog enriched in brain (Rheb) and activating
the mTOR pathway to promote the Warburg effect in OS. Furthermore,
it enhances E26 transformation-specific 1 (ETS1) mRNA expression
and facilitates the osteogenic differentiation of mesenchymal stem
cells derived from human bone marrow (96). Similarly, HOTAIRM1 promotes PH
domain leucine-rich repeat protein phosphatase 1 upregulation in GC
cells by sponging miR-29b-1-5p, thus demonstrating a classic ceRNA
mechanism (55). Wang et al
(97) reported that the
HOTAIRM1/miR-519a-3p axis is markedly involved in the
proliferation, apoptosis, inflammation and oxidative stress of
neuroblastoma cells when exposed to the toxic metabolite
1-methyl-4-phenylpyridinium. HOTAIRM1 directly regulates mRNA
stability. Specifically, its binding to insulin-like growth factor
binding protein 2 (IGFBP2) mRNA increases IGFBP2 expression, thus
promoting glioma cell proliferation, migration, invasion and
vasculogenic mimicry formation (27).
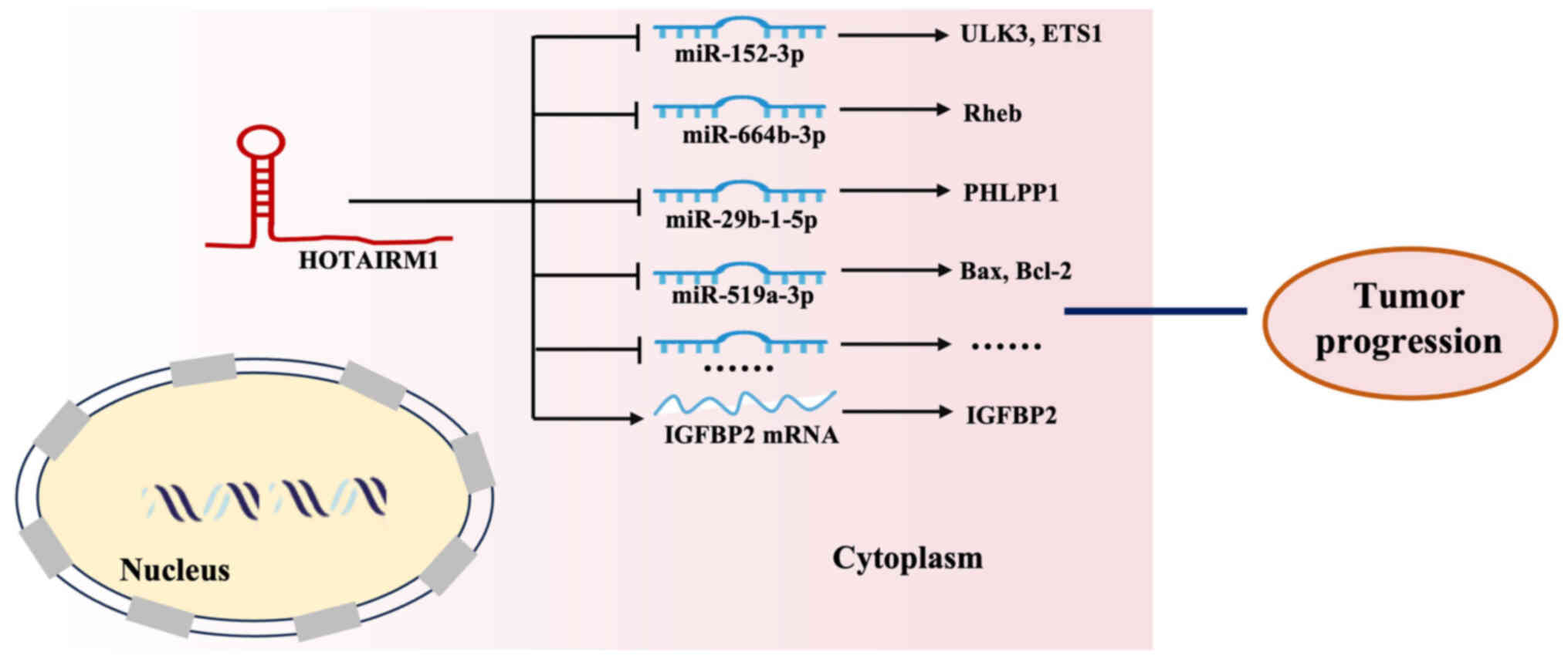 | Figure 3.HOTAIRM1 interaction with RNA to
exert a regulatory role in tumor progression. Interaction with
specific miRNAs (e.g., miR-152-3p, miR-664b-3p, miR-29b-1-5p,
miR-519a-3p): HOTAIRM1 functions as a competitive endogenous RNA or
‘molecular sponge’ by binding to these miRNAs, leading to the
upregulation of genes like ULK3, ETS1, Rheb, PHLPP1, Bax, and
Bcl-2, which in turn modulates tumor progression. Interaction with
IGFBP2 mRNA: HOTAIRM1 directly binds to IGFBP2 mRNA to promote
tumor progression. HOTAIRM1, HOXA transcript antisense RNA
myeloid-specific 1; miR, microRNA; ULK3, Unc-51 like kinase 3;
ETS1, E26 transformation-specific 1; Rheb, Ras homolog enriched in
brain; PHLPP1, PH domain leucine-rich repeat protein phosphatase 1;
IGFBP2, insulin-like growth factor binding protein 2. |
Interaction with proteins
Several lncRNAs including HOTAIRM1 are involved in
the molecular regulation of proteins by directly binding to them
(Fig. 4). For example, HOTAIRM1
interacts with heat shock protein family A (Hsp70) member 5 (HSPA5)
and transcriptionally regulates its expression, with the resulting
effects on proliferation being partly dependent on this chaperone
(98). Han et al (98) demonstrated that HOTAIRM1 forms a
complex with polypyrimidine tract-binding protein 1 and
insulin-like growth factor 2 mRNA-binding protein 2, strengthening
their interaction and facilitating their recruitment to the serine
hydroxymethyltransferase 2 (SHMT2) mRNA. Therefore, the stability
of SHMT2 mRNA has improved, leading to an increase in SHMT2 protein
expression. This ultimately induces mitochondrial activity and
promotes the malignant advancement of glioma. Liu et al
(28) reported that HOTAIRM1 binds
to the RNA-binding protein fused in sarcoma (FUS), thereby
regulating E2F transcription factor 7 expression, promoting the
proliferation, migration and invasion of glioma stem
cell-transformed mesenchymal stem cells. In addition, Chen et
al (99) demonstrated that the
regulation of programmed cell death-ligand 1 expression in lung
alveolar epithelial cells is controlled by HOTAIRM1 through its
interaction with the key transcription factor HOXA1, having
alleviated lung injury and improved survival of mice. Jing et
al (36) demonstrated that the
interaction of HOTAIRM1 with early growth response 1 (EGR1) and
murine double minute 2 homolog (MDM2) suggests that it serves as a
scaffold facilitating MDM2 recruitment to EGR1, thereby promoting
autophagy and proliferation in leukemia cells.
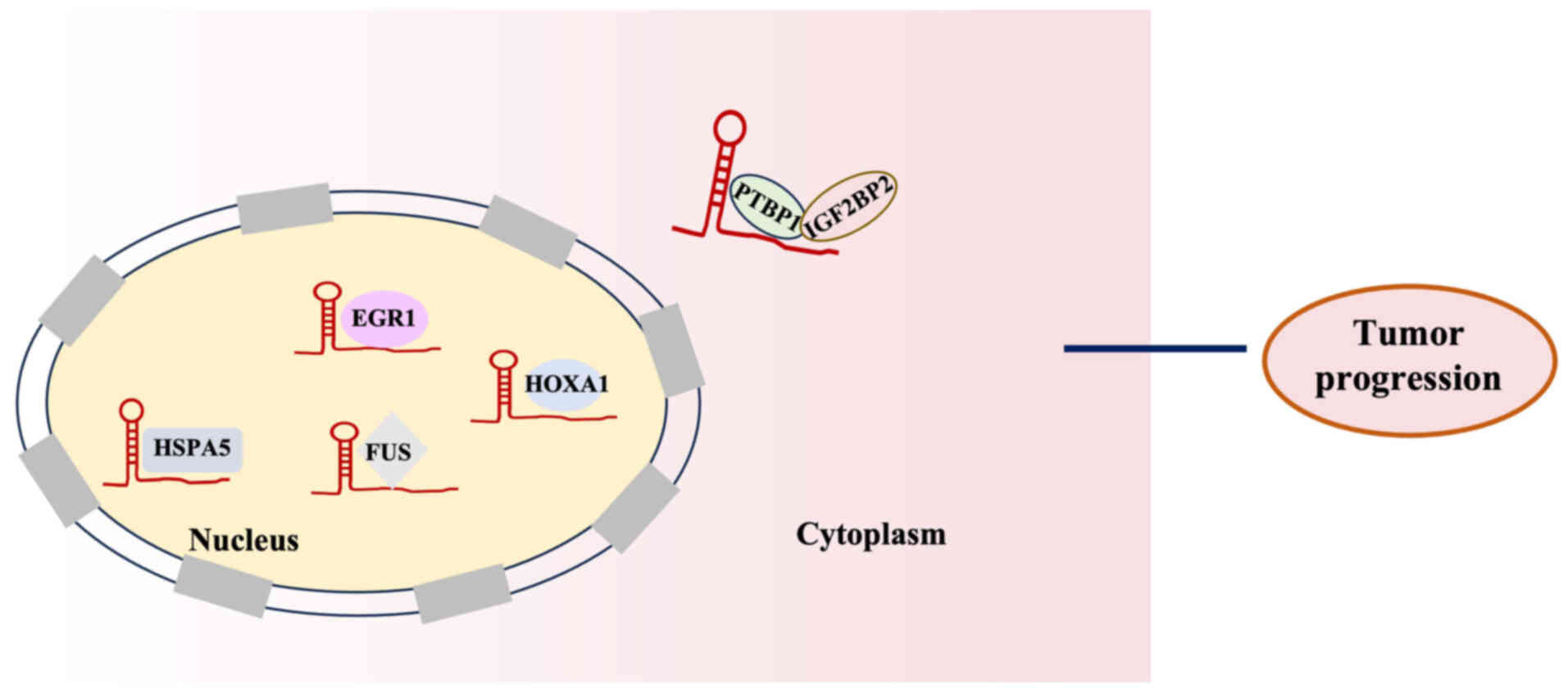 | Figure 4.HOTAIRM1 interaction with proteins to
exert a regulatory role in tumor progression. Interaction with
HSPA5: HOTAIRM1 interacts with HSPA5 to transcriptionally regulate
its expression, influencing tumor progression. Interaction with
PTBP1 and IGF2BP2: HOTAIRM1 forms a complex with PTBP1 and IGF2BP2
to enhance its stability, thereby promoting tumor progression.
Interaction with FUS: HOTAIRM1 binds to the RNA-binding protein FUS
to promote tumor progression. Interaction with HOXA1: HOTAIRM1
interacts with HOXA1 to regulate tumor progression. Interaction
with EGR1: HOTAIRM1 acts as a scaffold, facilitating the EGR1 to
promote tumor progression. HOTAIRM1, HOXA transcript antisense RNA
myeloid-specific 1; IGF2BP2, insulin-like growth factor 2
mRNA-binding protein 2; EGR1, early growth response 1; FUS, fused
in sarcoma; HSPA2, heat shock protein family A (Hsp70) member 2;
PTBP1, polypyrimidine tract-binding protein 1; HOXA1, homeobox
A1. |
HOTAIRM1 regulates various hallmarks of
cancer
Cancer is complex disease characterized by multiple
genetic abnormalities, including epigenetic alterations,
chromosomal translocations and gene deletions or amplifications.
The human genome encodes several lncRNAs (such as PVT1, HOTAIR and
MALAT1), most of which are not translated into proteins. Despite
their lack of coding potential, lncRNAs exert key regulatory
functions among different cellular processes (11,100).
Among them, HOTAIRM1 modulates multiple signaling pathways (Wnt,
Caspase signaling pathways) and is associated with hallmark cancer
traits such as cell proliferation, apoptosis, invasion, metastasis,
metabolic reprogramming and angiogenesis (Fig. 5).
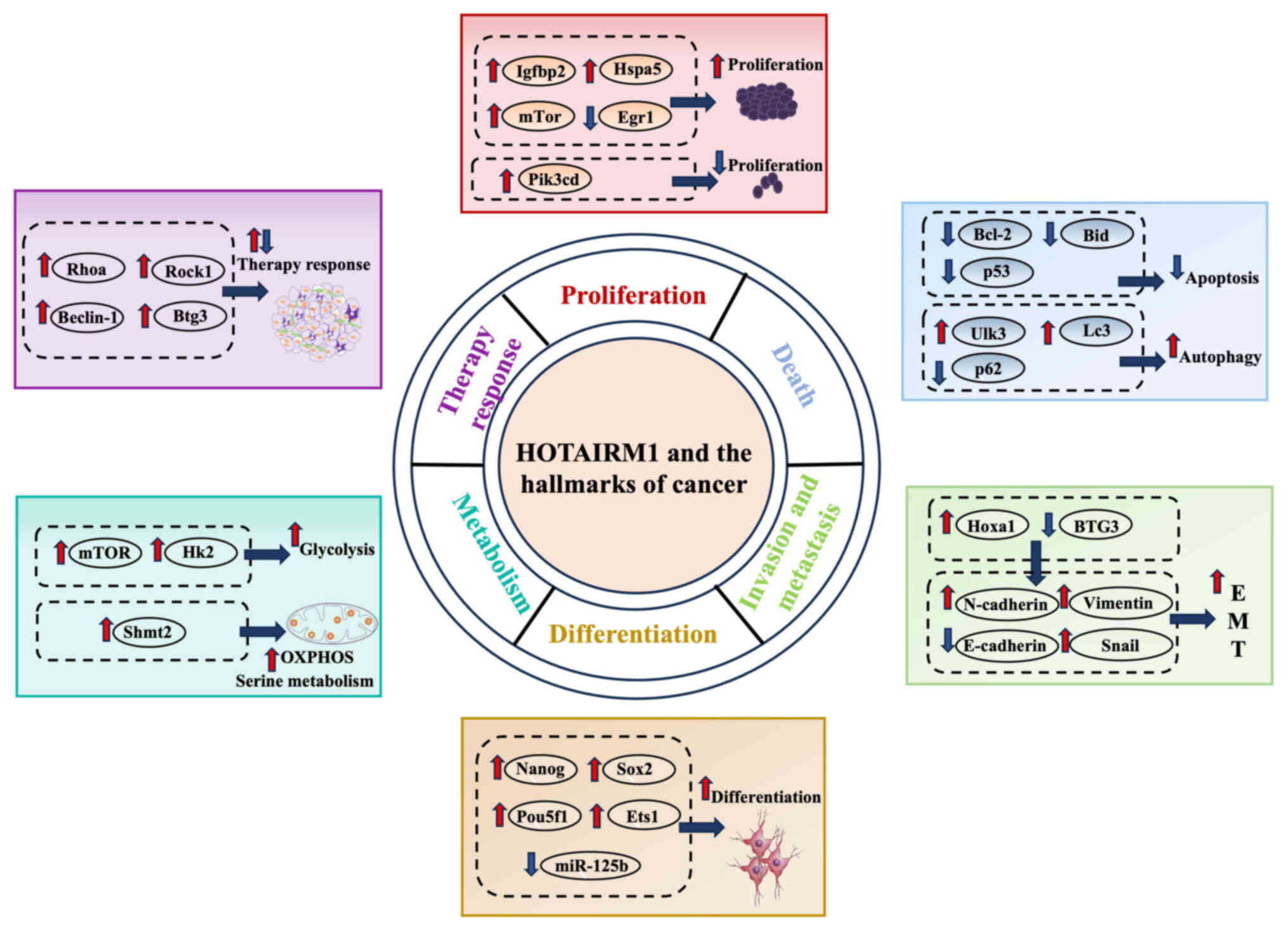 | Figure 5.HOTAIRM1 regulates various hallmarks
of cancer. HOTAIRM1 promotes cell proliferation by upregulating
Igfbp2/Hspa5/mTor or downregulating Egr1, while also upregulating
Pik3cd to exert an inhibitory effect on proliferation; HOTAIRM1
suppresses apoptosis by downregulating Bcl-2/Bid/p53 and promotes
autophagy by upregulating Ulk3/LC3 while downregulating p62;
HOTAIRM1 promotes EMT by upregulating Hoxa1 and downregulating
BTG3, leading to increased N-cadherin/Vimentin/Snail and decreased
E-cadherin; HOTAIRM1 promotes cell differentiation by upregulating
Nanog/Sox2/Pou5f1/Ets1 and downregulating miR-125b; HOTAIRM1
promotes metabolic reprogramming by upregulating mTOR/Hk2 to
enhance glycolysis and increasing Shmt2 to stimulate serine
metabolism; HOTAIRM1 modulates therapy response through
upregulating RhoA/ROCK1/Beclin-1/BTG3 expression. HOTAIRM1, HOXA
transcript antisense RNA myeloid-specific 1; Igfbp2, insulin-like
growth factor binding protein 2; Egr1, early growth response 1;
miR, microRNA; Ulk3, Unc-51 like kinase 3; Hoxa1, homeobox A1;
Ets1, E26 transformation-specific 1; Shmt2, serine
hydroxymethyltransferase 2; Hk2, hexokinase 2; Btg3, B-cell
translocation gene 3; Rock1, ρ-associated coiled-coil containing
protein kinase 1; Rhoa, Ras homolog family member A; Bid, BH3
interacting-domain death agonist; Pik3cd,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
∆. |
Proliferation
Dysregulated cell proliferation resulting in
uncontrolled growth represents a fundamental hallmark of
tumorigenesis (101). Aberrant
expression of HOTAIRM1 has been reported in multiple tumor types
and is closely associated with enhanced tumor cell proliferation.
For example, Wu et al (33)
identified an increase in HOTAIRM1 expression in both glioma
tissues and cells. Furthermore, HOTAIRM1 overexpression increases
the proliferation of glioma cells. Zhou et al (102) identified a key functional
connection among HOXA4, HOTAIRM1 and HSPA5, forming a novel
regulatory circuit that governs HUVEC proliferation. HOTAIRM1
promotes OS cell proliferation by sponging miR-664b-3p, thereby
activating the mTOR pathway (39).
On the other hand, HOTAIRM1 also suppresses the proliferative
ability of granulosa cells (103).
Furthermore, Jing et al (36) demonstrated that HOTAIRM1 markedly
promotes the proliferation of AML cells harboring NPM1
mutations.
Cell death
Apoptosis is a programmed form of cell death that
maintains cellular balance. The induction of apoptosis in cancer
cells is a key strategy in clinical cancer therapy. Dahariya et
al (104) reported that
HOTAIRM1 functions as a molecular decoy for miR-125b regulating
apoptosis to facilitate the terminal differentiation of
megakaryocytes, thereby regulating key processes including
apoptosis, cyclin D1-dependent cell cycling, and reactive oxygen
species production. HOTAIRM1 also induces Jurkat cell apoptosis
through the KIT/AKT signaling pathway (105). Liu et al (106) demonstrated that the loss of
HOTAIRM1 activity causes an abnormal increase in chondrocyte
apoptosis. Similarly, Ye et al (49) reported that HOTAIRM1 suppression
increases the expression of pro-apoptotic factors, including Bad
and Bax, while it reduces the expression of BH3 interacting-domain
death agonist and Bcl-2 (anti-apoptotic factors) in ovarian cancer
cells.
Autophagy is a self-degradative process in which
cytoplasmic proteins and organelles are delivered to lysosomes to
maintain cellular metabolism and homeostasis. A previous study has
reported that HOTAIRM1 induces autophagy by upregulating ULK3
expression, leading to an increased expression of autophagy-related
proteins including microtubule-associated protein LC3 II and a
concomitant decrease in the autophagy substrate p62, thereby
promoting cell proliferation and exerting a pro-tumorigenic effect
(36).
Invasion and metastasis
Metastasis is a complex, multistep process involving
EMT, invasion, intravasation, cell survival in the bloodstream,
extravasation and the formation of secondary tumor colonies. The
initial step in cancer metastasis is the EMT, during which
epithelial cells lose their polarity and intercellular adhesion,
acquire mesenchymal characteristics and gain enhanced migratory and
invasive abilities. The upregulation of HOXA1 expression by
HOTAIRM1 enhances the ability of type I EC cells to undergo EMT and
metastasis, resulting in a decrease in the epithelial marker
E-cadherin expression and an increase in the mesenchymal marker
N-cadherin (40). Ren et al
(58) examined the impact of
HOTAIRM1 dysregulation in 5-FU-resistant CRC cells and identified
that the increased expression of HOTAIRM1 reduces cell invasion and
migration according to EMT assay.
Cell differentiation
Cell differentiation is the process through which
cells acquire specialized function and form distinct tissues. By
contrast, carcinogenesis represents a breakdown of normal
differentiation, resulting in the uncontrolled proliferation of
immature or aberrant cells. Tollis et al (107) used a model framework of spinal
motor neurons to demonstrate that neuronal HOTAIRM1, a specific
isoform of HOTAIRM1, influences cell fate between motor neurons and
interneurons by promoting motor neuron differentiation. HOTAIRM1
contributes to proper progression during the early stages of
neuronal differentiation. It also influences the core pluripotency
network composed of NANOG, POU class 5 homeobox 1 and Sox2, thereby
helping to maintain cells in an undifferentiated state (108). Wang et al (73) identified HOTAIRM1 as a novel
regulator of the osteogenic differentiation of bone marrow-derived
mesenchymal stem cells. This regulation occurs through the
miR-152-3p/ETS1 axis, suggesting that HOTAIRM1 may represent a
potential therapeutic target for osteoporosis. Furthermore,
Dahariya et al (104)
demonstrated that HOTAIRM1 regulates the p53-mediated control of
cyclin D1 expression during megakaryocytopoiesis. The function of
HOTAIRM1 is to promote megakaryocyte maturation by acting as a
molecular sponge for miR-125b.
Metabolism
Tumor cells undergo notable metabolic reprogramming
to support their rapid proliferation. Unlike regular cells, they
preferentially use aerobic glycolysis, an energetically inefficient
pathway with a considerably higher turnover rate, even under
oxygen-sufficient conditions, a phenomenon known as the Warburg
effect. The miR-664b-3p/Rheb/mTOR axis markedly improves the
Warburg effect in OS cells, due to the notable enhancement by
HOTAIRM1 (39). HOTAIRM1 knockdown
attenuates the Warburg effect in NSCLC by reducing glucose uptake
and lactate production. This metabolic suppression is associated
with a marked decrease in hexokinase 2 protein expression, a key
glycolytic enzyme that phosphorylates glucose to initiate
glycolysis and sustain cancer cell energy metabolism (43). Han et al (98) reported that HOTAIRM1 increases SHMT2
protein expression by improving the lifespan of its mRNA, leading
to the stimulation of mitochondrial activity through oxidative
phosphorylation and serine metabolism.
Therapy response
Despite notable advances in antitumor therapies,
resistance to chemotherapy, radiotherapy, targeted therapy and
immunotherapy still represents a major obstacle to an effective
cancer treatment (109). HOTAIRM1
is involved in the response to therapy by cancer cells. According
to previous research, HOTAIRM1 interacts with the inhibitory region
of ρ GTPase-activating protein 18 and suppresses its expression,
resulting in the activation of the Ras homolog family member
A/ρ-associated coiled-coil containing protein kinase 1 signaling
pathway and enhancing leukemia cell resistance to glucocorticoid
treatment (110). Gu et al
(111) reported that HOTAIRM1
upregulation leads to lenvatinib resistance by reducing miR-34a
expression and increasing Beclin-1 expression in HCC cancer cells.
In addition, Chen et al (112) revealed that HOTAIRM1 suppression
increases the effectiveness of cytarabine in killing cells by
controlling the Wnt/β-catenin/platelet-type isoform of
phosphofructokinase signaling pathway. In vitro and in
vivo, HOTAIRM1 functions as a tumor suppressor in
5-FU-resistant CRC cells by reducing the activity of the
miR-17-5p/B-cell translocation gene 3 (BTG3) pathway and preventing
the development of multi-drug resistance (58).
Conclusion and prospects
Cancer remains a major global threat to human
health. Although recent advances have slightly reduced the overall
mortality rates, notable obstacles persist in the early diagnosis
and effective management. Numerous patients are still diagnosed at
advanced stages of the disease, which notably worsens their
prognosis (113,114). Hence, it is key to identify novel
biomarkers and examine the different molecular pathways involved in
cancer for its timely detection and management.
Several studies have indicated the abnormal
expression of lncRNAs in several diseases and their ability to
function as tumor suppressors or oncogenes (115,116). HOTAIRM1 exhibits an abnormal
expression in both tumor tissues and cells of different types of
cancer and it serves as a standalone indicator for unfavorable
prognosis in a wide range of carcinomas. HOTAIRM1 upregulation
promotes tumor cell proliferation in several neoplasms, including
glioma, AML, OS, EC, TC, NSCLC, OSCC, PCa, PDAC and OC. However,
HOTAIRM1 upregulation suppresses cancer cell proliferation in PTC,
OC, HNT, HCC, ADC, GC and CRC. Furthermore, a strong association
exists between atypical HOTAIRM1 expression and several clinical
and pathological characteristics of tumors, including age, tumor
size, invasion of blood vessels, metastasis, overall survival and
recurrence. These findings support the idea that the cellular
function of HOTAIRM1 is not static but context-dependent.
Initially, HOTAIRM1 triggers H3K27me3 by interacting
with EZH2/PRC2 and also participates in the alteration of chromatin
structure by directing the recruitment of chromatin-modifying
enzymes to a specific gene location. Furthermore, HOTAIRM1 acts as
a molecular sponge for miRNAs, exerting its regulatory effects
through the HOTAIRM1-miRNA-mRNA pathway. In addition, HOTAIRM1
binds to functional proteins within both the nucleus and cytoplasm,
thereby modulating their expression and markedly influencing tumor
progression. Other potential molecular mechanisms underlying the
biological functions of HOTAIRM1 warrant further investigation.
HOTAIRM1 potentially regulates tumors and impacts
multiple aspects of cancer, such as cell proliferation, death,
invasion, spread, cellular differentiation, metabolism and
resistance to chemotherapy. Nevertheless, in certain contexts, such
as drug resistance, the exact function of HOTAIRM1 remains to be
elucidated, as current studies report conflicting results that
require further detailed investigation. Liang et al
(110) revealed that HOTAIRM1
promotes GC resistance by inhibiting apoptosis in leukemia cells.
Notably, Ren et al (58)
reported that HOTAIRM1 suppresses the miR-17-5p/BTG3 pathway,
leading to the inhibition of multi-drug resistance. Currently,
further in-depth research is required to explore the potential
ability of HOTAIRM1 to control additional characteristics of
cancer, including the evasion of immune surveillance, genome
instability and mutation, non-mutational epigenetic reprogramming,
unlocking phenotypic plasticity and polymorphic microbiomes.
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science Foundation of
Beijing Municipal (grant no. 7254399) and supported by the
Fundamental Research Funds for the Central Universities (grant no.
APL24100310010301071027).
Availability of data and materials
Not applicable.
Authors' contributions
YJ and YG conceptualized the present review. YJ and
XL prepared the original draft. YJ and YG reviewed and edited the
manuscript. YJ and YG provided supervision and project
administration. YJ obtained funding for the present review. All
authors read and approved the final manuscript. Data authentication
is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
HOTAIRM1
|
HOXA transcript antisense RNA
myeloid-specific 1
|
|
AML
|
acute myeloid leukemia
|
|
OS
|
osteosarcoma
|
|
EC
|
endometrial cancer
|
|
EMT
|
epithelial-mesenchymal transition
|
|
TC
|
thyroid cancer
|
|
NSCLC
|
non-small cell lung cancer
|
|
OSCC
|
oral squamous cell carcinoma
|
|
PCa
|
prostate cancer
|
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
HCC
|
hepatocellular carcinoma
|
|
ADC
|
lung adenocarcinoma
|
|
GC
|
gastric cancer
|
|
CRC
|
colorectal cancer
|
|
PRC2
|
polycomb repressive complex 2
|
|
H3K27me3
|
lysine residue 27 of histone 3
|
References
|
1
|
Vaghari-Tabari M, Ferns GA, Qujeq D,
Andevari AN, Sabahi Z and Moein ZS: Signaling, metabolism, and
cancer: An important relationship for therapeutic intervention. J
Cell Physiol. 236:5512–5532. 2021. View Article : Google Scholar
|
|
2
|
Giunta S: Decoding human cancer with whole
genome sequencing: A review of PCAWG Project studies published in
February 2020. Cancer Metastasis Rev. 40:909–924. 2021. View Article : Google Scholar
|
|
3
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar
|
|
4
|
Ramezankhani R, Solhi R, Es HA, Vosough M
and Hassan M: Novel molecular targets in gastric adenocarcinoma.
Pharmacol Ther. 220:1077142021. View Article : Google Scholar
|
|
5
|
Guo N, Liu JB, Li W, Ma YS and Fu D: The
power and the promise of CRISPR/Cas9 genome editing for clinical
application with gene therapy. J Adv Res. 40:135–152. 2022.
View Article : Google Scholar
|
|
6
|
Traba J, Sack MN, Waldmann TA and Anton
OM: Immunometabolism at the nexus of cancer therapeutic efficacy
and resistance. Front Immunol. 12:6572932021. View Article : Google Scholar
|
|
7
|
Ghafouri-Fard S, Khoshbakht T, Taheri M
and Hajiesmaeili M: Long intergenic non-protein coding RNA 460:
Review of its role in carcinogenesis. Pathol Res Pract.
225:1535562021. View Article : Google Scholar
|
|
8
|
Hovhannisyan H and Gabaldón T: The long
non-coding RNA landscape of Candida yeast pathogens. Nat Commun.
12:73172021. View Article : Google Scholar
|
|
9
|
Kovalenko TF, Yadav B, Anufrieva KS,
Rubtsov YP, Zatsepin TS, Shcherbinina EY, Solyus EM, Staroverov DB,
Larionova TD, Latyshev YA, et al: Functions of long non-coding RNA
ROR in patient-derived glioblastoma cells. Biochimie. 200:131–139.
2022. View Article : Google Scholar
|
|
10
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar
|
|
11
|
Herman AB, Tsitsipatis D and Gorospe M:
Integrated lncRNA function upon genomic and epigenomic regulation.
Mol Cell. 82:2252–2266. 2022. View Article : Google Scholar
|
|
12
|
Sun KK, Zu C, Wu XY, Wang QH, Hua P, Zhang
YF, Shen XJ and Wu YY: Identification of lncRNA and mRNA regulatory
networks associated with gastric cancer progression. Front Oncol.
13:11404602023. View Article : Google Scholar
|
|
13
|
Pierce JB, Zhou H, Simion V and Feinberg
MW: Long noncoding RNAs as therapeutic targets. Adv Exp Med Biol.
1363:161–175. 2022. View Article : Google Scholar
|
|
14
|
Sun W, Xu J, Wang L, Jiang Y, Cui J, Su X,
Yang F, Tian L, Si Z and Xing Y: Non-coding RNAs in cancer
therapy-induced cardiotoxicity: Mechanisms, biomarkers, and
treatments. Front Cardiovasc Med. 9:9461372022. View Article : Google Scholar
|
|
15
|
Zhu Y, Ren J, Wu X, Zhang Y, Wang Y, Xu J,
Tan Q, Jiang Y and Li Y: lncRNA ENST00000422059 promotes cell
proliferation and inhibits cell apoptosis in breast cancer by
regulating the miR-145-5p/KLF5 axis. Acta Biochim Biophys Sin
(Shanghai). 55:1892–1901. 2023.
|
|
16
|
Bah I, Youssef D, Yao ZQ, McCall CE and El
Gazzar M: Hotairm1 controls S100A9 protein phosphorylation in
myeloid-derived suppressor cells during sepsis. J Clin Cell
Immunol. 14:10006912023.
|
|
17
|
Zhang X, Weissman SM and Newburger PE:
Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle
progression during myeloid maturation in NB4 human promyelocytic
leukemia cells. RNA Biol. 11:777–787. 2014. View Article : Google Scholar
|
|
18
|
Basyegit H, Tatar BG, Kose S, Gunduz C,
Ozmen Yelken B and Yilmaz Susluer S: Exploring the role of long
non-coding RNAs in predicting outcomes for hepatitis B patients.
Asian Pac J Cancer Prev. 25:4313–4321. 2024. View Article : Google Scholar
|
|
19
|
Bagheri-Mohammadi S, Karamivandishi A,
Mahdavi SA and Siahposht-Khachaki A: New sights on long non-coding
RNAs in glioblastoma: A review of molecular mechanism. Heliyon.
10:e397442024. View Article : Google Scholar
|
|
20
|
Nekoeian S, Rostami T, Norouzy A, Hussein
S, Tavoosidana G, Chahardouli B, Rostami S, Asgari Y and Azizi Z:
Identification of lncRNAs associated with the progression of acute
lymphoblastic leukemia using a competing endogenous RNAs network.
Oncol Res. 30:259–268. 2023. View Article : Google Scholar
|
|
21
|
Chen R, Zhou D, Chen Y, Chen M and Shuai
Z: Understanding the role of exosomal lncRNAs in rheumatic
diseases: A review. PeerJ. 11:e164342023. View Article : Google Scholar
|
|
22
|
Ramos TAR, Urquiza-Zurich S, Kim SY,
Gillette TG, Hill JA, Lavandero S, Rêgo TG and Maracaja-Coutinho V:
Single-cell transcriptional landscape of long non-coding RNAs
orchestrating mouse heart development. Cell Death Dis. 14:8412023.
View Article : Google Scholar
|
|
23
|
Hu C, Dai Q, Zhang R, Yang H, Wang M, Gu
K, Yang J, Meng W, Chen P and Xu M: Case report: Identification of
a novel LYN::LINC01900 transcript with promyelocytic phenotype and
TP53 mutation in acute myeloid leukemia. Front Oncol.
13:13224032023. View Article : Google Scholar
|
|
24
|
Wang D, Zhao X, Li S, Guo H, Li S and Yu
D: The impact of LncRNA-SOX2-OT/let-7c-3p/SKP2 Axis on head and
neck squamous cell carcinoma progression: Insights from
bioinformatics analysis and experimental validation. Cell Signal.
115:1110182024. View Article : Google Scholar
|
|
25
|
Yu M, He X, Liu T and Li J: lncRNA
GPRC5D-AS1 as a ceRNA inhibits skeletal muscle aging by regulating
miR-520d-5p. Aging (Albany NY). 15:13980–13997. 2023. View Article : Google Scholar
|
|
26
|
Zhang X, Zhong Y, Liu L, Jia C, Cai H,
Yang J, Wu B and Lv Z: Fasting regulates mitochondrial function
through lncRNA PRKCQ-AS1-mediated IGF2BPs in papillary thyroid
carcinoma. Cell Death Dis. 14:8272023. View Article : Google Scholar
|
|
27
|
Wu Z, Lin Y and Wei N:
N6-methyladenosine-modified HOTAIRM1 promotes vasculogenic mimicry
formation in glioma. Cancer Sci. 114:129–141. 2023. View Article : Google Scholar
|
|
28
|
Liu L, Zhou Y, Dong X, Li S, Cheng S, Li
H, Li Y, Yuan J, Wang L and Dong J: HOTAIRM1 maintained the
malignant phenotype of tMSCs transformed by GSCs via E2F7 by
binding to FUS. J Oncol. 2022:77344132022.
|
|
29
|
Shi T, Guo D, Xu H, Su G, Chen J, Zhao Z,
Shi J, Wedemeyer M, Attenello F, Zhang L and Lu W: HOTAIRM1, an
enhancer lncRNA, promotes glioma proliferation by regulating
long-range chromatin interactions within HOXA cluster genes. Mol
Biol Rep. 47:2723–2733. 2020. View Article : Google Scholar
|
|
30
|
Wang H, Li H, Jiang Q, Dong X, Li S, Cheng
S, Shi J, Liu L, Qian Z and Dong J: HOTAIRM1 promotes malignant
progression of transformed fibroblasts in glioma stem-like cells
remodeled microenvironment via regulating miR-133b-3p/TGFβ Axis.
Front Oncol. 11:6031282021. View Article : Google Scholar
|
|
31
|
Xia H, Liu Y, Wang Z, Zhang W, Qi M, Qi B
and Jiang X: Long noncoding RNA HOTAIRM1 maintains tumorigenicity
of glioblastoma stem-like cells through regulation of HOX gene
expression. Neurotherapeutics. 17:754–764. 2020. View Article : Google Scholar
|
|
32
|
Hao Y, Li X, Chen H, Huo H, Liu Z and Chai
E: Over-expression of long noncoding RNA HOTAIRM1 promotes cell
proliferation and invasion in human glioblastoma by up-regulating
SP1 via sponging miR-137. Neuroreport. 31:109–117. 2020. View Article : Google Scholar
|
|
33
|
Wu Z and Wei N: METTL3-mediated HOTAIRM1
promotes vasculogenic mimicry icontributionsn glioma via regulating
IGFBP2 expression. J Transl Med. 21:8552023. View Article : Google Scholar
|
|
34
|
Xie P, Li X, Chen R, Liu Y, Liu DC, Liu W,
Cui G and Xu J: Upregulation of HOTAIRM1 increases migration and
invasion by glioblastoma cells. Aging (Albany NY). 13:2348–2364.
2020. View Article : Google Scholar
|
|
35
|
Ahmadov U, Picard D, Bartl J, Silginer M,
Trajkovic-Arsic M, Qin N, Blümel L, Wolter M, Lim JKM, Pauck D, et
al: The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness
and radiotherapy resistance in glioblastoma. Cell Death Dis.
12:8852021. View Article : Google Scholar
|
|
36
|
Jing Y, Jiang X, Lei L, Peng M, Ren J,
Xiao Q, Tao Y, Tao Y, Huang J, Wang L, et al: Mutant NPM1-regulated
lncRNA HOTAIRM1 promotes leukemia cell autophagy and proliferation
by targeting EGR1 and ULK3. J Exp Clin Cancer Res. 40:3122021.
View Article : Google Scholar
|
|
37
|
Wei S, Zhao M, Wang X, Li Y and Wang K:
PU.1 controls the expression of long noncoding RNA HOTAIRM1 during
granulocytic differentiation. J Hematol Oncol. 9:442016. View Article : Google Scholar
|
|
38
|
Hu N, Chen L, Li Q and Zhao H: LncRNA
HOTAIRM1 is involved in the progression of acute myeloid leukemia
through targeting miR-148b. RSC Adv. 9:10352–10359. 2019.
View Article : Google Scholar
|
|
39
|
Yu X, Duan W, Wu F, Yang D, Wang X, Wu J,
Zhou D and Shen Y: LncRNA-HOTAIRM1 promotes aerobic glycolysis and
proliferation in osteosarcoma via the miR-664b-3p/Rheb/mTOR
pathway. Cancer Sci. 114:3537–3552. 2023. View Article : Google Scholar
|
|
40
|
Li X, Pang L, Yang Z, Liu J, Li W and Wang
D: LncRNA HOTAIRM1/HOXA1 axis promotes cell proliferation,
migration and invasion in endometrial cancer. Onco Targets Ther.
12:10997–11015. 2019. View Article : Google Scholar
|
|
41
|
Zhang L, Zhang J, Li S, Zhang Y, Liu Y,
Dong J, Zhao W, Yu B, Wang H and Liu J: Genomic amplification of
long noncoding RNA HOTAIRM1 drives anaplastic thyroid cancer
progression via repressing miR-144 biogenesis. RNA Biol.
18:547–562. 2021. View Article : Google Scholar
|
|
42
|
Li C, Chen X, Liu T and Chen G: lncRNA
HOTAIRM1 regulates cell proliferation and the metastasis of thyroid
cancer by targeting Wnt10b. Oncol Rep. 45:1083–1093. 2021.
View Article : Google Scholar
|
|
43
|
Chen D, Li Y, Wang Y and Xu J: LncRNA
HOTAIRM1 knockdown inhibits cell glycolysis metabolism and tumor
progression by miR-498/ABCE1 axis in non-small cell lung cancer.
Genes Genomics. 43:183–194. 2021. View Article : Google Scholar
|
|
44
|
Xiong F, Yin H, Zhang H, Zhu C, Zhang B,
Chen S, Ling C and Chen X: Clinicopathologic features and the
prognostic implications of long noncoding RNA HOTAIRM1 in non-small
cell lung cancer. Genet Test Mol Biomarkers. 24:47–53. 2020.
View Article : Google Scholar
|
|
45
|
Yu Y, Niu J, Zhang X, Wang X, Song H, Liu
Y, Jiao X and Chen F: Identification and validation of HOTAIRM1 as
a novel biomarker for oral squamous cell carcinoma. Front Bioeng
Biotechnol. 9:7985842022. View Article : Google Scholar
|
|
46
|
Wang L, Wang L, Wang Q, Yosefi B, Wei S,
Wang X and Shen D: The function of long noncoding RNA HOTAIRM1 in
the progression of prostate cancer cells. Andrologia.
53:e138972021. View Article : Google Scholar
|
|
47
|
Luo Y, He Y, Ye X, Song J, Wang Q, Li Y
and Xie X: High expression of long noncoding RNA HOTAIRM1 is
associated with the proliferation and migration in pancreatic
ductal adenocarcinoma. Pathol Oncol Res. 25:1567–1577. 2019.
View Article : Google Scholar
|
|
48
|
Zhou Y, Gong B, Jiang ZL, Zhong S, Liu XC,
Dong K, Wu HS, Yang HJ and Zhu SK: Microarray expression profile
analysis of long non-coding RNAs in pancreatic ductal
adenocarcinoma. Int J Oncol. 48:670–680. 2016. View Article : Google Scholar
|
|
49
|
Ye L, Meng X, Xiang R, Li W and Wang J:
Investigating function of long noncoding RNA of HOTAIRM1 in
progression of SKOV3 ovarian cancer cells. Drug Dev Res.
82:1162–1168. 2021. View Article : Google Scholar
|
|
50
|
Chao H, Zhang M, Hou H, Zhang Z and Li N:
HOTAIRM1 suppresses cell proliferation and invasion in ovarian
cancer through facilitating ARHGAP24 expression by sponging
miR-106a-5p. Life Sci. 243:1172962020. View Article : Google Scholar
|
|
51
|
Li D, Chai L, Yu X, Song Y, Zhu X, Fan S,
Jiang W, Qiao T, Tong J, Liu S, et al: The HOTAIRM1/miR-107/TDG
axis regulates papillary thyroid cancer cell proliferation and
invasion. Cell Death Dis. 11:2272020. View Article : Google Scholar
|
|
52
|
Zheng M, Liu X, Zhou Q and Liu G: HOTAIRM1
competed endogenously with miR-148a to regulate DLGAP1 in head and
neck tumor cells. Cancer Med. 7:3143–3156. 2018. View Article : Google Scholar
|
|
53
|
Zhang Y, Mi L, Xuan Y, Gao C, Wang YH,
Ming HX and Liu J: LncRNA HOTAIRM1 inhibits the progression of
hepatocellular carcinoma by inhibiting the Wnt signaling pathway.
Eur Rev Med Pharmacol Sci. 22:4861–4868. 2018.
|
|
54
|
Chen TJ, Gao F, Yang T, Li H, Li Y, Ren H
and Chen MW: LncRNA HOTAIRM1 inhibits the proliferation and
invasion of lung adenocarcinoma cells via the miR-498/WWOX Axis.
Cancer Manag Res. 12:4379–4390. 2020. View Article : Google Scholar
|
|
55
|
Xu F, Chen M, Chen H, Wu N, Qi Q, Jiang X,
Fang D, Feng Q, Jin R and Jiang L: The curcumin analog Da0324
inhibits the proliferation of gastric cancer cells via
HOTAIRM1/miR-29b-1-5p/PHLPP1 Axis. J Cancer. 13:2644–2655. 2022.
View Article : Google Scholar
|
|
56
|
Lu R, Zhao G, Yang Y, Jiang Z, Cai J,
Zhang Z and Hu H: Long noncoding RNA HOTAIRM1 inhibits cell
progression by regulating miR-17-5p/PTEN axis in gastric cancer. J
Cell Biochem. 120:4952–4965. 2019. View Article : Google Scholar
|
|
57
|
Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M
and Zhang H: HOTAIRM1 as a potential biomarker for diagnosis of
colorectal cancer functions the role in the tumour suppressor. J
Cell Mol Med. 20:2036–2044. 2016. View Article : Google Scholar
|
|
58
|
Ren T, Hou J, Liu C, Shan F, Xiong X, Qin
A, Chen J and Ren W: The long non-coding RNA HOTAIRM1 suppresses
cell progression via sponging endogenous miR-17-5p/B-cell
translocation gene 3 (BTG3) axis in 5-fluorouracil resistant
colorectal cancer cells. Biomed Pharmacother. 117:1091712019.
View Article : Google Scholar
|
|
59
|
Zhang W, Dang R, Liu H, Dai L, Liu H,
Adegboro AA, Zhang Y, Li W, Peng K, Hong J and Li X: Machine
learning-based investigation of regulated cell death for predicting
prognosis and immunotherapy response in glioma patients. Sci Rep.
14:41732024. View Article : Google Scholar
|
|
60
|
Zafar N, Ghias K and Fadoo Z: Genetic
aberrations involved in relapse of pediatric acute myeloid
leukemia: A literature review. Asia Pac J Clin Oncol. 17:e135–e141.
2021. View Article : Google Scholar
|
|
61
|
Hayatigolkhatmi K, Valzelli R, El Menna O
and Minucci S: Epigenetic alterations in AML: Deregulated functions
leading to new therapeutic options. Int Rev Cell Mol Biol.
387:27–75. 2024. View Article : Google Scholar
|
|
62
|
Ding Y and Chen Q: Wnt/β-catenin signaling
pathway: An attractive potential therapeutic target in
osteosarcoma. Front Oncol. 14:14569592025. View Article : Google Scholar
|
|
63
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The Eighth Edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
“personalized” approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017.
|
|
64
|
Ding S, Hao Y, Qi Y, Wei H, Zhang J and Li
H: Molecular mechanism of tumor-infiltrating immune cells
regulating endometrial carcinoma. Genes Dis. 12:1014422024.
View Article : Google Scholar
|
|
65
|
Menendez-Santos M, Gonzalez-Baerga C,
Taher D, Waters R, Virarkar M and Bhosale P: Endometrial cancer:
2023 Revised FIGO staging system and the role of imaging. Cancers
(Basel). 16:18692024. View Article : Google Scholar
|
|
66
|
Chen DW, Lang BHH, McLeod DSA, Newbold K
and Haymart MR: Thyroid cancer. Lancet. 401:1531–1544. 2023.
View Article : Google Scholar
|
|
67
|
Boucai L, Zafereo M and Cabanillas ME:
Thyroid cancer: A review. JAMA. 331:425–435. 2024. View Article : Google Scholar
|
|
68
|
Meyer ML, Fitzgerald BG, Paz-Ares L,
Cappuzzo F, Jänne PA, Peters S and Hirsch FR: New promises and
challenges in the treatment of advanced non-small-cell lung cancer.
Lancet. 404:803–822. 2024. View Article : Google Scholar
|
|
69
|
Badwelan M, Muaddi H, Ahmed A, Lee KT and
Tran SD: Oral squamous cell carcinoma and concomitant primary
tumors, what do we know? A review of the literature. Curr Oncol.
30:3721–3734. 2023. View Article : Google Scholar
|
|
70
|
Jagadeesan D, Sathasivam KV, Fuloria NK,
Balakrishnan V, Khor GH, Ravichandran M, Solyappan M, Fuloria S,
Gupta G, Ahlawat A, et al: Comprehensive insights into oral
squamous cell carcinoma: Diagnosis, pathogenesis, and therapeutic
advances. Pathol Res Pract. 261:1554892024. View Article : Google Scholar
|
|
71
|
Almeeri MNE, Awies M and Constantinou C:
Prostate cancer, pathophysiology and recent developments in
management: A narrative review. Curr Oncol Rep. 26:1511–1519. 2024.
View Article : Google Scholar
|
|
72
|
Wilson TK and Zishiri OT: Prostate cancer:
A review of genetics, current biomarkers and personalised
treatments. Cancer Rep (Hoboken). 7:e700162024.
|
|
73
|
Dhillon J and Betancourt M: Pancreatic
ductal adenocarcinoma. Monogr Clin Cytol. 26:74–91. 2020.
View Article : Google Scholar
|
|
74
|
Konstantinopoulos PA and Matulonis UA:
Clinical and translational advances in ovarian cancer therapy. Nat
Cancer. 4:1239–1257. 2023. View Article : Google Scholar
|
|
75
|
Chow LQM: Head and neck cancer. N Engl J
Med. 382:60–72. 2020. View Article : Google Scholar
|
|
76
|
Kudrimoti A and Kudrimoti MR: Head and
neck cancers. Prim Care. 52:139–155. 2025. View Article : Google Scholar
|
|
77
|
Da BL, Suchman KI, Lau L, Rabiee A, He AR,
Shetty K, Yu H, Wong LL, Amdur RL, Crawford JM, et al: Pathogenesis
to management of hepatocellular carcinoma. Genes Cancer. 13:72–87.
2022. View Article : Google Scholar
|
|
78
|
Akhras A, Beran A, Guardiola J,
Bhavsar-Burke I, Reyes B and Ur Rahman A: S2808 direct oral
anticoagulants versus warfarin in elderly patients with child-pugh
C cirrhosis and atrial fibrillation: A real-world perspective. Am J
Gastroenterol. 120((10S2)): pS6032025.
|
|
79
|
Tawfiq RK, de Camargo Correia GS, Li S,
Zhao Y, Lou Y and Manochakian R: Targeting lung cancer with
precision: The ADC therapeutic revolution. Curr Oncol Rep.
27:669–686. 2025. View Article : Google Scholar
|
|
80
|
Merle G, Friedlaender A, Desai A and Addeo
A: Antibody drug conjugates in lung cancer. Cancer J. 28:429–435.
2022. View Article : Google Scholar
|
|
81
|
López MJ, Carbajal J, Alfaro AL, Saravia
LG, Zanabria D, Araujo JM, Quispe L, Zevallos A, Buleje JL, Cho CE,
et al: Characteristics of gastric cancer around the world. Crit Rev
Oncol Hematol. 181:1038412023. View Article : Google Scholar
|
|
82
|
Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh
Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M and Saad A:
Colorectal cancer epidemiology: Recent trends and impact on
outcomes. Curr Drug Targets. 22:998–1009. 2021. View Article : Google Scholar
|
|
83
|
Mahmoud NN: Colorectal cancer:
Preoperative evaluation and staging. Surg Oncol Clin N Am.
31:127–141. 2022. View Article : Google Scholar
|
|
84
|
Salido-Guadarrama I, Romero-Cordoba SL and
Rueda-Zarazua B: Multi-Omics Mining of lncRNAs with biological and
clinical relevance in cancer. Int J Mol Sci. 24:166002023.
View Article : Google Scholar
|
|
85
|
Mahato RK, Bhattacharya S, Khullar N,
Sidhu IS, Reddy PH, Bhatti GK and Bhatti JS: Targeting long
non-coding RNAs in cancer therapy using CRISPR-Cas9 technology: A
novel paradigm for precision oncology. J Biotechnol. 379:98–119.
2024. View Article : Google Scholar
|
|
86
|
Yang Q, Fu Y, Wang J, Yang H and Zhang X:
Roles of lncRNA in the diagnosis and prognosis of triple-negative
breast cancer. J Zhejiang Univ Sci B. 24:1123–1140. 2023.(In
English, Chinese). View Article : Google Scholar
|
|
87
|
Alharthi NS, Al-Zahrani MH, Hazazi A,
Alhuthali HM, Gharib AF, Alzahrani S, Altalhi W, Almalki WH and
Khan FR: Exploring the lncRNA-VEGF axis: Implications for cancer
detection and therapy. Pathol Res Pract. 253:1549982023. View Article : Google Scholar
|
|
88
|
Mehmandar-Oskuie A, Jahankhani K,
Rostamlou A, Mardafkan N, Karamali N, Razavi ZS and Mardi A:
Molecular mechanism of lncRNAs in pathogenesis and diagnosis of
auto-immune diseases, with a special focus on lncRNA-based
therapeutic approaches. Life Sci. 336:1223222024. View Article : Google Scholar
|
|
89
|
Li Q, Dong C, Cui J, Wang Y and Hong X:
Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion
through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away
from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer
Res. 37:2652018. View Article : Google Scholar
|
|
90
|
Kim CY, Oh JH, Lee JY and Kim MH: The
LncRNA HOTAIRM1 promotes tamoxifen resistance by mediating HOXA1
expression in ER+ breast cancer cells. J Cancer. 11:3416–3423.
2020. View Article : Google Scholar
|
|
91
|
Wang XQ and Dostie J: Reciprocal
regulation of chromatin state and architecture by HOTAIRM1
contributes to temporal collinear HOXA gene activation. Nucleic
Acids Res. 45:1091–1104. 2017.
|
|
92
|
Usman M, Li A, Wu D, Qinyan Y, Yi LX, He G
and Lu H: The functional role of lncRNAs as ceRNAs in both ovarian
processes and associated diseases. Noncoding RNA Res. 9:165–177.
2023. View Article : Google Scholar
|
|
93
|
Kim S: LncRNA-miRNA-mRNA regulatory
networks in skin aging and therapeutic potentials. Front Physiol.
14:13031512023. View Article : Google Scholar
|
|
94
|
Lin Y, Wen H, Yang B, Wang C and Liang R:
Integrated bioinformatics and validation to construct
lncRNA-miRNA-mRNA ceRNA network in status epilepticus. Heliyon.
9:e222052023. View Article : Google Scholar
|
|
95
|
Jiang M, Wang Z, Lu T, Li X, Yang K, Zhao
L, Zhang D, Li J and Wang L: Integrative analysis of long noncoding
RNAs dysregulation and synapse-associated ceRNA regulatory axes in
autism. Transl Psychiatry. 13:3752023. View Article : Google Scholar
|
|
96
|
Wang X, Liu Y and Lei P: LncRNA HOTAIRM1
promotes osteogenic differentiation of human bone marrow-derived
mesenchymal stem cells by targeting miR-152-3p/ETS1 axis. Mol Biol
Rep. 50:5597–5608. 2023. View Article : Google Scholar
|
|
97
|
Wang G, Yu Y and Wang Y: Effects of
propofol on neuroblastoma cells via the HOTAIRM1/miR-519a-3p axis.
Transl Neurosci. 13:57–69. 2022. View Article : Google Scholar
|
|
98
|
Han W, Wang S, Qi Y, Wu F, Tian N, Qiang B
and Peng X: Targeting HOTAIRM1 ameliorates glioblastoma by
disrupting mitochondrial oxidative phosphorylation and serine
metabolism. iScience. 25:1048232022. View Article : Google Scholar
|
|
99
|
Chen W, Liu J, Ge F, Chen Z, Qu M, Nan K,
Gu J, Jiang Y, Gao S and Liao Y: Long noncoding RNA HOTAIRM1
promotes immunosuppression in sepsis by inducing T cell exhaustion.
J Immunol. 208:618–632. 2022. View Article : Google Scholar
|
|
100
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar
|
|
101
|
Ge T, Gu X, Jia R, Ge S, Chai P, Zhuang A
and Fan X: Crosstalk between metabolic reprogramming and
epigenetics in cancer: Updates on mechanisms and therapeutic
opportunities. Cancer Commun (Lond). 42:1049–1082. 2022. View Article : Google Scholar
|
|
102
|
Zhou Y, Wu Q, Long X, He Y and Huang J:
lncRNA HOTAIRM1 Activated by hoxa4 drives huvec proliferation
through direct interaction with protein partner HSPA5.
Inflammation. 47:421–437. 2024. View Article : Google Scholar
|
|
103
|
Guo H, Li T and Sun X: LncRNA HOTAIRM1,
miR-433-5p and PIK3CD function as a ceRNA network to exacerbate the
development of PCOS. J Ovarian Res. 14:192021. View Article : Google Scholar
|
|
104
|
Dahariya S, Raghuwanshi S, Thamodaran V,
Velayudhan SR and Gutti RK: Role of long non-coding RNAs in
human-induced pluripotent stem cells derived megakaryocytes: A p53,
HOX antisense intergenic RNA Myeloid 1, and miR-125b interaction
study. J Pharmacol Exp Ther. 384:92–101. 2023. View Article : Google Scholar
|
|
105
|
Li YR, Yan WJ, Cai LL and Deng XL: Effect
of down-regulation of LncRNA-HOTAIRM1 to proliferation, apoptosis
and KIT/AKT pathway of jurkat cells. Zhongguo Shi Yan Xue Ye Xue Za
Zhi. 29:1123–1128. 2021.(In Chinese).
|
|
106
|
Liu WB, Li GS, Shen P, Li YN and Zhang FJ:
Long non-coding RNA HOTAIRM1-1 silencing in cartilage tissue
induces osteoarthritis through microRNA-125b. Exp Ther Med.
22:9332021. View Article : Google Scholar
|
|
107
|
Tollis P, Vitiello E, Migliaccio F,
D'Ambra E, Rocchegiani A, Garone MG, Bozzoni I, Rosa A, Carissimo
A, Laneve P and Caffarelli E: The long noncoding RNA nHOTAIRM1 is
necessary for differentiation and activity of iPSC-derived spinal
motor neurons. Cell Death Dis. 14:7412023. View Article : Google Scholar
|
|
108
|
Segal D, Coulombe S, Sim J and Josée
Dostie J: A conserved HOTAIRM1-HOXA1 regulatory axis contributes
early to neuronal differentiation. RNA Biol. 20:1523–1539. 2023.
View Article : Google Scholar
|
|
109
|
Zamame Ramirez JA, Romagnoli GG and Kaneno
R: Inhibiting autophagy to prevent drug resistance and improve
anti-tumor therapy. Life Sci. 265:1187452021. View Article : Google Scholar
|
|
110
|
Liang L, Gu W, Li M, Gao R, Zhang X, Guo C
and Mi S: The long noncoding RNA HOTAIRM1 controlled by AML1
enhances glucocorticoid resistance by activating RHOA/ROCK1 pathway
through suppressing ARHGAP18. Cell Death Dis. 12:7022021.
View Article : Google Scholar
|
|
111
|
Gu D, Tong M, Wang J, Zhang B, Liu J, Song
G and Zhu B: Overexpression of the lncRNA HOTAIRM1 promotes
lenvatinib resistance by downregulating miR-34a and activating
autophagy in hepatocellular carcinoma. Discov Oncol. 14:662023.
View Article : Google Scholar
|
|
112
|
Chen L, Hu N, Wang C and Zhao H: HOTAIRM1
knockdown enhances cytarabine-induced cytotoxicity by suppression
of glycolysis through the Wnt/β-catenin/PFKP pathway in acute
myeloid leukemia cells. Arch Biochem Biophys. 680:1082442020.
View Article : Google Scholar
|
|
113
|
Gustafson MP, Ligon JA, Bersenev A, McCann
CD, Shah NN and Hanley PJ: Emerging frontiers in immuno- and gene
therapy for cancer. Cytotherapy. 25:20–32. 2023. View Article : Google Scholar
|
|
114
|
Kaur R, Bhardwaj A and Gupta S: Cancer
treatment therapies: Traditional to modern approaches to combat
cancers. Mol Biol Rep. 50:9663–9676. 2023. View Article : Google Scholar
|
|
115
|
Hashemi M, Moosavi MS, Abed HM, Dehghani
M, Aalipour M, Heydari EA, Behroozaghdam M, Entezari M,
Salimimoghadam S, Gunduz ES, et al: Long non-coding RNA (lncRNA)
H19 in human cancer: From proliferation and metastasis to therapy.
Pharmacol Res. 184:1064182022. View Article : Google Scholar
|
|
116
|
McCabe EM and Rasmussen TP: lncRNA
involvement in cancer stem cell function and epithelial-mesenchymal
transitions. Semin Cancer Biol. 75:38–48. 2021. View Article : Google Scholar
|