|
1
|
Giaquinto AN, Sung H, Newman LA, Freedman
RA, Smith RA, Star J, Jemal A and Siegel RL: Breast cancer
statistics 2024. CA Cancer J Clin. 74:477–495. 2024.
|
|
2
|
Xiong X, Zheng LW, Ding Y, Chen YF, Cai YW
and Wang LP: Huang L, Liu CC, Shao ZM and Yu KD: Breast cancer:
Pathogenesis and treatments. Signal Transduct Target Ther.
10:492025. View Article : Google Scholar
|
|
3
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar
|
|
4
|
Yin Y, Song L, Shi D, Liu B, Li X, Yang M,
Liu B, Wang D and Qin J: Identification of recurrent insertions and
deletions in exon 18 and 19 of human epidermal growth factor
receptor 2 as potential drivers in non-small-cell lung cancer and
other cancer types. JCO Precis Oncol. 6:e21003252022. View Article : Google Scholar
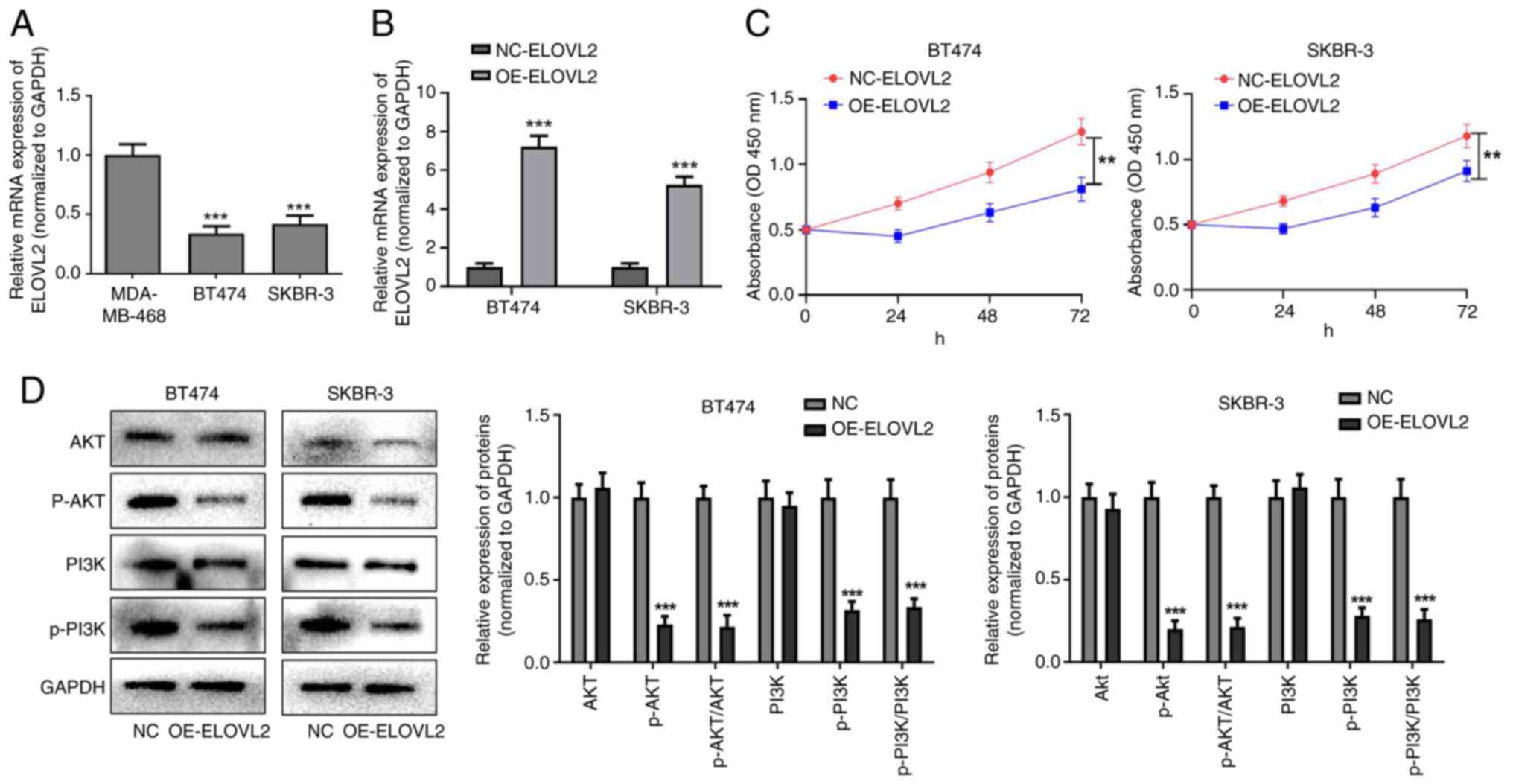
|
|
5
|
Cheng X: A comprehensive review of HER2 in
cancer biology and therapeutics. Genes (Basel). 15:9032024.
View Article : Google Scholar
|
|
6
|
Marchiò C, Annaratone L, Marques A,
Casorzo L, Berrino E and Sapino A: Evolving concepts in HER2
evaluation in breast cancer: Heterogeneity, HER2-low carcinomas and
beyond. Semin Cancer Biol. 72:123–135. 2021. View Article : Google Scholar
|
|
7
|
Chen Z, Yang L, Yang Z, Wang Z, He W and
Zhang W: Ultrasonic-responsive piezoelectric stimulation enhances
sonodynamic therapy for HER2-positive breast cancer. J
Nanobiotechnology. 22:3692024. View Article : Google Scholar
|
|
8
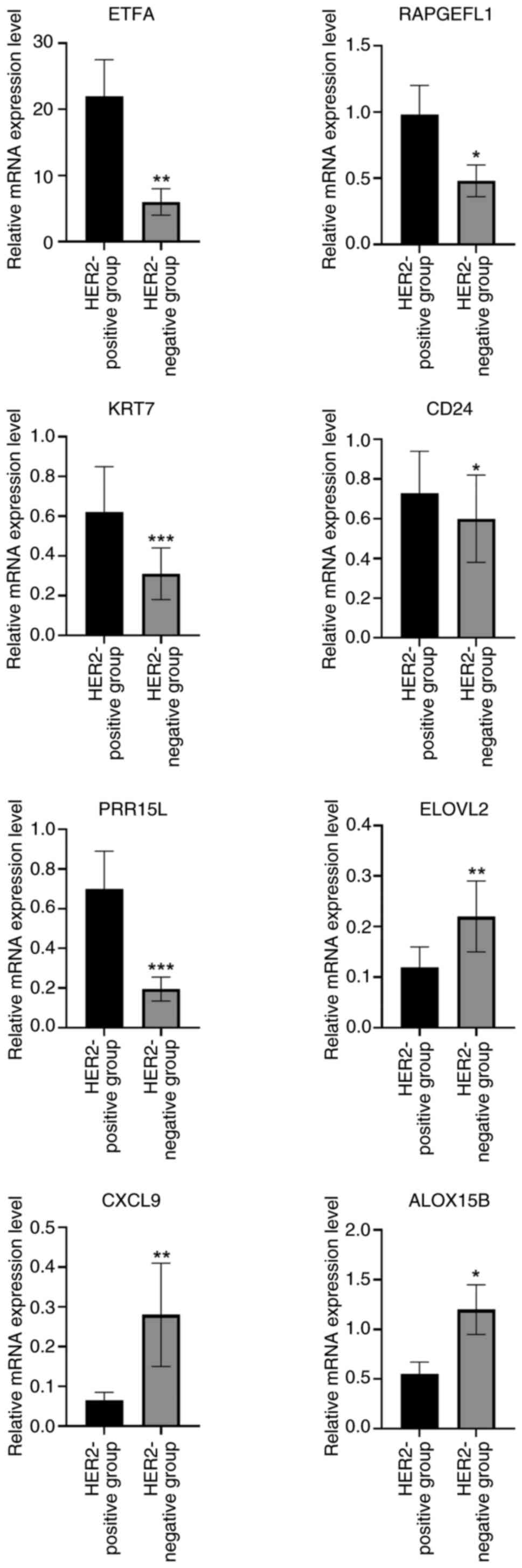
|
Ocaña A, Amir E and Pandiella A: HER2
heterogeneity and resistance to anti-HER2 antibody-drug conjugates.
Breast Cancer Res. 22:152020. View Article : Google Scholar
|
|
9
|
Kang S and Kim SB: HER2-low breast cancer:
Now and in the future. Cancer Res Treat. 56:700–720. 2024.
View Article : Google Scholar
|
|
10
|
Shi Z, Liu Y, Fang X, Liu X, Meng J and
Zhang J: Efficacy and prognosis of HER2-low and HER2-zero in
triple-negative breast cancer after neoadjuvant chemotherapy. Sci
Rep. 14:168992024. View Article : Google Scholar
|
|
11
|
Rizvi AA, Karaesmen E, Morgan M, Preus L,
Wang J, Sovic M, Hahn T and Sucheston-Campbell LE: gwasurvivr: An R
package for genome-wide survival analysis. Bioinformatics.
35:1968–1970. 2019. View Article : Google Scholar
|
|
12
|
Chen B, Khodadoust MS, Liu CL, Newman AM
and Alizadeh AA: Profiling tumor infiltrating immune cells with
CIBERSORT. Methods Mol Biol. 1711:243–259. 2018. View Article : Google Scholar
|
|
13
|
Xiao B, Liu L, Li A, Xiang C, Wang P, Li H
and Xiao T: Identification and verification of immune-related gene
prognostic signature based on ssGSEA for osteosarcoma. Front Oncol.
10:6076222020. View Article : Google Scholar
|
|
14
|
Aran D: Cell-type enrichment analysis of
bulk transcriptomes using xCell. Methods Mol Biol. 2120:263–276.
2020. View Article : Google Scholar
|
|
15
|
Hu D, Zhou M and Zhu X: Deciphering
immune-associated genes to predict survival in clear cell renal
cell cancer. Biomed Res Int. 2019:25068432019. View Article : Google Scholar
|
|
16
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:1747–1756. 2018.
View Article : Google Scholar
|
|
17
|
Geeleher P, Cox N and Huang RS:
pRRophetic: An R package for prediction of clinical
chemotherapeutic response from tumor gene expression levels. PLoS
One. 9:e1074682014. View Article : Google Scholar
|
|
18
|
Reimand J, Isserlin R, Voisin V, Kucera M,
Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, et
al: Pathway enrichment analysis and visualization of omics data
using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc.
14:482–517. 2019. View Article : Google Scholar
|
|
19
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar
|
|
20
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar
|
|
21
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar
|
|
22
|
Zhang S, Tong YX, Zhang XH, Zhang YJ, Xu
XS, Xiao AT, Chao TF and Gong JP: A novel and validated nomogram to
predict overall survival for gastric neuroendocrine neoplasms. J
Cancer. 10:5944–5954. 2019. View Article : Google Scholar
|
|
23
|
Wang L, Peng F, Li ZH, Deng YF, Ruan MN,
Mao ZG and Li L: Identification of AKI signatures and
classification patterns in ccRCC based on machine learning. Front
Med (Lausanne). 10:11956782023. View Article : Google Scholar
|
|
24
|
Gustavsson EK, Zhang D, Reynolds RH,
Garcia-Ruiz S and Ryten M: ggtranscript: An R package for the
visualization and interpretation of transcript isoforms using
ggplot2. Bioinformatics. 38:3844–3846. 2022. View Article : Google Scholar
|
|
25
|
Zhang H, Meltzer P and Davis S: RCircos:
An R package for Circos 2D track plots. BMC Bioinformatics.
14:2442013. View Article : Google Scholar
|
|
26
|
Pal B, Chen Y, Vaillant F, Capaldo BD,
Joyce R, Song X, Bryant VL, Penington JS, Di Stefano L, Tubau
Ribera N, et al: A single-cell RNA expression atlas of normal,
preneoplastic and tumorigenic states in the human breast. EMBO J.
40:e1073332021. View Article : Google Scholar
|
|
27
|
Han Y, Wang Y, Dong X, Sun D, Liu Z, Yue
J, Wang H, Li T and Wang C: TISCH2: Expanded datasets and new tools
for single-cell transcriptome analyses of the tumor
microenvironment. Nucleic Acids Res. 51(D1): D1425–D1431. 2023.
View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Kokot A, Gadakh S, Saha I, Gajda E,
Łaźniewski M, Rakshit S, Sengupta K, Mollah AF, Denkiewicz M,
Górczak K, et al: Unveiling the molecular mechanism of trastuzumab
resistance in SKBR3 and BT474 cell lines for HER2 positive breast
cancer. Curr Issues Mol Biol. 46:2713–2740. 2024. View Article : Google Scholar
|
|
30
|
Ogitani Y, Hagihara K, Oitate M, Naito H
and Agatsuma T: Bystander killing effect of DS-8201a, a novel
anti-human epidermal growth factor receptor 2 antibody-drug
conjugate, in tumors with human epidermal growth factor receptor 2
heterogeneity. Cancer Sci. 107:1039–1046. 2016. View Article : Google Scholar
|
|
31
|
Jeong D, Ham J, Kim HW, Kim H, Ji HW, Yun
SH, Park JE, Lee KS, Jo H, Han JH, et al: ELOVL2: A novel tumor
suppressor attenuating tamoxifen resistance in breast cancer. Am J
Cancer Res. 11:2568–2589. 2021.
|
|
32
|
Hu T, Zhang H, Du Y, Luo S, Yang X, Zhang
H, Feng J, Chen X, Tu X, Wang C and Zhang Y: ELOVL2 restrains cell
proliferation, migration, and invasion of prostate cancer via
regulation of the tumor suppressor INPP4B. Cell Signal.
96:1103732022. View Article : Google Scholar
|
|
33
|
Vranić S, Bešlija S and Gatalica Z:
Targeting HER2 expression in cancer: New drugs and new indications.
Bosn J Basic Med Sci. 21:1–4. 2021.
|
|
34
|
Rittler D, Baranyi M, Molnár E, Garay T,
Jalsovszky I, Varga IK, Hegedűs L, Aigner C, Tóvári J, Tímár J and
Hegedűs B: The antitumor effect of lipophilic bisphosphonate
BPH1222 in melanoma models: The role of the PI3K/Akt pathway and
the small G protein Rheb. Int J Mol Sci. 20:49172019. View Article : Google Scholar
|
|
35
|
Soltani A, Torki S, Ghahfarokhi MS, Jami
MS and Ghatrehsamani M: Targeting the phosphoinositide 3-kinase/AKT
pathways by small molecules and natural compounds as a therapeutic
approach for breast cancer cells. Mol Biol Rep. 46:4809–4816. 2019.
View Article : Google Scholar
|
|
36
|
Ye X, Liu J, Yuan X, Yang S, Huang Y and
Chen Y: Molecular mechanism of Salvia miltiorrhiza bunge in
treating cerebral infarction. Evid Based Complement Alternat Med.
2022:59923942022. View Article : Google Scholar
|
|
37
|
Wu X, Pan J, Yu JJ, Kang J, Hou S, Cheng
M, Xu L, Gong L and Li Y: DiDang decoction improves mitochondrial
function and lipid metabolism via the HIF-1 signaling pathway to
treat atherosclerosis and hyperlipidemia. J Ethnopharmacol.
308:1162892023. View Article : Google Scholar
|
|
38
|
Kulkoyluoglu-Cotul E, Arca A and
Madak-Erdogan Z: Crosstalk between estrogen signaling and breast
cancer metabolism. Trends Endocrinol Metab. 30:25–38. 2019.
View Article : Google Scholar
|
|
39
|
Hinshaw DC and Shevde LA: The tumor
microenvironment innately modulates cancer progression. Cancer Res.
79:4557–4566. 2019. View Article : Google Scholar
|
|
40
|
de Visser KE and Joyce JA: The evolving
tumor microenvironment: From cancer initiation to metastatic
outgrowth. Cancer Cell. 41:374–403. 2023. View Article : Google Scholar
|
|
41
|
Mao M, Yu Q, Huang R, Lu Y, Wang Z and
Liao L: Stromal score as a prognostic factor in primary gastric
cancer and close association with tumor immune microenvironment.
Cancer Med. 9:4980–4990. 2020. View Article : Google Scholar
|
|
42
|
Song J, Yang R, Wei R, Du Y, He P and Liu
X: Pan-cancer analysis reveals RIPK2 predicts prognosis and
promotes immune therapy resistance via triggering cytotoxic T
lymphocytes dysfunction. Mol Med. 28:472022. View Article : Google Scholar
|
|
43
|
Xia Y, Rao L, Yao H, Wang Z, Ning P and
Chen X: Engineering macrophages for cancer immunotherapy and drug
delivery. Adv Mater. 32:e20020542020. View Article : Google Scholar
|
|
44
|
Sun Y, Lian Y, Mei X, Xia J, Feng L, Gao
J, Xu H, Zhang X, Yang H, Hao X and Feng Y: Cinobufagin inhibits
M2-like tumor-associated macrophage polarization to attenuate the
invasion and migration of lung cancer cells. Int J Oncol.
65:1022024. View Article : Google Scholar
|
|
45
|
Zhang Y, Chen S, You L, He Z, Xu P and
Huang W: LINC00161 upregulated by M2-like tumor-associated
macrophages promotes hepatocellular carcinoma progression by
methylating HACE1 promoters. Cytotechnology. 76:777–793. 2024.
View Article : Google Scholar
|
|
46
|
Yamaguchi T, Fushida S, Yamamoto Y,
Tsukada T, Kinoshita J, Oyama K, Miyashita T, Tajima H, Ninomiya I,
Munesue S, et al: Tumor-associated macrophages of the M2 phenotype
contribute to progression in gastric cancer with peritoneal
dissemination. Gastric Cancer. 19:1052–1065. 2016. View Article : Google Scholar
|
|
47
|
Wu Q, Wu W, Jacevic V, Franca TCC, Wang X
and Kuca K: Selective inhibitors for JNK signalling: A potential
targeted therapy in cancer. J Enzyme Inhib Med Chem. 35:574–583.
2020. View Article : Google Scholar
|
|
48
|
Bubici C and Papa S: JNK signalling in
cancer: In need of new, smarter therapeutic targets. Br J
Pharmacol. 171:24–37. 2014. View Article : Google Scholar
|
|
49
|
Luo W, Han Y, Li X, Liu Z, Meng P and Wang
Y: Breast cancer prognosis prediction and immune pathway molecular
analysis based on mitochondria-related genes. Genet Res (Camb).
2022:22499092022. View Article : Google Scholar
|
|
50
|
Katoh M: FGFR inhibitors: Effects on
cancer cells, tumor microenvironment and whole-body homeostasis
(review). Int J Mol Med. 38:3–15. 2016. View Article : Google Scholar
|
|
51
|
Massaro F, Molica M and Breccia M:
Ponatinib: A review of efficacy and safety. Curr Cancer Drug
Targets. 18:847–856. 2018. View Article : Google Scholar
|
|
52
|
Katoh M and Nakagama H: FGF receptors:
Cancer biology and therapeutics. Med Res Rev. 34:280–300. 2014.
View Article : Google Scholar
|
|
53
|
Cicenas J, Kalyan K, Sorokinas A,
Stankunas E, Levy J, Meskinyte I, Stankevicius V, Kaupinis A and
Valius M: Roscovitine in cancer and other diseases. Ann Transl Med.
3:1352015.
|
|
54
|
Raynaud FI, Whittaker SR, Fischer PM,
McClue S, Walton MI, Barrie SE, Garrett MD, Rogers P, Clarke SJ,
Kelland LR, et al: In vitro and in vivo
pharmacokinetic-pharmacodynamic relationships for the
trisubstituted aminopurine cyclin-dependent kinase inhibitors
olomoucine, bohemine and CYC202. Clin Cancer Res. 11:4875–4887.
2005. View Article : Google Scholar
|
|
55
|
De Azevedo WF, Leclerc S, Meijer L,
Havlicek L, Strnad M and Kim SH: Inhibition of cyclin-dependent
kinases by purine analogues: Crystal structure of human cdk2
complexed with roscovitine. Eur J Biochem. 243:518–526. 1997.
View Article : Google Scholar
|
|
56
|
Lambert LA, Qiao N, Hunt KK, Lambert DH,
Mills GB, Meijer L and Keyomarsi K: Autophagy: A novel mechanism of
synergistic cytotoxicity between doxorubicin and roscovitine in a
sarcoma model. Cancer Res. 68:7966–7974. 2008. View Article : Google Scholar
|
|
57
|
Chen W, Di Z, Chen Z, Nan K, Gu J, Ge F,
Liu J, Zhang H and Miao C: NBPF4 mitigates progression in
colorectal cancer through the regulation of EZH2-associated ETFA. J
Cell Mol Med. 25:9038–9050. 2021. View Article : Google Scholar
|
|
58
|
Zhou C, Feng X, Yuan F, Ji J, Shi M, Yu Y,
Zhu Z and Zhang J: Difference of molecular alterations in
HER2-positive and HER2-negative gastric cancers by whole-genome
sequencing analysis. Cancer Manag Res. 10:3945–3954. 2018.
View Article : Google Scholar
|
|
59
|
An Q, Liu T, Wang MY, Yang YJ, Zhang ZD,
Liu ZJ and Yang B: KRT7 promotes epithelial-mesenchymal transition
in ovarian cancer via the TGF-β/Smad2/3 signaling pathway. Oncol
Rep. 45:481–492. 2021. View Article : Google Scholar
|
|
60
|
Song J, Ruze R, Chen Y, Xu R, Yin X, Wang
C, Xu Q and Zhao Y: Construction of a novel model based on
cell-in-cell-related genes and validation of KRT7 as a biomarker
for predicting survival and immune microenvironment in pancreatic
cancer. BMC Cancer. 22:8942022. View Article : Google Scholar
|
|
61
|
Altevogt P, Sammar M, Hüser L and
Kristiansen G: Novel insights into the function of CD24: A driving
force in cancer. Int J Cancer. 148:546–559. 2021. View Article : Google Scholar
|
|
62
|
Li X, Xiong W, Wang Y, Li Y, Cheng X and
Liu W: p53 activates the lipoxygenase activity of ALOX15B via
inhibiting SLC7A11 to induce ferroptosis in bladder cancer cells.
Lab Invest. 103:1000582023. View Article : Google Scholar
|
|
63
|
Seitz S, Dreyer TF, Stange C, Steiger K,
Bräuer R, Scheutz L, Multhoff G, Weichert W, Kiechle M, Magdolen V
and Bronger H: CXCL9 inhibits tumour growth and drives anti-PD-L1
therapy in ovarian cancer. Br J Cancer. 126:1470–1480. 2022.
View Article : Google Scholar
|
|
64
|
Mizuguchi Y, Sakamoto T, Hashimoto T,
Tsukamoto S, Iwasa S, Saito Y and Sekine S: Identification of a
novel PRR15L-RSPO2 fusion transcript in a sigmoid colon cancer
derived from superficially serrated adenoma. Virchows Arch.
475:659–663. 2019. View Article : Google Scholar
|
|
65
|
Wu J, Zhang H, Li L, Hu M, Chen L, Xu B
and Song Q: A nomogram for predicting overall survival in patients
with low-grade endometrial stromal sarcoma: A population-based
analysis. Cancer Commun (Lond). 40:301–312. 2020. View Article : Google Scholar
|
|
66
|
Wang F, Fang L, Fu B and Fan C:
Construction of a prognostic risk assessment model for HER2 +
breast cancer based on autophagy-related genes. Breast Cancer.
30:478–488. 2023. View Article : Google Scholar
|
|
67
|
Zhao L, Lin Z, Nong S, Li C, Li J, Lin C,
Safi SZ, Huang S and Ismail ISB: Development and validation of a
prognostic nomogram model for HER2-positive male breast cancer
patients. Asian Pac J Cancer Prev. 25:3199–3207. 2024. View Article : Google Scholar
|
|
68
|
Lin J, Zhao A and Fu D: Evaluating the
tumor immune profile based on a three-gene prognostic risk model in
HER2 positive breast cancer. Sci Rep. 12:93112022. View Article : Google Scholar
|