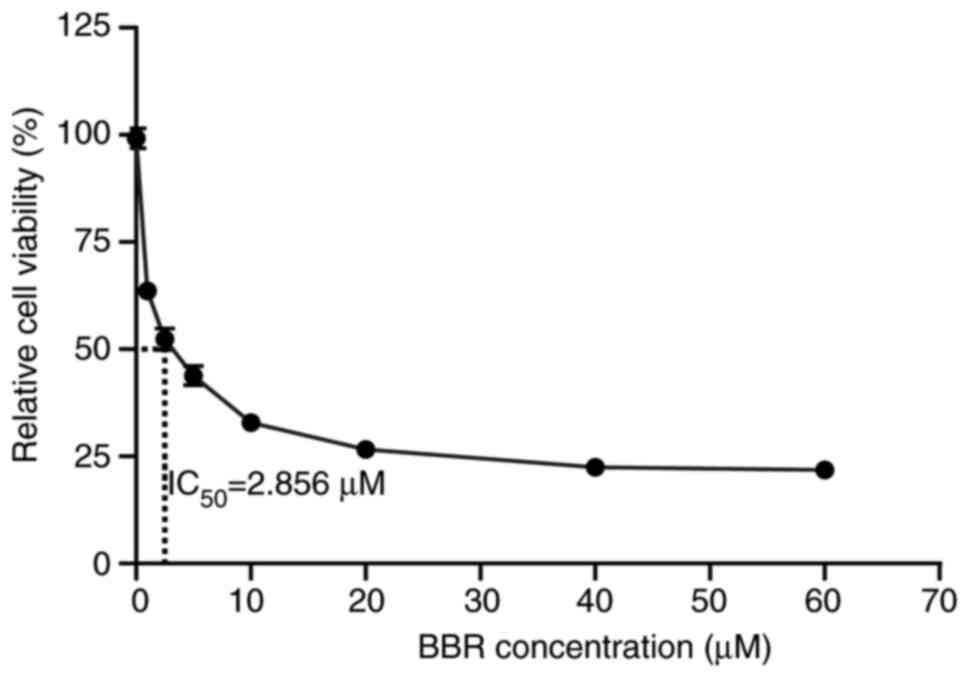
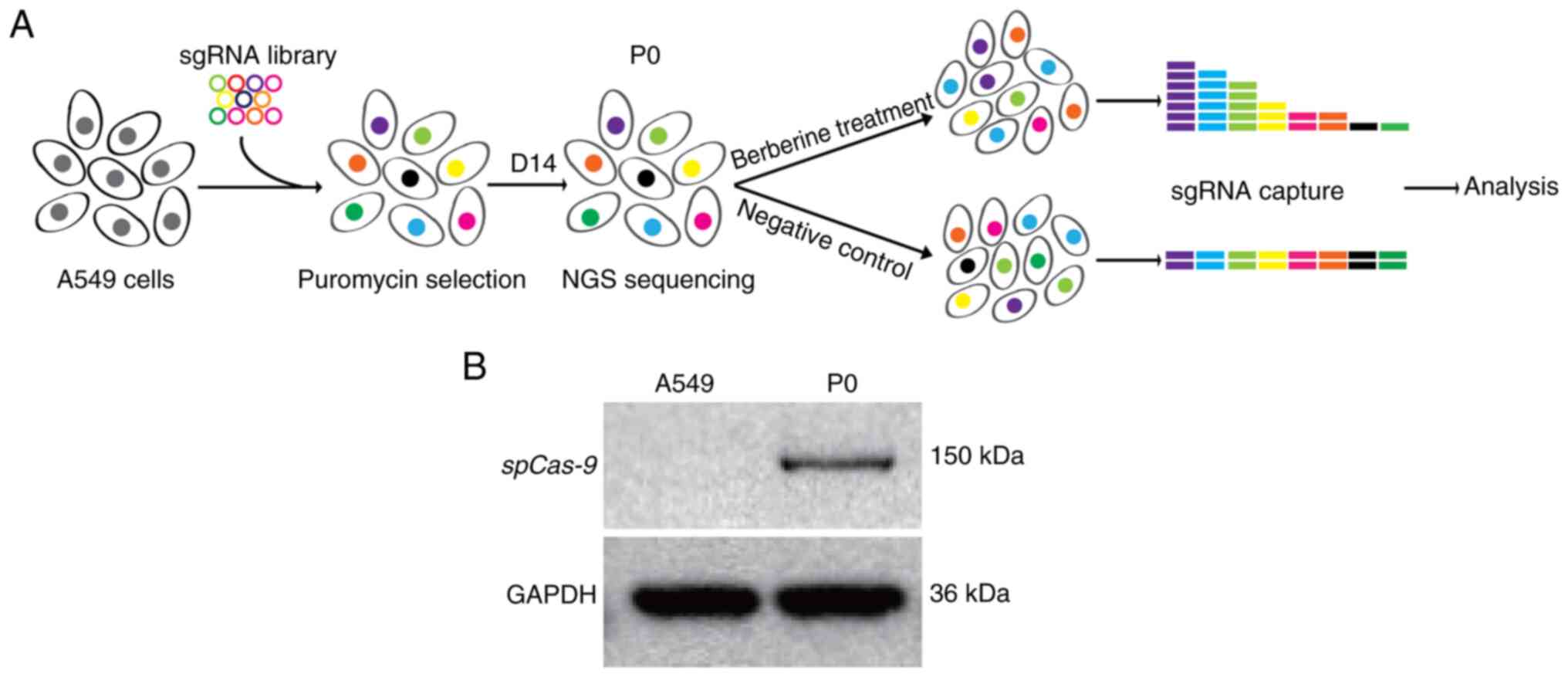
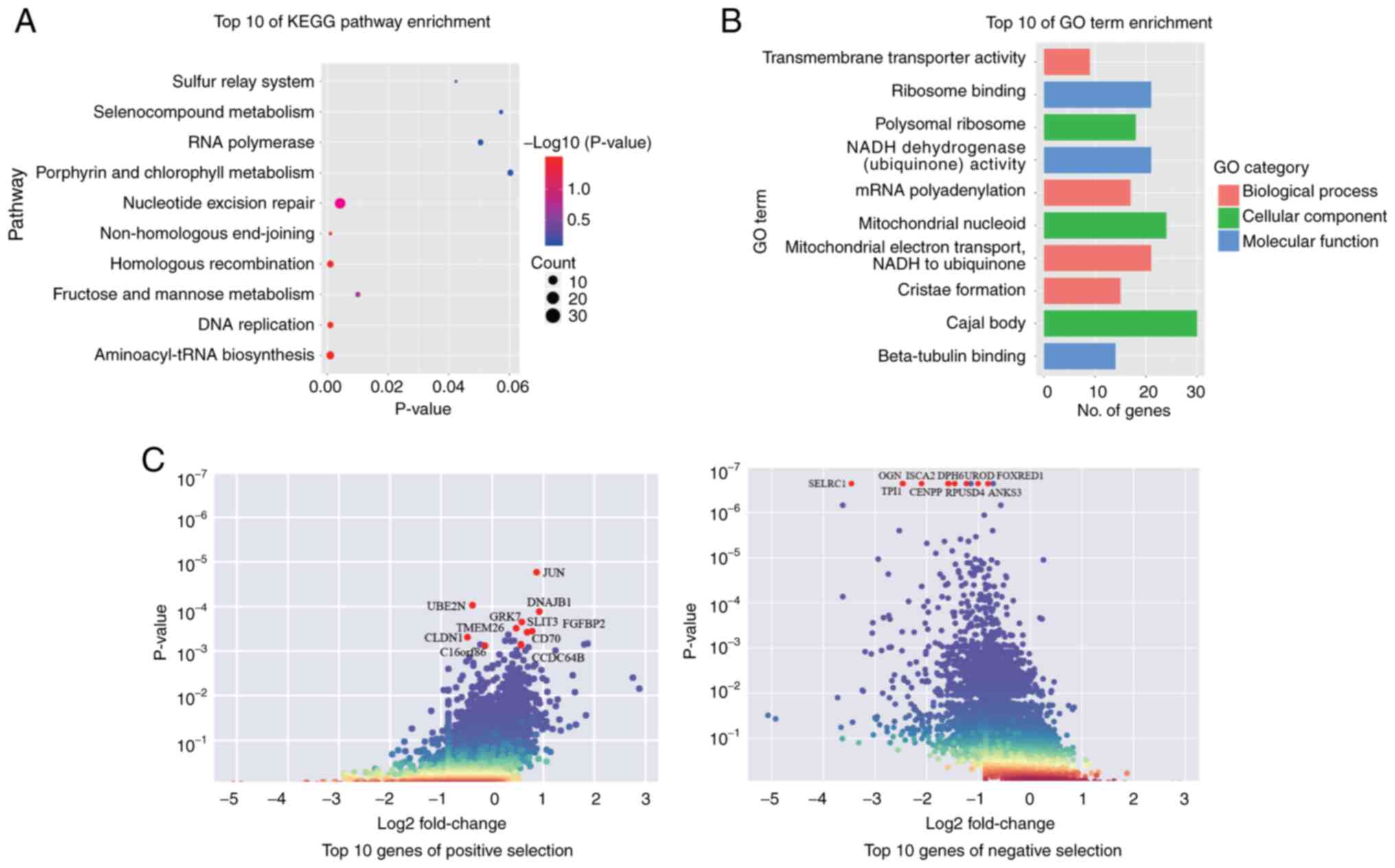
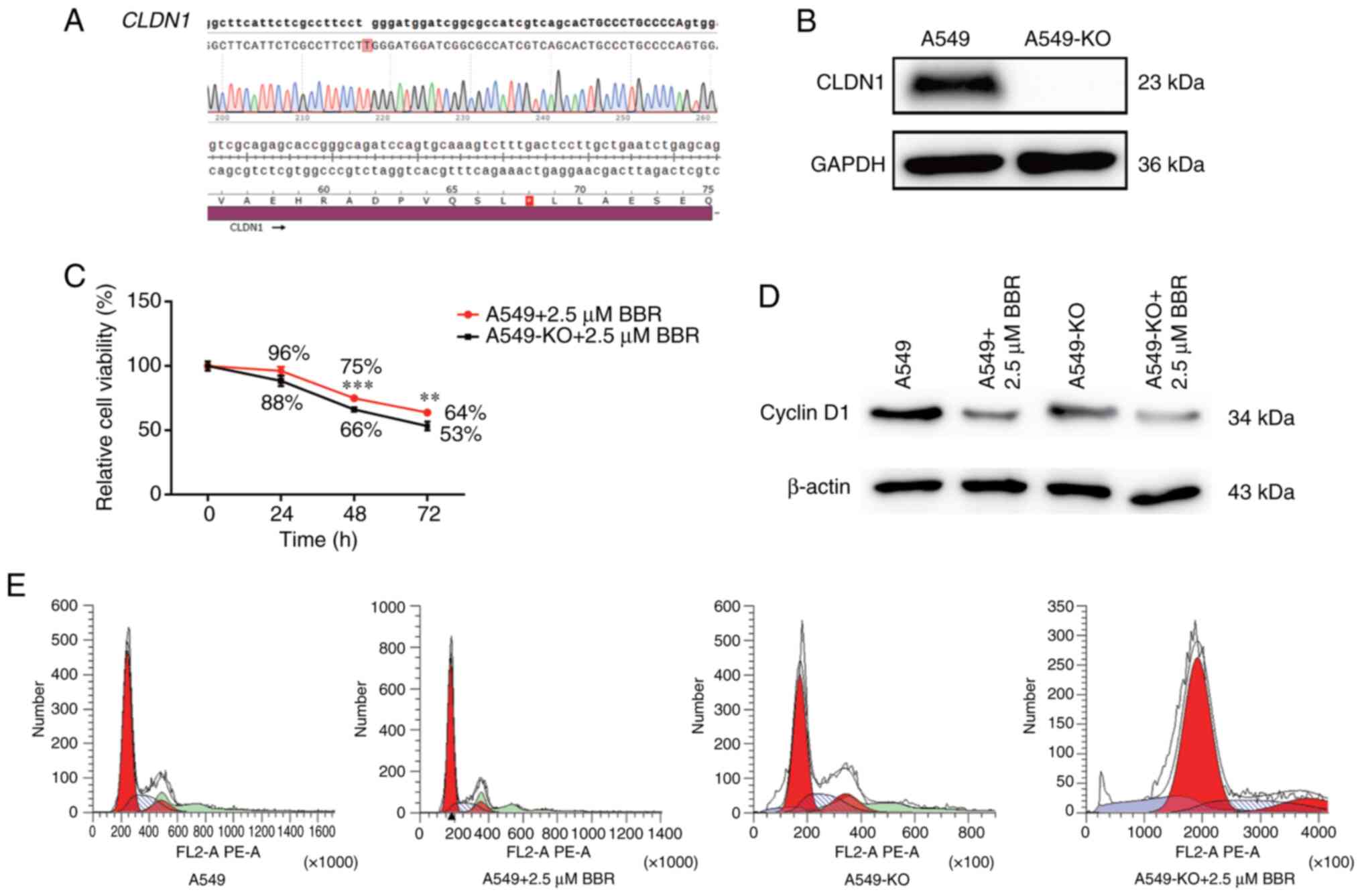
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
|
|
2
|
Phillips I, Stares M, Allan L, Sayers J,
Skipworth R and Laird B: Optimising outcomes in non small cell lung
cancer: Targeting cancer cachexia. Front Biosci (Landmark Ed).
27:1292022. View Article : Google Scholar
|
|
3
|
Su PL, Furuya N, Asrar A, Rolfo C, Li Z,
Carbone DP and He K: Recent advances in therapeutic strategies for
non-small cell lung cancer. J Hematol Oncol. 18:352025. View Article : Google Scholar
|
|
4
|
Riely GJ, Wood DE, Ettinger DS, Aisner DL,
Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, et
al: Non-small cell lung cancer, version 4.2024, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
22:249–274. 2024. View Article : Google Scholar
|
|
5
|
Wang JL, Qiu TY and Ren SX: Updates to the
2024 CSCO advanced non-small cell lung cancer guidelines. Cancer
Biol Med. 22:77–82. 2025.
|
|
6
|
Huang Q, Li Y, Huang Y, Wu J, Bao W, Xue
C, Li X, Dong S, Dong Z and Hu S: Advances in molecular pathology
and therapy of non-small cell lung cancer. Signal Transduct Tar.
10:1862025. View Article : Google Scholar
|
|
7
|
Li S, Chen X, Shi H, Yi M, Xiong B and Li
T: Tailoring traditional Chinese medicine in cancer therapy. Mol
Cancer. 24:272025. View Article : Google Scholar
|
|
8
|
Chen JF, Wu SW, Shi ZM and Hu B:
Traditional Chinese medicine for colorectal cancer treatment:
potential targets and mechanisms of action. Chin Med. 18:142023.
View Article : Google Scholar
|
|
9
|
Paolini GV, Shapland RH, van Hoorn WP,
Mason JS and Hopkins AL: Global mapping of pharmacological space.
Nat Biotechnol. 24:805–815. 2006. View
Article : Google Scholar
|
|
10
|
Mohamad Zobir SZ, Mohd Fauzi F, Liggi S,
Drakakis G, Fu X, Fan TP and Bender A: Global Mapping of
Traditional Chinese Medicine into Bioactivity Space and Pathways
Annotation Improves Mechanistic Understanding and Discovers
Relationships between Therapeutic Action (Sub)classes. Evid Based
Complement Alternat Med. 2016:21064652016. View Article : Google Scholar
|
|
11
|
Wang J, Wong YK and Liao F: What has
traditional Chinese medicine delivered for modern medicine? Expert
Rev Mol Med. 20:e42018. View Article : Google Scholar
|
|
12
|
Koshelev YA: Affinity chromatography and
proteomic screening as the effective method for S100A4 new protein
targets discovery. Mol Biol (Mosk). 48:868–872. 2014.(In Russian).
View Article : Google Scholar
|
|
13
|
Niphakis MJ and Cravatt BF: Enzyme
inhibitor discovery by activity-based protein profiling. Annu Rev
Biochem. 83:341–377. 2014. View Article : Google Scholar
|
|
14
|
Lomenick B, Hao R, Jonai N, Chin RM,
Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al:
Target identification using drug affinity responsive target
stability (DARTS). Proc Natl Acad Sci USA. 106:21984–21989. 2009.
View Article : Google Scholar
|
|
15
|
Dearmond PD, Xu Y, Strickland EC, Daniels
KG and Fitzgerald MC: Thermodynamic analysis of protein-ligand
interactions in complex biological mixtures using a shotgun
proteomics approach. J Proteome Res. 10:4948–4958. 2011. View Article : Google Scholar
|
|
16
|
Martinez Molina D, Jafari R,
Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y and
Nordlund P: Monitoring drug target engagement in cells and tissues
using the cellular thermal shift assay. Science. 341:84–87. 2013.
View Article : Google Scholar
|
|
17
|
Savitski MM, Reinhard FB, Franken H,
Werner T, Savitski MF, Eberhard D, Martinez Molina D, Jafari R,
Dovega RB, Klaeger S, et al: Tracking cancer drugs in living cells
by thermal profiling of the proteome. Science. 346:12557842014.
View Article : Google Scholar
|
|
18
|
Nabet B, Roberts JM, Buckley DL, Paulk J,
Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al:
The dTAG system for immediate and target-specific protein
degradation. Nat Chem Biol. 14:431–441. 2018. View Article : Google Scholar
|
|
19
|
Wittrup A, Ai A, Liu X, Hamar P, Trifonova
R, Charisse K, Manoharan M, Kirchhausen T and Lieberman J:
Visualizing Lipid-formulated siRNA release from endosomes and
target gene knockdown. Nat Biotechnol. 33:870–876. 2015. View Article : Google Scholar
|
|
20
|
Schirle M, Bantscheff M and Kuster B: Mass
Spectrometry-based proteomics in preclinical drug discovery. Chem
Biol. 19:72–84. 2012. View Article : Google Scholar
|
|
21
|
Koike-Yusa H, Li Y, Tan EP,
Velasco-Herrera Mdel C and Yusa K: Genome-wide recessive genetic
screening in mammalian cells with a lentiviral CRISPR-guide RNA
library. Nat Biotechnol. 32:267–273. 2014. View Article : Google Scholar
|
|
22
|
Hou P, Wu C, Wang Y, Qi R, Bhavanasi D,
Zuo Z, Dos Santos C, Chen S, Chen Y, Zheng H, et al: A Genome-Wide
CRISPR screen identifies genes critical for resistance to FLT3
inhibitor AC220. Cancer Res. 77:4402–4413. 2017. View Article : Google Scholar
|
|
23
|
Song CQ, Li Y, Mou H, Moore J, Park A,
Pomyen Y, Hough S, Kennedy Z, Fischer A, Yin H, et al: Genome-Wide
CRISPR screen identifies regulators of Mitogen-activated protein
kinase as suppressors of liver tumors in mice. Gastroenterology.
152:1161–1173.e1. 2017. View Article : Google Scholar
|
|
24
|
Parnas O, Jovanovic M, Eisenhaure TM,
Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I,
Sanjana NE, et al: A Genome-wide CRISPR screen in primary immune
cells to dissect regulatory networks. Cell. 162:675–686. 2015.
View Article : Google Scholar
|
|
25
|
Patel SJ, Sanjana NE, Kishton RJ,
Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM,
Yamamoto TN, et al: Identification of essential genes for cancer
immunotherapy. Nature. 548:537–542. 2017. View Article : Google Scholar
|
|
26
|
Kurata M, Yamamoto K, Moriarity BS,
Kitagawa M and Largaespada DA: CRISPR/Cas9 library screening for
drug target discovery. J Hum Genet. 63:179–186. 2018. View Article : Google Scholar
|
|
27
|
Kwon JW, Oh JS, Seok SH, An HW, Lee YJ,
Lee NY, Ha T, Kim HA, Yoon GM, Kim SE, et al: Combined inhibition
of Bcl-2 family members and YAP induces synthetic lethality in
metastatic gastric cancer with RASA1 and NF2 deficiency. Mol
Cancer. 22:1562023. View Article : Google Scholar
|
|
28
|
Och A, Podgorski R and Nowak R: Biological
activity of Berberine-A summary update. Toxins (Basel). 12:7132020.
View Article : Google Scholar
|
|
29
|
Habtemariam S: Recent advances in
berberine inspired anticancer approaches: From drug combination to
novel formulation technology and derivatization. Molecules.
25:14262020. View Article : Google Scholar
|
|
30
|
Samadi P, Sarvarian P, Gholipour E,
Asenjan KS, Aghebati-Maleki L, Motavalli R, Hojjat-Farsangi M and
Yousefi M: Berberine: A novel therapeutic strategy for cancer.
IUBMB Life. 72:2065–2079. 2020. View Article : Google Scholar
|
|
31
|
Paudel KR, Mehta M, Yin GHS, Yen LL,
Malyla V, Patel VK, Panneerselvam J, Madheswaran T, MacLoughlin R,
Jha NK, et al: Berberine-loaded liquid crystalline nanoparticles
inhibit non-small cell lung cancer proliferation and migration in
vitro. Environ Sci Pollut Res Int. 29:46830–46847. 2022. View Article : Google Scholar
|
|
32
|
Li Q, Zhao H, Chen W and Huang P:
Berberine induces apoptosis and arrests the cell cycle in multiple
cancer cell lines. Arch Med Sci. 19:1530–1537. 2023. View Article : Google Scholar
|
|
33
|
Shen F, Zheng YS, Dong L, Cao Z and Cao J:
Enhanced tumor suppression in colorectal cancer via
berberine-loaded PEG-PLGA nanoparticles. Front Pharmacol.
15:15007312024. View Article : Google Scholar
|
|
34
|
Sun L, Lan J, Li Z, Zeng R, Shen Y, Zhang
T and Ding Y: Transforming cancer treatment with nanotechnology:
The role of berberine as a star natural compound. Int J
Nanomedicine. 19:8621–8640. 2024. View Article : Google Scholar
|
|
35
|
Haque S, Mathkor DM, Bhat SA, Musayev A,
Khituova L, Ramniwas S, Phillips E, Swamy N, Kumar S, Yerer MB, et
al: A comprehensive review highlighting the prospects of
phytonutrient berberine as an anticancer agent. J Biochem Mol
Toxicol. 39:e700732025. View Article : Google Scholar
|
|
36
|
Shalem O, Sanjana NE and Zhang F:
High-throughput functional genomics using CRISPR-Cas9. Nat Rev
Genet. 16:299–311. 2015. View Article : Google Scholar
|
|
37
|
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang
F, Irizarry RA, Liu JS, Brown M and Liu XS: MAGeCK enables robust
identification of essential genes from genome-scale CRISPR/Cas9
knockout screens. Genome Biol. 15:5542014. View Article : Google Scholar
|
|
38
|
Estoppey D, Schutzius G, Kolter C, Salathe
A, Wunderlin T, Meyer A, Nigsch F, Bouwmeester T, Hoepfner D and
Kirkland S: Genome-wide CRISPR-Cas9 screens identify mechanisms of
BET bromodomain inhibitor sensitivity. Iscience. 24:1033232021.
View Article : Google Scholar
|
|
39
|
Zhao Y, Wei L, Tagmount A, Loguinov A,
Sobh A, Hubbard A, McHale CM, Chang CJ, Vulpe CD and Zhang L:
Applying genome-wide CRISPR to identify known and novel genes and
pathways that modulate formaldehyde toxicity. Chemosphere.
269:1287012021. View Article : Google Scholar
|
|
40
|
Yan YQ, Fu YJ, Wu S, Qin HQ, Zhen X, Song
BM, Weng YS, Wang PC, Chen XY and Jiang ZY: Anti-influenza activity
of berberine improves prognosis by reducing viral replication in
mice. Phytother Res. 32:2560–2567. 2018. View Article : Google Scholar
|
|
41
|
Kalaiarasi A, Anusha C, Sankar R,
Rajasekaran S, John Marshal J, Muthusamy K and Ravikumar V: Plant
isoquinoline alkaloid berberine exhibits chromatin remodeling by
modulation of histone deacetylase to induce growth arrest and
apoptosis in the A549 cell line. J Agric Food Chem. 64:9542–9550.
2016. View Article : Google Scholar
|
|
42
|
Qi HW, Xin LY, Xu X, Ji XX and Fan LH:
Epithelial-to-mesenchymal transition markers to predict response of
Berberine in suppressing lung cancer invasion and metastasis. J
Transl Med. 12:222014. View Article : Google Scholar
|
|
43
|
Zheng F, Wu J, Tang Q, Xiao Q, Wu W and
Hann SS: The enhancement of combination of berberine and metformin
in inhibition of DNMT1 gene expression through interplay of SP1 and
PDPK1. J Cell Mol Med. 22:600–612. 2018. View Article : Google Scholar
|
|
44
|
Chen QQ, Shi JM, Ding Z, Xia Q, Zheng TS,
Ren YB, Li M and Fan LH: Berberine induces apoptosis in
non-small-cell lung cancer cells by upregulating miR-19a targeting
tissue factor. Cancer Manag Res. 11:9005–9015. 2019. View Article : Google Scholar
|
|
45
|
Kumar R, Awasthi M, Sharma A, Padwad Y and
Sharma R: Berberine induces dose-dependent quiescence and apoptosis
in A549 cancer cells by modulating cell cyclins and inflammation
independent of mTOR pathway. Life Sci. 244:1173462020. View Article : Google Scholar
|
|
46
|
Pope JL, Bhat AA, Sharma A, Ahmad R,
Krishnan M, Washington MK, Beauchamp RD, Singh AB and Dhawan P:
Claudin-1 regulates intestinal epithelial homeostasis through the
modulation of Notch-signalling. Gut. 63:622–634. 2014. View Article : Google Scholar
|
|
47
|
Singh AB, Uppada SB and Dhawan P: Claudin
proteins, outside-in signaling, and carcinogenesis. Pflugers Arch.
469:69–75. 2017. View Article : Google Scholar
|
|
48
|
Turksen K: Claudins and cancer stem cells.
Stem Cell Rev Rep. 7:797–798. 2011. View Article : Google Scholar
|
|
49
|
Zhou B, Flodby P, Luo J, Castillo DR, Liu
Y, Yu FX, McConnell A, Varghese B, Li G, Chimge NO, et al:
Claudin-18-mediated YAP activity regulates lung stem and progenitor
cell homeostasis and tumorigenesis. J Clin Invest. 128:970–984.
2018. View Article : Google Scholar
|
|
50
|
Zhao Z, Li J, Jiang Y, Xu W, Li X and Jing
W: CLDN1 increases drug resistance of Non-small cell lung cancer by
activating autophagy via Up-regulation of ULK1 phosphorylation. Med
Sci Monit. 23:2906–2916. 2017. View Article : Google Scholar
|
|
51
|
Cai W, Shu LZ, Liu DJ, Zhou L, Wang MM and
Deng H: Targeting cyclin D1 as a therapeutic approach for papillary
thyroid carcinoma. Front Oncol. 13:11450822023. View Article : Google Scholar
|
|
52
|
O'Kane GM and Leighl NB: Are immune
checkpoint blockade monoclonal antibodies active against CNS
metastases from NSCLC?-current evidence and future perspectives.
Transl Lung Cancer Res. 5:628–636. 2016. View Article : Google Scholar
|
|
53
|
Economopoulou P and Mountzios G: Non-small
cell lung cancer (NSCLC) and central nervous system (CNS)
metastases: Role of tyrosine kinase inhibitors (TKIs) and evidence
in favor or against their use with concurrent cranial radiotherapy.
Transl Lung Cancer Res. 5:588–598. 2016. View Article : Google Scholar
|
|
54
|
Lin JJ and Shaw AT: Resisting resistance:
Targeted therapies in lung cancer. Trends Cancer. 2:350–364. 2016.
View Article : Google Scholar
|
|
55
|
Wang Y, Liu Y, Du X, Ma H and Yao J: The
Anti-cancer mechanisms of berberine: A review. Cancer Manag Res.
12:695–702. 2020. View Article : Google Scholar
|
|
56
|
Liao LX, Song XM, Wang LC, Lv HN, Chen JF,
Liu D, Fu G, Zhao MB, Jiang Y, Zeng KW and Tu PF: Highly selective
inhibition of IMPDH2 provides the basis of antineuroinflammation
therapy. Proc Natl Acad Sci USA. 114:E5986–E5994. 2017. View Article : Google Scholar
|
|
57
|
Luo P, Zhang Q, Zhong TY, Chen JY, Zhang
JZ, Tian Y, Zheng LH, Yang F, Dai LY, Zou C, et al: Celastrol
mitigates inflammation in sepsis by inhibiting the PKM2-dependent
Warburg effect. Mil Med Res. 9:222022.
|
|
58
|
Reinhard FB, Eberhard D, Werner T, Franken
H, Childs D, Doce C, Savitski MF, Huber W, Bantscheff M, Savitski
MM and Drewes G: Thermal proteome profiling monitors ligand
interactions with cellular membrane proteins. Nat Methods.
12:1129–1131. 2015. View Article : Google Scholar
|
|
59
|
Chehelgerdi M, Chehelgerdi M,
Khorramian-Ghahfarokhi M, Shafieizadeh M, Mahmoudi E, Eskandari F,
Rashidi M, Arshi A and Mokhtari-Farsani A: Comprehensive review of
CRISPR-based gene editing: Mechanisms, challenges, and applications
in cancer therapy. Mol Cancer. 23:92024. View Article : Google Scholar
|
|
60
|
Masarwy R, Breier D, Stotsky-Oterin L,
Ad-El N, Qassem S, Naidu GS, Aitha A, Ezra A, Goldsmith M,
Hazan-Halevy I and Peer D: Targeted CRISPR/Cas9 lipid nanoparticles
elicits therapeutic genome editing in head and neck cancer. Adv Sci
(Weinh). 12:e24110322025. View Article : Google Scholar
|
|
61
|
Xu X, He Y and Liu J: Berberine: A
multifaceted agent for lung cancer treatment-from molecular insight
to clinical applications. Gene. 934:1490212025. View Article : Google Scholar
|
|
62
|
Fujiwara-Tani R, Mori S, Ogata R, Sasaki
R, Ikemoto A, Kishi S, Kondoh M and Kuniyasu H: Claudin-4: A new
molecular target for epithelial cancer therapy. Int J Mol Sci.
24:54942023. View Article : Google Scholar
|
|
63
|
Tojjari A, Idrissi YA and Saeed A:
Emerging targets in gastric and pancreatic cancer: Focus on claudin
18.2. Cancer Lett. 611:2173622024. View Article : Google Scholar
|
|
64
|
Nagaoka Y, Oshiro K, Yoshino Y, Matsunaga
T, Endo S and Ikari A: Activation of the TGF-beta1/EMT signaling
pathway by claudin-1 overexpression reduces doxorubicin sensitivity
in small cell lung cancer SBC-3 cells. Arch Biochem Biophys.
751:1098242024. View Article : Google Scholar
|