Introduction
Cancer remains a major health burden. Lung cancer
ranks as the leading cause of cancer-related mortality worldwide,
accounting for ~1.8 million new cases and ~1.6 million deaths
annually (1). Non-small cell lung
cancer, which accounts for ~85% of all lung canc er cases, is of
particular concern as the majority of patients are diagnosed at
advanced or metastatic stages (2).
Although notable progress has been made in surgery, chemotherapy,
radiotherapy, targeted therapy and immunotherapy, resistance to
treatment and relapse remain major challenges, thus highlighting
the need for novel therapeutic strategies (3–6).
Traditional Chinese Medicine (TCM) offers unique
advantages due to its multi-target and multi-component nature, as
well as its relative low toxicity. Emerging evidence has
highlighted its role in cancer therapy, with research reporting
improvements in patient quality of life and an increase in the
efficacy and reduction of adverse effects of conventional therapies
(7,8). TCM exerts its therapeutic effects via
targeting several biomolecules, modulating downstream signaling
pathways and disrupting disease-associated networks (9,10).
Therefore, identifying the molecular targets of bioactive compounds
in TCM is a pivotal step toward understanding its underlying
therapeutic mechanisms and advancing their modern clinical
applications (11).
Despite marked progress in drug discovery,
elucidating the molecular targets of TCM compounds remains a major
challenge due to the lack of effective tools for investigating
small molecule-protein interactions. Conventional methods include
affinity chromatography (12),
activity-based protein profiling (13) and label-free approaches such as drug
affinity responsive target stability (DARTS) (14), oxidative rate-dependent protein
stability analysis (15) and
thermal proteome profiling (16–19).
For example, oxidative rate-dependent protein stability leverages
changes in protein oxidation kinetics upon ligand binding to
identify potential targets (15).
However, label-free methods commonly lack sufficient sensitivity,
whilst label-based techniques frequently require structural
modifications that can compromise compound activity (20). These limitations have hindered the
comprehensive understanding of the mechanisms underlying TCM and
slowed their broader integration into modern therapeutic
applications.
The advent of clustered regularly interspaced short
palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)
genome-editing technology in 2013 has revolutionized functional
genomics research (21).
Genome-wide CRISPR libraries enable unbiased and high-throughput
screening of functional genes, thus offering transformative
potential in drug resistance (22),
tumor biology (23), immune
responses (24) and cancer
immunotherapy (25). Previous
applications of CRISPR-based screening in drug discovery have
supported its potential in identifying molecular targets of
bioactive compounds (26,27), thus providing novel insights into
its adoption in TCM research.
Berberine (BBR), a natural alkaloid extracted from
Coptis chinensis and related medicinal plants, exhibits
several biological activities, including anti-inflammatory,
antimicrobial and anticancer properties (28–30).
Notably, a previous study reported that BBR could inhibit lung
cancer cell proliferation (31).
However, its particular molecular mechanisms remain largely
unknown, thus limiting its therapeutic application. Recent studies
have demonstrated the anticancer versatility of BBR. For instance,
Li et al (32) reported that
BBR induced apoptosis and cell-cycle arrest across different cancer
cell lines. Additionally, Shen et al (33) reported that BBR delivered via
polyethylene glycol-poly-lactide co-glycolide acid nanocarriers
could enhance transcriptomic changes in colorectal cancer cells,
thus modulating the Wnt, TGF-β, Hippo and mammalian target of
rapamycin signaling pathways. Consistently, Sun et al
(34) reported the multifaceted
antitumor activities of BBR, including anti-inflammatory activity,
inhibition of angiogenesis and reversal of drug resistance. In
2024, a review article further summarized the effects of BBR on
nuclear factor κB, mitogen-activated protein kinase and
phosphoinositide 3-kinase/protein kinase B signaling, as well as
its synergistic effects with conventional therapies (35).
Therefore, the present study aimed to systematically
identify genes responsive to BBR in A549 lung cancer cells via
employing a genome-wide CRISPR/Cas9 screening library. This
approach could provide a powerful platform for identifying
molecular targets of TCM, thus contributing to a deeper
understanding of their mechanisms of action and accelerating their
integration into modern oncological practice.
Materials and methods
Reagents and instruments
BBR (purity ≥98%; cat. no. 2086-83-1) was purchased
from MedChemExpress. F-12K medium, fetal bovine serum (FBS) and
trypsin were purchased from Gibco (Thermo Fisher Scientific, Inc.).
RIPA lysis buffer, protease inhibitor cocktail, western blot
reagents, Cell Counting Kit 8 (CCK-8) assay and BCA protein assay
kits were purchased from Beyotime Biotechnology. The cell culture
plates and microplate reader were purchased from Falcon (Corning
Life Sciences) and Thermo Fisher Scientific, Inc.,
respectively.
Cell culture
The A549 human lung adenocarcinoma cell line was
purchased from Shanghai Hanyu Biotechnology Co., Ltd. Cells were
maintained in F-12K medium supplemented with 10% FBS at 37°C in a
humidified incubator with 5% CO2. Cells cultured at a
maximum of 5–20 passages were used for all experiments. Routine
mycoplasma testing was performed and confirmed negative. Culture
medium was replaced every 2–3 days. The incubator was maintained at
~95% relative humidity to ensure optimal growth conditions. All
experiments, including CCK-8 assays, CRISPR screening, western
blotting and flow cytometry, were performed in at least three
independent biological replicates, each using separately cultured
cell batches. For all assays, cells in the logarithmic growth phase
were digested, counted and seeded as single-cell suspensions at a
density of 2,000 cells/well in 12-well plates, to ensure uniform
distribution. Edge wells were filled with sterile water to minimize
evaporation.
Cell viability assay
The effects of BBR on cell viability were assessed
using a CCK-8 assay. Briefly, cells were treated with 1–60 µM BBR
or with the corresponding control reagent (DMSO). Following
incubation for 72 h, each well was supplemented with 10 µl CCK-8
reagent, followed by incubation for an additional 2 h. Finally, to
quantify cell viability, the absorbance in each well was measured
at a wavelength of 450 nm using a microplate reader.
CRISPR/Cas9 genome-wide knockout
screening
A genome-wide CRISPR knockout library was generated
using the GeCKO v2.0 platform (36), encompassing 123,411 single-guide
(sg)RNAs targeting 19,050 human genes. The sgRNA plasmid library
(Addgene plasmid #1000000048) used in the present study may be
obtained through Addgene, Inc. (https://www.addgene.org/pooled-library/zhang-human-gecko-v2/),
and its use and construction have been previously described
(36). Each gene was targeted by
six independent sgRNAs distributed across constitutive exons to
maximize knockout efficiency.
To achieve comprehensive genome coverage,
~1.6×108 A549 cells were infected with lentiviral
particles at a multiplicity of infection of 0.3-0.5. Cells were
seeded in the presence of 8 µg/ml polybrene and subjected to spin
infection (450 × g at 37°C for 2 h), followed by replacement with
fresh medium. Cells were then selected with puromycin (1 µg/ml) for
14 days. To confirm stable expression, the expression levels of
Cas9 in A549 cells were assessed using western blot analysis. The
screening employed a single-vector lentiviral system (lentiCRISPR
v2; cat. no. 52961; Addgene, Inc.), in which both Cas9 and sgRNA
expression cassettes are integrated within the same backbone;
therefore, no additional Cas9 transfection was required.
Library quality was evaluated using next-generation
sequencing (NGS), which verified uniform sgRNA representation, with
>200-fold coverage per sgRNA. Genomic DNA was amplified using
the NEBNext High-Fidelity 2X PCR Master Mix (cat. no. M0541; New
England Biolabs, Inc.) and purified using the QIAquick Gel
Extraction Kit (cat. no. 28704; Qiagen GmbH). The integrity of the
200- to 300-bp amplified fragments was verified by 1.5% agarose gel
electrophoresis prior to sequencing. Sequencing was performed on an
Illumina NovaSeq 6000 platform using the NovaSeq 6000 S4 Reagent
Kit v1.5 (cat. no. 20028312; Illumina, Inc.), generating paired-end
150 bp (PE150) reads at an average depth exceeding 300 times. The
final pooled library was loaded at a concentration of 1.457 µg/µl
(2.18×10−9 mol/µl), quantified using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). To maintain
library complexity, a total of 4×107 cells were
collected every 3 days and stored at −80°C. For NGS, genomic DNA
was extracted on day 14, and candidate genes responsive to BBR were
identified using MAGeCKFlute (version 0.5) analysis (37). Genes were ranked based on the
changes in their sgRNA abundances between treatment and control
groups. Those significantly enriched or reduced (P<0.05) were
subsequently subjected to Gene Ontology (GO) classification and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis using the Omicsbean proteomics toolkit (www.omicsbean.cn).
Treatment of genome-wide edited cell
libraries with BBR
A549 cells harboring the CRISPR knockout library
were expanded and divided into the following three groups: i)
Untreated (control); ii) BBR-treated; and iii) vehicle-treated
(DMSO) groups. Cells in the experimental group were exposed to 10
µM BBR at 37°C in a humidified 5% CO2 incubator for 3
days, whilst those in the control group were cultured under routine
conditions with regular medium changes and passaging. When cell
viability in the BBR-treated group reached ~10% of that of the
untreated group, genomic DNA was extracted for NGS to identify
sgRNA enrichment patterns.
Claudin (CLDN)-1 knockout and
lentiviral transduction
A second-generation lentiviral packaging system was
used for transduction. The lentiviral vector encompassing sgRNA
sequences targeting CLDN1 was constructed after cloning
oligonucleotide sequences (sgRNA-Z9, 5′-AGCGAGTCATGGCCAACGCG-3′;
sgRNA-Z10, 5′-GGCGCCGATCCATCCCAGGA-3′) into the lentiCRISPR v2
plasmid (cat. no. 52961; Addgene, Inc.). Prior to transduction, the
successful construction of plasmids was verified by Sanger
sequencing, which was performed by Sangon Biotech Co., Ltd.
(Shanghai, China) using an Applied Biosystems (Thermo Fisher
Scientific) platform. Lentiviral particles were produced in 293T
cells (ATCC CRL-3216) by co-transfecting the transfer plasmid
(lentiCRISPR v2) with the packaging plasmids psPAX2 (cat. no.
12260; Addgene, Inc.) and the envelope plasmid pMD2.G (cat. no.
12259; Addgene, Inc.) at a mass ratio of 2:1:1, using
polyethylenimine as the transfection reagent. Transfection was
performed at 37°C in a humidified incubator with 5% CO2.
The viral supernatant was collected 48 and 72 h post-transfection,
pooled, filtered through a 0.45-µm filter and concentrated by
ultracentrifugation. A549 cells were seeded in 6-well plates at
~70% confluency and were then infected with lentiviral supernatant
at a multiplicity of infection of 5–10, containing 8 µg/ml
polybrene. Following incubation for 12 h at 37°C, the medium was
replaced with fresh F-12K supplemented with 10% FBS. Transduced
cells were subsequently selected with 2 µg/ml puromycin for 7 days
to establish stable CLDN1 knockout cell lines. A puromycin
concentration of 1 µg/ml was used for the maintenance of selected
cells. Following a 7-day recovery period after puromycin selection,
the cells were used for subsequent experimentation. Genomic DNA
from puromycin-resistant A549 cells was extracted using a genomic
DNA purification kit (Tiangen Biotech, Co., Ltd.), and the
CLDN1 target region encompassing the Z10 site was amplified
using PCR performed with PrimeSTAR HS DNA Polymerase (Takara Bio
Inc.) and the following primers: Forward,
5′-GCAACCGCAGCTTCTAGTATC-3′; and reverse,
5′-TGCACACTTGAGAAGTTACC-3′. The following thermocycling conditions
were applied: Initial denaturation at 98°C for 2 min; 35 cycles of
98°C for 10 sec, 58°C for 10 sec and 72°C for 15 sec; with a final
extension at 72°C for 5 min. PCR products were separated on a 1.2%
agarose gel stained with GoldView (Takara Bio Inc.) and visualized
using a GelDoc XR+ imaging system (Bio-Rad Laboratories, Inc.). PCR
products were purified and subjected to Sanger sequencing by Sangon
Biotech Co., Ltd. (Shanghai, China) to verify the successful genome
editing of CLDN1.
Western blot analysis
Protein lysates were prepared using NP-40 lysis
buffer supplemented with protease inhibitors. The protein
concentration was measured using a BCA kit (Thermo Fisher
Scientific, Inc.). Equal amounts of protein extract (20 µg per
lane) were first separated using 10% SDS-PAGE and were then
transferred to PVDF membranes. Following blocking with 3% bovine
serum albumin (Beyotime Biotechnology) at room temperature for 2 h,
the membranes were incubated overnight at 4°C with the following
primary antibodies: Anti-CLDN1 (1:1,000 dilution; cat. no.
28674-1-AP; Proteintech Group, Inc.), anti-cyclin D1 (1:1,000
dilution; cat. no. 26939-1-AP; Proteintech Group, Inc.), anti-Cas9
(1:1,000 dilution; cat. no. 26758-1-AP; Proteintech Group, Inc.),
anti-GAPDH (1:5,000 dilution; cat. no. 10494-1-AP; Proteintech
Group, Inc.) and anti-β-actin (1:5,000 dilution; cat. no.
66009-1-Ig; Proteintech Group, Inc.). Subsequently, the membranes
were incubated with corresponding HRP-conjugated secondary
antibodies (goat anti-rabbit IgG; 1:5,000 dilution; cat. no.
SA00001-2; Proteintech Group, Inc.) for 1 h at room temperature,
and the immunoreactive bands were visualized using enhanced
chemiluminescence (BeyoECL Plus; cat. no. P0018S; Beyotime
Biotechnology).
Flow cytometry analysis
To assess the effects of BBR on cell cycle
progression, A549 and CLDN1-knockout A549 cells were treated
with 2.5 µM BBR, a sublethal dose slightly below the calculated
half-maximal inhibitory concentration (IC50; 2.856 µM).
This concentration was selected to minimize cytotoxicity whilst
allowing the detection of sensitization effects associated with
CLDN1 knockout. This was consistent with standard
pharmacological studies aiming to evaluate differential cellular
responses using sub-lethal or slightly sub-IC50
concentrations (38,39). Following incubation for 72 h, cells
were fixed with 70% ethanol, washed with PBS and stained with
propidium iodide. Samples were incubated at 37°C in the dark for 30
min prior analysis on a flow cytometer (CytoFLEX; Beckman Coulter,
Inc.) equipped with a 488-nm laser. Data analysis was performed
using FlowJo software (version 8.0; FlowJo LLC).
Statistical analysis
The experimental data were statistically analyzed
using GraphPad Prism 5.0 software (Dotmatics). Groups were compared
using an unpaired two-tailed Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Effects of BBR on A549 cell
proliferation
Previous studies have reported that BBR can inhibit
the growth of lung cancer cells, including that of A549 cells, in a
dose-dependent manner. However, effective concentrations vary among
different cell lines (40–42). Therefore, to determine the optimal
concentration of BBR in A549 cells, the cells were treated with
increasing concentrations of BBR (1, 2.5, 5, 10, 20, 40 and 60 µM)
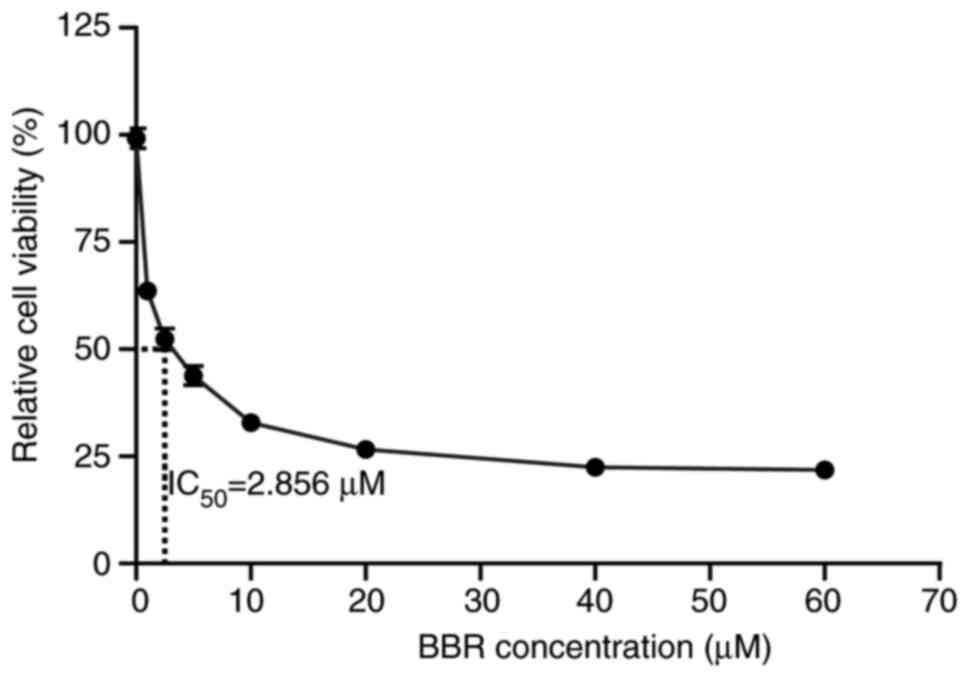
and cell viability was then assessed using a CCK-8 assay. The
results revealed that BBR significantly reduced cell viability in a
dose-dependent manner, with only 21.5% of cells remaining viable at
a concentration of 60 µM (Fig. 1).
Analysis using GraphPad Prism software revealed that the
IC50 was 2.856 µM. These findings indicated that BBR
could effectively suppresses A549 cell proliferation, thus
providing a foundation for subsequent mechanistic experiments.
Construction of A549 genome-wide
knockout library and BBR target screening
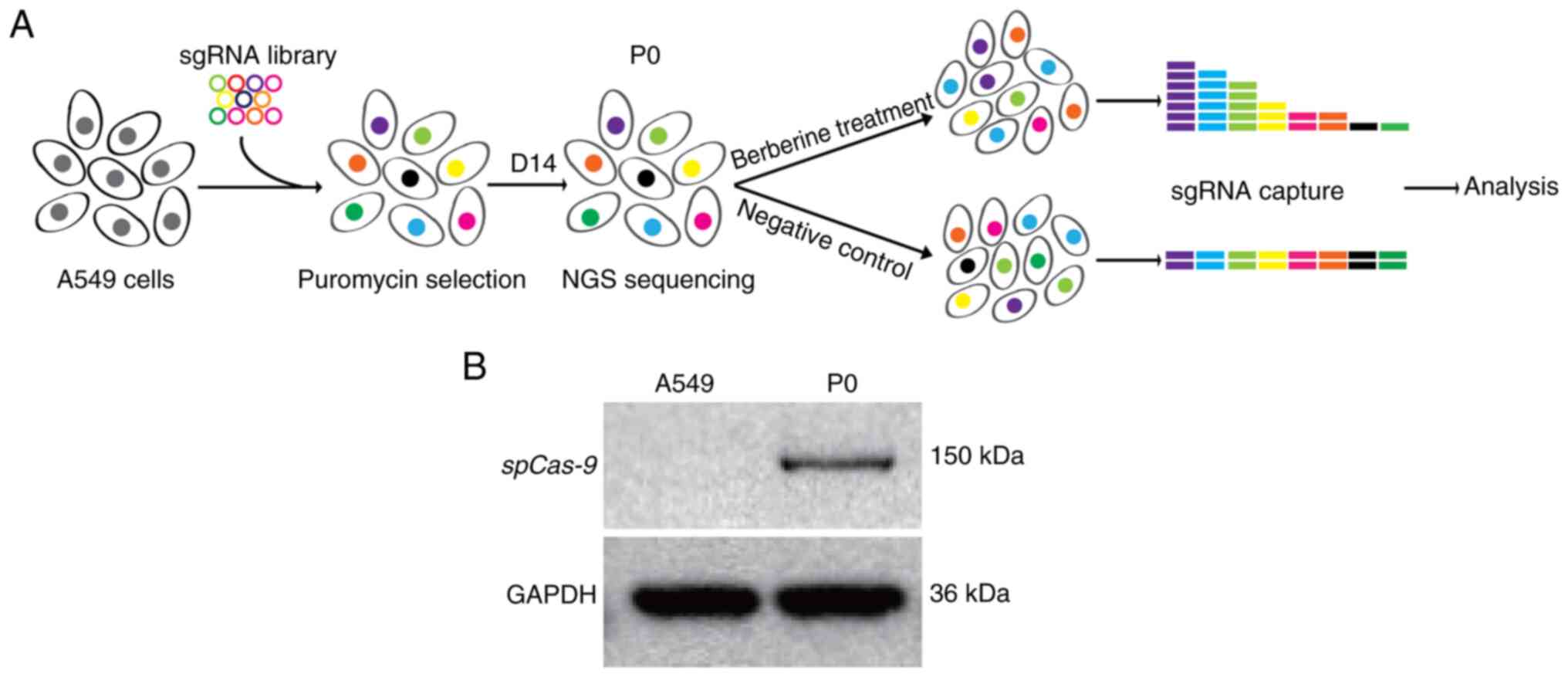
To comprehensively identify genes responsive to BBR
treatment, a genome-wide CRISPR/Cas9 knockout library was
constructed in A549 cells (Fig.
2A). Library generation followed established protocols
(36), with lentiviral transduction
enabling stable expression of Streptococcus pyogenes
(sp)Cas9 in A549 cells. Western blot analysis was performed
to confirm the successful expression of spCas9 protein at the
expected molecular weight of 140–160 kDa (Fig. 2B). The resulting knockout library
was designated as P0. To assess the capacity of the library in
identifying BBR targets, P0 cells were treated with BBR until cell
viability reduced to ~10% of the untreated cell population.
Subsequently, genomic DNA was extracted from surviving cells, and
sgRNA sequence representation was determined using NGS. Candidate
genes were ranked based on positive (pos|score) or negative
(neg|score) selection values using the MAGeCK algorithm, with a
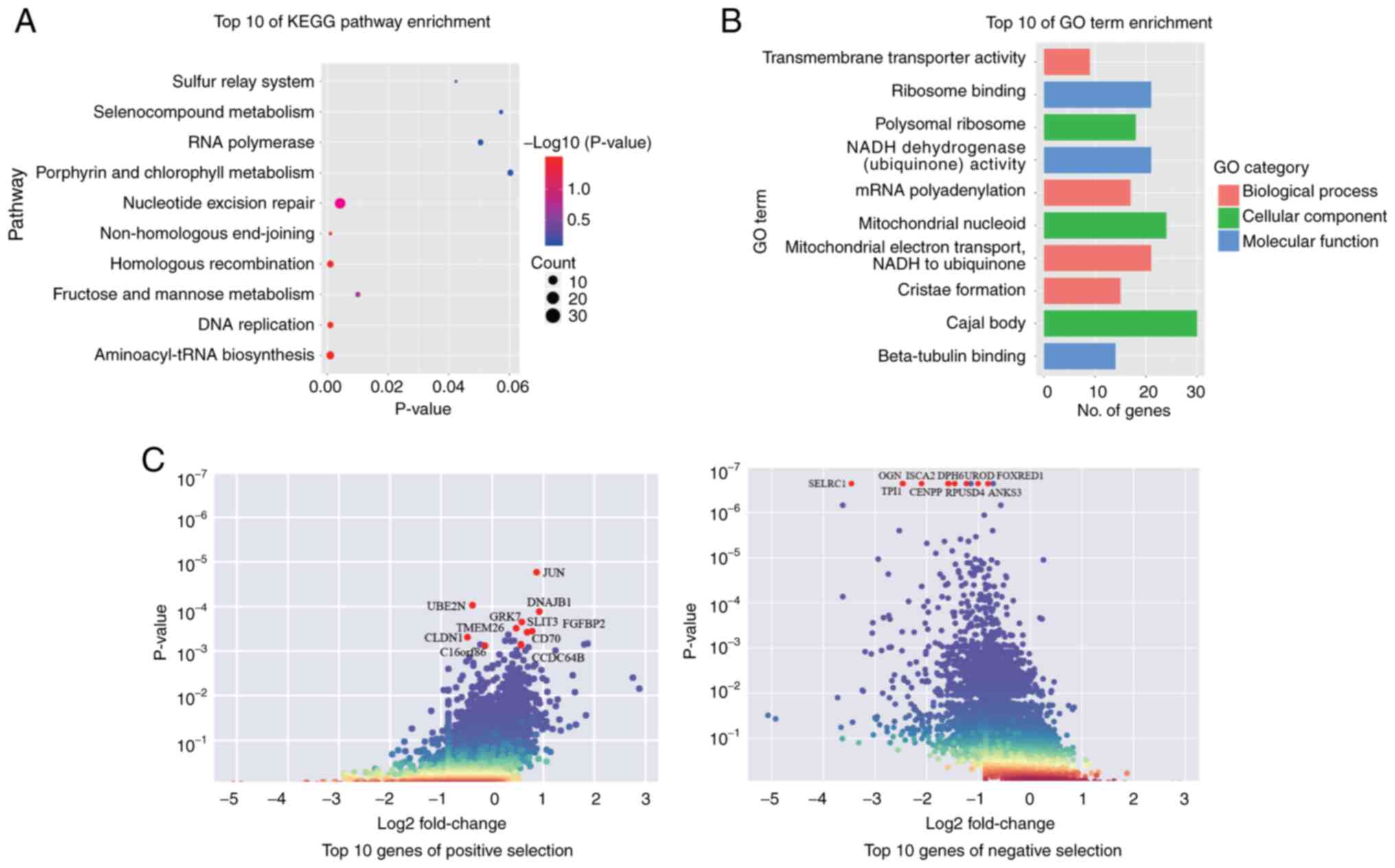
significance threshold of P<0.05. A total of 3,873 genes
exhibited significant differential selection, including 749
positively selected genes (associated with resistance) and 3,124
negatively selected ones (associated with sensitivity) (Tables SI and SII).
Enrichment analysis of resistance and
sensitivity genes
To elucidate the biological pathways underlying BBR
response, KEGG pathway and GO enrichment analyses were performed on
the differentially selected genes. KEGG analysis revealed that the
selected genes were mainly enriched in the ‘aminoacyl-tRNA
biosynthesis’, ‘homologous recombination’, ‘DNA replication’ and
‘non-homologous end-joining’ pathways (Fig. 3A). Notably, several DNA damage
repair-related pathways were highlighted, thus suggesting that BBR
could be associated with genomic stability and cell cycle
regulation. GO analysis further revealed that the selected genes
were enriched into the following molecular functions: ‘ribosome
binding’, ‘beta-tubulin binding’ and ‘NADH dehydrogenase activity’;
cellular components: ‘mitochondrial nucleoid’, ‘Cajal body’ and
‘polysomal ribosome’; and biological processes: ‘mitochondrial
electron transport’, ‘mRNA polyadenylation’ and ‘transmembrane
transporter activity’ (Fig. 3B).
These findings indicate that BBR could potentially disrupt cellular
energy metabolism and fundamental biological processes. Among the
top-ranked candidate genes (Fig.
3C), slit guidance ligand 3, DNAJ heat shock protein family
(Hsp40) member B1, ubiquitin conjugating enzyme E2 N and
CLDN1 were of particular interest due to their established
roles in lung cancer cell proliferation, invasion and drug
resistance (43–50). Notably, CLDN1, a member of
the CLDN family involved in tight junction integrity, has been
associated with tumor progression and chemoresistance in A549 cells
(50), highlighting its potential
as a compelling target for further investigation.
Functional validation of CLDN1 in BBR
response
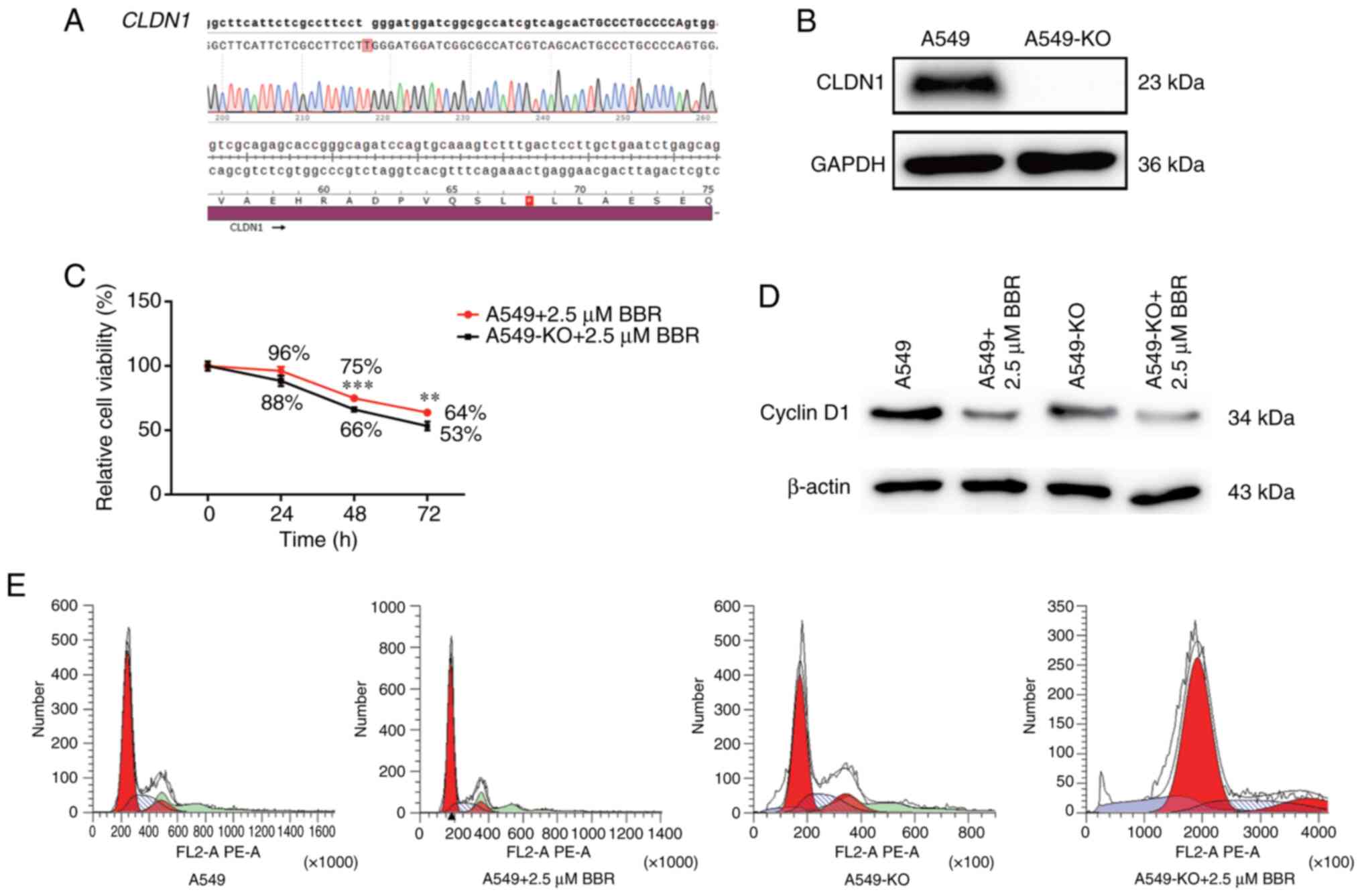
To assess the role of CLDN1 in mediating
response to BBR, two sgRNAs targeting the CLDN1 coding
region (sgRNA-Z9, 5′-AGCGAGTCATGGCCAACGCG-3′; sgRNA-Z10,
5′-GGCGCCGATCCATCCCAGGA-3′) were designed, and
CLDN1-knockout A549 cells were established via lentiviral
transduction. Sanger sequencing confirmed a single-nucleotide
insertion at the Z10 target site, introducing a premature stop
codon at amino acid position 68 (Fig.
4A). Additionally, western blot analysis demonstrated the
complete absence of CLDN1 protein expression in knockout cells
(A549-KO cells; Fig. 4B).
Furthermore, A549-KO cell treatment with BBR significantly reduced
cell viability compared with that in wildtype A549 cells, with
notable differences observed at 48 and 72 h, thus supporting the
dose- and time-dependent sensitization effects of BBR (Fig. 4C). Additionally, flow cytometry
revealed a marked reduction in the S-phase population and a
concomitant increase in the G1-phase population in A549-KO cells
following BBR treatment, indicative of G1-phase cell cycle arrest
(Fig. 4E). Consistent with the
aforementioned findings, cyclin D1, a key regulator of G1/S
transition (51), was notably
downregulated in BBR-treated A549-KO cells compared with that in
wild-type cells (Fig. 4D). These
results indicate that CLDN1 could be involved in BBR-induced
cell cycle arrest and growth suppression in A549 cells.
Discussion
Lung cancer, accounting for ~1.8 million new cases
and ~1.6 million deaths annually, remains a global health challenge
due to its low 5-year survival rate, ranging from 4–17% depending
on the stage of diagnosis and geographical region (52,53).
Despite advances in chemotherapy, radiotherapy, targeted therapies
and immunotherapies, drug resistance still poses a critical
barrier, notably limiting long-term efficacy (54). In this context, TCM offers a
promising complementary approach, owning to its multi-target and
multi-component characteristics. Among TCM-derived compounds, BBR
has shown potent anticancer properties via modulating key cellular
processes, such as proliferation, apoptosis, autophagy and immune
modulation within the tumor microenvironment (55). However, the particular molecular
targets and pathways underlying its anticancer effects remain
insufficiently characterized.
The identification of molecular targets of bioactive
compounds is essential for the modernization of TCM and the
development of novel cancer therapeutics (9,11).
Although label-based approaches, such as chemical probe-assisted
affinity capture, have successfully identified key targets, their
application is limited due to chemical modification-induced
activity loss and structural constraints (56,57).
By contrast, label-free methods such as DARTS, cellular thermal
shift assay and thermal proteome profiling offer non-invasive
alternatives. However, their use is commonly hindered by limited
sensitivity to weak or low-abundance protein interactions (14,15,17,58).
These limitations highlight the urgent need for scalable and
precise strategies to enable systematic target identification.
In the present study, a CRISPR/Cas9 genome-wide
knockout library was constructed in A549 lung adenocarcinoma cells
to systematically investigate the molecular targets and resistance
mechanisms associated with BBR treatment. The unbiased screening
identified a total of 3,873 genes with differential selection under
BBR exposure, including 749 positive-selection genes associated
with resistance and 3,124 negative-selection genes essential for
cellular viability. Among the resistance-related genes, CLDN1, a
tight junction protein of the CLDN family, could serve as a pivotal
mediator of BBR resistance. In a previous study, CLDN1
overexpression was associated with poor prognosis and resistance to
cisplatin in patients with lung cancer (50). Consistently, the results of the
present study demonstrated that CLDN1 knockout enhanced the
sensitivity of A549 cells to BBR, characterized by reduced cell
proliferation, decreased S-phase entry and G1-phase arrest. The
CRISPR-based identification of CLDN1 as a mediator of the
anticancer effects of BBR aligned with broader advances in CRISPR
therapeutic strategies. Previous comprehensive reviews summarized
the potential of CRISPR in targeting oncogenes, precise gene
modulation and overcoming drug resistance (59). Notably, an in vivo study
reported that CRISPR-lipid nanoparticle platforms could eliminate
≤50% of tumors in mouse models (60). Furthermore, BBR could inhibit tumor
growth in lung cancer models via several pathways, including
epigenetic and DNA-repair modulation (61). Taken together, the results of the
present study could extend current therapeutic paradigms by
positioning CLDN1 as a critical regulator of BBR
responsiveness. Notably, the functional assays in the present study
were performed at 2.5 µM BBR, slightly below the IC50
value (2.856 µM), thus ensuring that the observed differences were
due to genetic manipulation rather than cytotoxicity. However,
future studies are needed to refine this analysis using a
concentration gradient (such as 2.0, 2.5 and 3.0 µM) to further
validate the robustness of the findings. Nevertheless, the results
strongly suggest that CLDN1 could serve a key role in
mediating resistance to BBR via modulating cell cycle
progression.
Beyond CLDN1, other CLDN family members have
also been involved in cancer progression and therapeutic targeting.
For example, a study reported that CLDN4 could act as a
molecular target in epithelial cancers, thus being involved in
epithelial-to-mesenchymal transition (EMT) and YAP-mediated
proliferation, with ongoing preclinical efforts exploring
CLDN4-directed antibody therapies (62). Additionally, CLDN18.2 could serve as
a clinically actionable target in gastric and pancreatic cancers,
with monoclonal antibody approaches, such as zolbetuximab,
demonstrating promising antitumor activity (63). Collectively, these findings could
frame CLDN1 within a broader CLDN-triggered regulatory
network and highlight the potential of multi-CLDN targeting
approaches in cancer therapy.
Beyond its role in cell cycle regulation,
CLDN1 has also been involved in additional signaling
pathways associated with cancer biology. A previous study reported
that CLDN1 overexpression could activate the TGF-β1/EMT
signaling pathway, thus mitigating the sensitivity of lung cancer
cells to doxorubicin (64). These
findings indicated that CLDN1 could act as a multifunctional
regulator linking junctional integrity to oncogenic signaling.
Whilst the present study primarily established CLDN1 as a
mediator of BBR sensitivity, future research is needed to further
investigate EMT- and TGF-β1-associated mechanisms downstream of
CLDN1 to provide a more comprehensive mechanistic framework.
Notably, the identification of CLDN1 as a BBR-resistance
gene underscored the broader utility of the current CRISPR/Cas9
screening platform. Compared with conventional methods, this
genome-wide approach could enable unbiased and high-throughput
target identification, thus providing a powerful framework for
elucidating the molecular mechanisms of TCM-derived compounds. The
discovery of CLDN1 could not only deepen the understanding
of the pharmacological effects of BBR, but also highlight its
potential as a therapeutic target for overcoming drug resistance in
lung cancer.
Moving forward, the development of specific
CLDN1 inhibitors could provide novel opportunities for
combination therapies, enhancing the efficacy of BBR and that of
other anticancer agents. In addition, the present study established
a proof-of-concept for applying CRISPR-based screening in TCM
research, thus offering novel insights into the systematic
exploration of bioactive compounds and their molecular targets.
This integrative approach could not only support the modernization
and global acceptance of TCM, but it could be also involved into
the broader advancement of precision oncology.
While the present study successfully identified
CLDN1 as a pivotal mediator of berberine (BBR) resistance
through a genome-wide CRISPR/Cas9 screening in A549 cells, several
aspects merit further investigation. The findings were derived from
a single lung adenocarcinoma model, and validation in additional
lung cancer subtypes or in vivo systems would strengthen the
generalizability of the results. Moreover, this study focused on
gene-level knockout screening; integration with transcriptomic or
proteomic data could provide a broader mechanistic perspective on
the BBR-CLDN1 network. In addition, although a sub-IC50
concentration of BBR was used to highlight genetic effects rather
than cytotoxicity, evaluation across a concentration gradient would
better characterize dose-dependent responses. Addressing these
aspects in future work will refine the mechanistic insights and
enhance the translational relevance of the findings.
In conclusion, the present study identifies
CLDN1 as a critical determinant of berberine responsiveness
in lung cancer cells and establishes a CRISPR/Cas9-based framework
for target discovery in traditional Chinese medicine. These
findings provide mechanistic insight into BBR resistance and offer
a promising direction for the development of CLDN1-targeted
combination therapies.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by the Natural Science Foundation
of Chongqing-Postdoctoral Science Foundation Project (grant nos.
cstc2021jcyj-bshX0016 and cstc2021jcyj-bshX0034), Chongqing
Scientific Research Institutions Performance Incentive and Guidance
Project (grant nos. cstc2023jxjl-jbky130009, zzkjjh2022012,
zzkjjh2022013 and CSTB2023TIAD-KPX0101) and the Project of Basic
Scientific Research Plan of Chongqing (grant nos. jbky20210030 and
jbky20200008).
Availability of data and materials
The NGS data generated in the present study may be
found in the NCBI BioProject database under accession number
PRJNA1345357 or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1345357.
All other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
XGW and XT contributed to the design of CRISPR/Cas9
screening strategies. YDW contributed to the design and
optimization of the functional validation experiments following the
primary CRISPR/Cas9 screen. XGW, BZM and YDW were involved in the
literature search and review. XJW, BZM, YJX and FL performed the
experiments. JFL and JYH performed the data analysis. BZM and YDW
provided key reagents, cell lines and instrumental resources. XGW,
YJX and YDZ wrote the manuscript. YDZ conceived and coordinated the
study, integrated the overall data analysis and manuscript
preparation, and was responsible for communication and revision
with the journal, with YDW assisting in correspondence and resource
support. XGW and YDZ confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BBR
|
berberine
|
|
CLDN1
|
claudin-1
|
|
CRISPR
|
clustered regularly interspaced short
palindromic repeats
|
|
DARTS
|
drug affinity responsive target
stability
|
|
sgRNA
|
single-guide RNA
|
|
KO
|
knockout
|
|
TCM
|
traditional Chinese medicine
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
|
|
2
|
Phillips I, Stares M, Allan L, Sayers J,
Skipworth R and Laird B: Optimising outcomes in non small cell lung
cancer: Targeting cancer cachexia. Front Biosci (Landmark Ed).
27:1292022. View Article : Google Scholar
|
|
3
|
Su PL, Furuya N, Asrar A, Rolfo C, Li Z,
Carbone DP and He K: Recent advances in therapeutic strategies for
non-small cell lung cancer. J Hematol Oncol. 18:352025. View Article : Google Scholar
|
|
4
|
Riely GJ, Wood DE, Ettinger DS, Aisner DL,
Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, et
al: Non-small cell lung cancer, version 4.2024, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
22:249–274. 2024. View Article : Google Scholar
|
|
5
|
Wang JL, Qiu TY and Ren SX: Updates to the
2024 CSCO advanced non-small cell lung cancer guidelines. Cancer
Biol Med. 22:77–82. 2025.
|
|
6
|
Huang Q, Li Y, Huang Y, Wu J, Bao W, Xue
C, Li X, Dong S, Dong Z and Hu S: Advances in molecular pathology
and therapy of non-small cell lung cancer. Signal Transduct Tar.
10:1862025. View Article : Google Scholar
|
|
7
|
Li S, Chen X, Shi H, Yi M, Xiong B and Li
T: Tailoring traditional Chinese medicine in cancer therapy. Mol
Cancer. 24:272025. View Article : Google Scholar
|
|
8
|
Chen JF, Wu SW, Shi ZM and Hu B:
Traditional Chinese medicine for colorectal cancer treatment:
potential targets and mechanisms of action. Chin Med. 18:142023.
View Article : Google Scholar
|
|
9
|
Paolini GV, Shapland RH, van Hoorn WP,
Mason JS and Hopkins AL: Global mapping of pharmacological space.
Nat Biotechnol. 24:805–815. 2006. View
Article : Google Scholar
|
|
10
|
Mohamad Zobir SZ, Mohd Fauzi F, Liggi S,
Drakakis G, Fu X, Fan TP and Bender A: Global Mapping of
Traditional Chinese Medicine into Bioactivity Space and Pathways
Annotation Improves Mechanistic Understanding and Discovers
Relationships between Therapeutic Action (Sub)classes. Evid Based
Complement Alternat Med. 2016:21064652016. View Article : Google Scholar
|
|
11
|
Wang J, Wong YK and Liao F: What has
traditional Chinese medicine delivered for modern medicine? Expert
Rev Mol Med. 20:e42018. View Article : Google Scholar
|
|
12
|
Koshelev YA: Affinity chromatography and
proteomic screening as the effective method for S100A4 new protein
targets discovery. Mol Biol (Mosk). 48:868–872. 2014.(In Russian).
View Article : Google Scholar
|
|
13
|
Niphakis MJ and Cravatt BF: Enzyme
inhibitor discovery by activity-based protein profiling. Annu Rev
Biochem. 83:341–377. 2014. View Article : Google Scholar
|
|
14
|
Lomenick B, Hao R, Jonai N, Chin RM,
Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al:
Target identification using drug affinity responsive target
stability (DARTS). Proc Natl Acad Sci USA. 106:21984–21989. 2009.
View Article : Google Scholar
|
|
15
|
Dearmond PD, Xu Y, Strickland EC, Daniels
KG and Fitzgerald MC: Thermodynamic analysis of protein-ligand
interactions in complex biological mixtures using a shotgun
proteomics approach. J Proteome Res. 10:4948–4958. 2011. View Article : Google Scholar
|
|
16
|
Martinez Molina D, Jafari R,
Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y and
Nordlund P: Monitoring drug target engagement in cells and tissues
using the cellular thermal shift assay. Science. 341:84–87. 2013.
View Article : Google Scholar
|
|
17
|
Savitski MM, Reinhard FB, Franken H,
Werner T, Savitski MF, Eberhard D, Martinez Molina D, Jafari R,
Dovega RB, Klaeger S, et al: Tracking cancer drugs in living cells
by thermal profiling of the proteome. Science. 346:12557842014.
View Article : Google Scholar
|
|
18
|
Nabet B, Roberts JM, Buckley DL, Paulk J,
Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al:
The dTAG system for immediate and target-specific protein
degradation. Nat Chem Biol. 14:431–441. 2018. View Article : Google Scholar
|
|
19
|
Wittrup A, Ai A, Liu X, Hamar P, Trifonova
R, Charisse K, Manoharan M, Kirchhausen T and Lieberman J:
Visualizing Lipid-formulated siRNA release from endosomes and
target gene knockdown. Nat Biotechnol. 33:870–876. 2015. View Article : Google Scholar
|
|
20
|
Schirle M, Bantscheff M and Kuster B: Mass
Spectrometry-based proteomics in preclinical drug discovery. Chem
Biol. 19:72–84. 2012. View Article : Google Scholar
|
|
21
|
Koike-Yusa H, Li Y, Tan EP,
Velasco-Herrera Mdel C and Yusa K: Genome-wide recessive genetic
screening in mammalian cells with a lentiviral CRISPR-guide RNA
library. Nat Biotechnol. 32:267–273. 2014. View Article : Google Scholar
|
|
22
|
Hou P, Wu C, Wang Y, Qi R, Bhavanasi D,
Zuo Z, Dos Santos C, Chen S, Chen Y, Zheng H, et al: A Genome-Wide
CRISPR screen identifies genes critical for resistance to FLT3
inhibitor AC220. Cancer Res. 77:4402–4413. 2017. View Article : Google Scholar
|
|
23
|
Song CQ, Li Y, Mou H, Moore J, Park A,
Pomyen Y, Hough S, Kennedy Z, Fischer A, Yin H, et al: Genome-Wide
CRISPR screen identifies regulators of Mitogen-activated protein
kinase as suppressors of liver tumors in mice. Gastroenterology.
152:1161–1173.e1. 2017. View Article : Google Scholar
|
|
24
|
Parnas O, Jovanovic M, Eisenhaure TM,
Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I,
Sanjana NE, et al: A Genome-wide CRISPR screen in primary immune
cells to dissect regulatory networks. Cell. 162:675–686. 2015.
View Article : Google Scholar
|
|
25
|
Patel SJ, Sanjana NE, Kishton RJ,
Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM,
Yamamoto TN, et al: Identification of essential genes for cancer
immunotherapy. Nature. 548:537–542. 2017. View Article : Google Scholar
|
|
26
|
Kurata M, Yamamoto K, Moriarity BS,
Kitagawa M and Largaespada DA: CRISPR/Cas9 library screening for
drug target discovery. J Hum Genet. 63:179–186. 2018. View Article : Google Scholar
|
|
27
|
Kwon JW, Oh JS, Seok SH, An HW, Lee YJ,
Lee NY, Ha T, Kim HA, Yoon GM, Kim SE, et al: Combined inhibition
of Bcl-2 family members and YAP induces synthetic lethality in
metastatic gastric cancer with RASA1 and NF2 deficiency. Mol
Cancer. 22:1562023. View Article : Google Scholar
|
|
28
|
Och A, Podgorski R and Nowak R: Biological
activity of Berberine-A summary update. Toxins (Basel). 12:7132020.
View Article : Google Scholar
|
|
29
|
Habtemariam S: Recent advances in
berberine inspired anticancer approaches: From drug combination to
novel formulation technology and derivatization. Molecules.
25:14262020. View Article : Google Scholar
|
|
30
|
Samadi P, Sarvarian P, Gholipour E,
Asenjan KS, Aghebati-Maleki L, Motavalli R, Hojjat-Farsangi M and
Yousefi M: Berberine: A novel therapeutic strategy for cancer.
IUBMB Life. 72:2065–2079. 2020. View Article : Google Scholar
|
|
31
|
Paudel KR, Mehta M, Yin GHS, Yen LL,
Malyla V, Patel VK, Panneerselvam J, Madheswaran T, MacLoughlin R,
Jha NK, et al: Berberine-loaded liquid crystalline nanoparticles
inhibit non-small cell lung cancer proliferation and migration in
vitro. Environ Sci Pollut Res Int. 29:46830–46847. 2022. View Article : Google Scholar
|
|
32
|
Li Q, Zhao H, Chen W and Huang P:
Berberine induces apoptosis and arrests the cell cycle in multiple
cancer cell lines. Arch Med Sci. 19:1530–1537. 2023. View Article : Google Scholar
|
|
33
|
Shen F, Zheng YS, Dong L, Cao Z and Cao J:
Enhanced tumor suppression in colorectal cancer via
berberine-loaded PEG-PLGA nanoparticles. Front Pharmacol.
15:15007312024. View Article : Google Scholar
|
|
34
|
Sun L, Lan J, Li Z, Zeng R, Shen Y, Zhang
T and Ding Y: Transforming cancer treatment with nanotechnology:
The role of berberine as a star natural compound. Int J
Nanomedicine. 19:8621–8640. 2024. View Article : Google Scholar
|
|
35
|
Haque S, Mathkor DM, Bhat SA, Musayev A,
Khituova L, Ramniwas S, Phillips E, Swamy N, Kumar S, Yerer MB, et
al: A comprehensive review highlighting the prospects of
phytonutrient berberine as an anticancer agent. J Biochem Mol
Toxicol. 39:e700732025. View Article : Google Scholar
|
|
36
|
Shalem O, Sanjana NE and Zhang F:
High-throughput functional genomics using CRISPR-Cas9. Nat Rev
Genet. 16:299–311. 2015. View Article : Google Scholar
|
|
37
|
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang
F, Irizarry RA, Liu JS, Brown M and Liu XS: MAGeCK enables robust
identification of essential genes from genome-scale CRISPR/Cas9
knockout screens. Genome Biol. 15:5542014. View Article : Google Scholar
|
|
38
|
Estoppey D, Schutzius G, Kolter C, Salathe
A, Wunderlin T, Meyer A, Nigsch F, Bouwmeester T, Hoepfner D and
Kirkland S: Genome-wide CRISPR-Cas9 screens identify mechanisms of
BET bromodomain inhibitor sensitivity. Iscience. 24:1033232021.
View Article : Google Scholar
|
|
39
|
Zhao Y, Wei L, Tagmount A, Loguinov A,
Sobh A, Hubbard A, McHale CM, Chang CJ, Vulpe CD and Zhang L:
Applying genome-wide CRISPR to identify known and novel genes and
pathways that modulate formaldehyde toxicity. Chemosphere.
269:1287012021. View Article : Google Scholar
|
|
40
|
Yan YQ, Fu YJ, Wu S, Qin HQ, Zhen X, Song
BM, Weng YS, Wang PC, Chen XY and Jiang ZY: Anti-influenza activity
of berberine improves prognosis by reducing viral replication in
mice. Phytother Res. 32:2560–2567. 2018. View Article : Google Scholar
|
|
41
|
Kalaiarasi A, Anusha C, Sankar R,
Rajasekaran S, John Marshal J, Muthusamy K and Ravikumar V: Plant
isoquinoline alkaloid berberine exhibits chromatin remodeling by
modulation of histone deacetylase to induce growth arrest and
apoptosis in the A549 cell line. J Agric Food Chem. 64:9542–9550.
2016. View Article : Google Scholar
|
|
42
|
Qi HW, Xin LY, Xu X, Ji XX and Fan LH:
Epithelial-to-mesenchymal transition markers to predict response of
Berberine in suppressing lung cancer invasion and metastasis. J
Transl Med. 12:222014. View Article : Google Scholar
|
|
43
|
Zheng F, Wu J, Tang Q, Xiao Q, Wu W and
Hann SS: The enhancement of combination of berberine and metformin
in inhibition of DNMT1 gene expression through interplay of SP1 and
PDPK1. J Cell Mol Med. 22:600–612. 2018. View Article : Google Scholar
|
|
44
|
Chen QQ, Shi JM, Ding Z, Xia Q, Zheng TS,
Ren YB, Li M and Fan LH: Berberine induces apoptosis in
non-small-cell lung cancer cells by upregulating miR-19a targeting
tissue factor. Cancer Manag Res. 11:9005–9015. 2019. View Article : Google Scholar
|
|
45
|
Kumar R, Awasthi M, Sharma A, Padwad Y and
Sharma R: Berberine induces dose-dependent quiescence and apoptosis
in A549 cancer cells by modulating cell cyclins and inflammation
independent of mTOR pathway. Life Sci. 244:1173462020. View Article : Google Scholar
|
|
46
|
Pope JL, Bhat AA, Sharma A, Ahmad R,
Krishnan M, Washington MK, Beauchamp RD, Singh AB and Dhawan P:
Claudin-1 regulates intestinal epithelial homeostasis through the
modulation of Notch-signalling. Gut. 63:622–634. 2014. View Article : Google Scholar
|
|
47
|
Singh AB, Uppada SB and Dhawan P: Claudin
proteins, outside-in signaling, and carcinogenesis. Pflugers Arch.
469:69–75. 2017. View Article : Google Scholar
|
|
48
|
Turksen K: Claudins and cancer stem cells.
Stem Cell Rev Rep. 7:797–798. 2011. View Article : Google Scholar
|
|
49
|
Zhou B, Flodby P, Luo J, Castillo DR, Liu
Y, Yu FX, McConnell A, Varghese B, Li G, Chimge NO, et al:
Claudin-18-mediated YAP activity regulates lung stem and progenitor
cell homeostasis and tumorigenesis. J Clin Invest. 128:970–984.
2018. View Article : Google Scholar
|
|
50
|
Zhao Z, Li J, Jiang Y, Xu W, Li X and Jing
W: CLDN1 increases drug resistance of Non-small cell lung cancer by
activating autophagy via Up-regulation of ULK1 phosphorylation. Med
Sci Monit. 23:2906–2916. 2017. View Article : Google Scholar
|
|
51
|
Cai W, Shu LZ, Liu DJ, Zhou L, Wang MM and
Deng H: Targeting cyclin D1 as a therapeutic approach for papillary
thyroid carcinoma. Front Oncol. 13:11450822023. View Article : Google Scholar
|
|
52
|
O'Kane GM and Leighl NB: Are immune
checkpoint blockade monoclonal antibodies active against CNS
metastases from NSCLC?-current evidence and future perspectives.
Transl Lung Cancer Res. 5:628–636. 2016. View Article : Google Scholar
|
|
53
|
Economopoulou P and Mountzios G: Non-small
cell lung cancer (NSCLC) and central nervous system (CNS)
metastases: Role of tyrosine kinase inhibitors (TKIs) and evidence
in favor or against their use with concurrent cranial radiotherapy.
Transl Lung Cancer Res. 5:588–598. 2016. View Article : Google Scholar
|
|
54
|
Lin JJ and Shaw AT: Resisting resistance:
Targeted therapies in lung cancer. Trends Cancer. 2:350–364. 2016.
View Article : Google Scholar
|
|
55
|
Wang Y, Liu Y, Du X, Ma H and Yao J: The
Anti-cancer mechanisms of berberine: A review. Cancer Manag Res.
12:695–702. 2020. View Article : Google Scholar
|
|
56
|
Liao LX, Song XM, Wang LC, Lv HN, Chen JF,
Liu D, Fu G, Zhao MB, Jiang Y, Zeng KW and Tu PF: Highly selective
inhibition of IMPDH2 provides the basis of antineuroinflammation
therapy. Proc Natl Acad Sci USA. 114:E5986–E5994. 2017. View Article : Google Scholar
|
|
57
|
Luo P, Zhang Q, Zhong TY, Chen JY, Zhang
JZ, Tian Y, Zheng LH, Yang F, Dai LY, Zou C, et al: Celastrol
mitigates inflammation in sepsis by inhibiting the PKM2-dependent
Warburg effect. Mil Med Res. 9:222022.
|
|
58
|
Reinhard FB, Eberhard D, Werner T, Franken
H, Childs D, Doce C, Savitski MF, Huber W, Bantscheff M, Savitski
MM and Drewes G: Thermal proteome profiling monitors ligand
interactions with cellular membrane proteins. Nat Methods.
12:1129–1131. 2015. View Article : Google Scholar
|
|
59
|
Chehelgerdi M, Chehelgerdi M,
Khorramian-Ghahfarokhi M, Shafieizadeh M, Mahmoudi E, Eskandari F,
Rashidi M, Arshi A and Mokhtari-Farsani A: Comprehensive review of
CRISPR-based gene editing: Mechanisms, challenges, and applications
in cancer therapy. Mol Cancer. 23:92024. View Article : Google Scholar
|
|
60
|
Masarwy R, Breier D, Stotsky-Oterin L,
Ad-El N, Qassem S, Naidu GS, Aitha A, Ezra A, Goldsmith M,
Hazan-Halevy I and Peer D: Targeted CRISPR/Cas9 lipid nanoparticles
elicits therapeutic genome editing in head and neck cancer. Adv Sci
(Weinh). 12:e24110322025. View Article : Google Scholar
|
|
61
|
Xu X, He Y and Liu J: Berberine: A
multifaceted agent for lung cancer treatment-from molecular insight
to clinical applications. Gene. 934:1490212025. View Article : Google Scholar
|
|
62
|
Fujiwara-Tani R, Mori S, Ogata R, Sasaki
R, Ikemoto A, Kishi S, Kondoh M and Kuniyasu H: Claudin-4: A new
molecular target for epithelial cancer therapy. Int J Mol Sci.
24:54942023. View Article : Google Scholar
|
|
63
|
Tojjari A, Idrissi YA and Saeed A:
Emerging targets in gastric and pancreatic cancer: Focus on claudin
18.2. Cancer Lett. 611:2173622024. View Article : Google Scholar
|
|
64
|
Nagaoka Y, Oshiro K, Yoshino Y, Matsunaga
T, Endo S and Ikari A: Activation of the TGF-beta1/EMT signaling
pathway by claudin-1 overexpression reduces doxorubicin sensitivity
in small cell lung cancer SBC-3 cells. Arch Biochem Biophys.
751:1098242024. View Article : Google Scholar
|