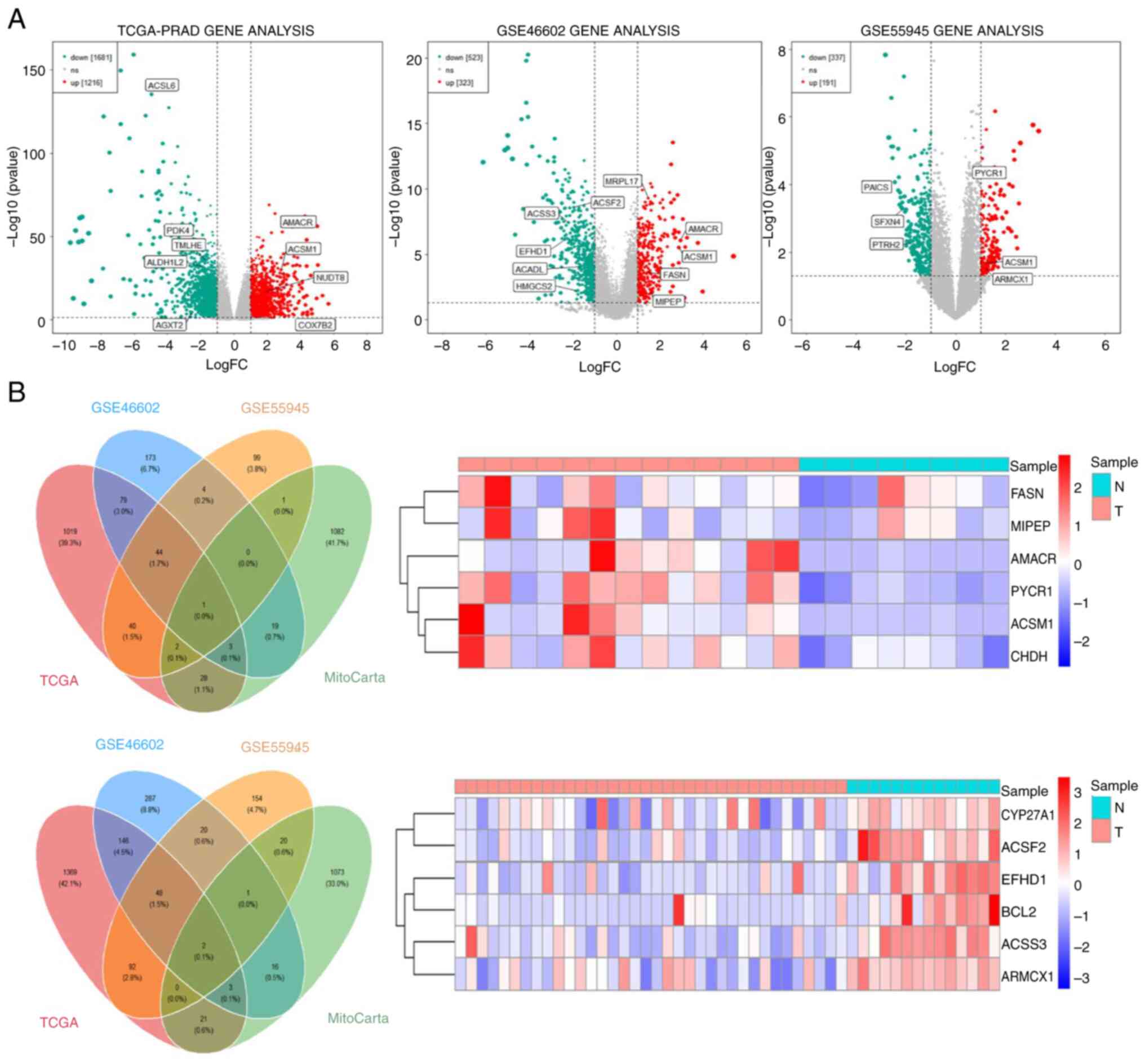
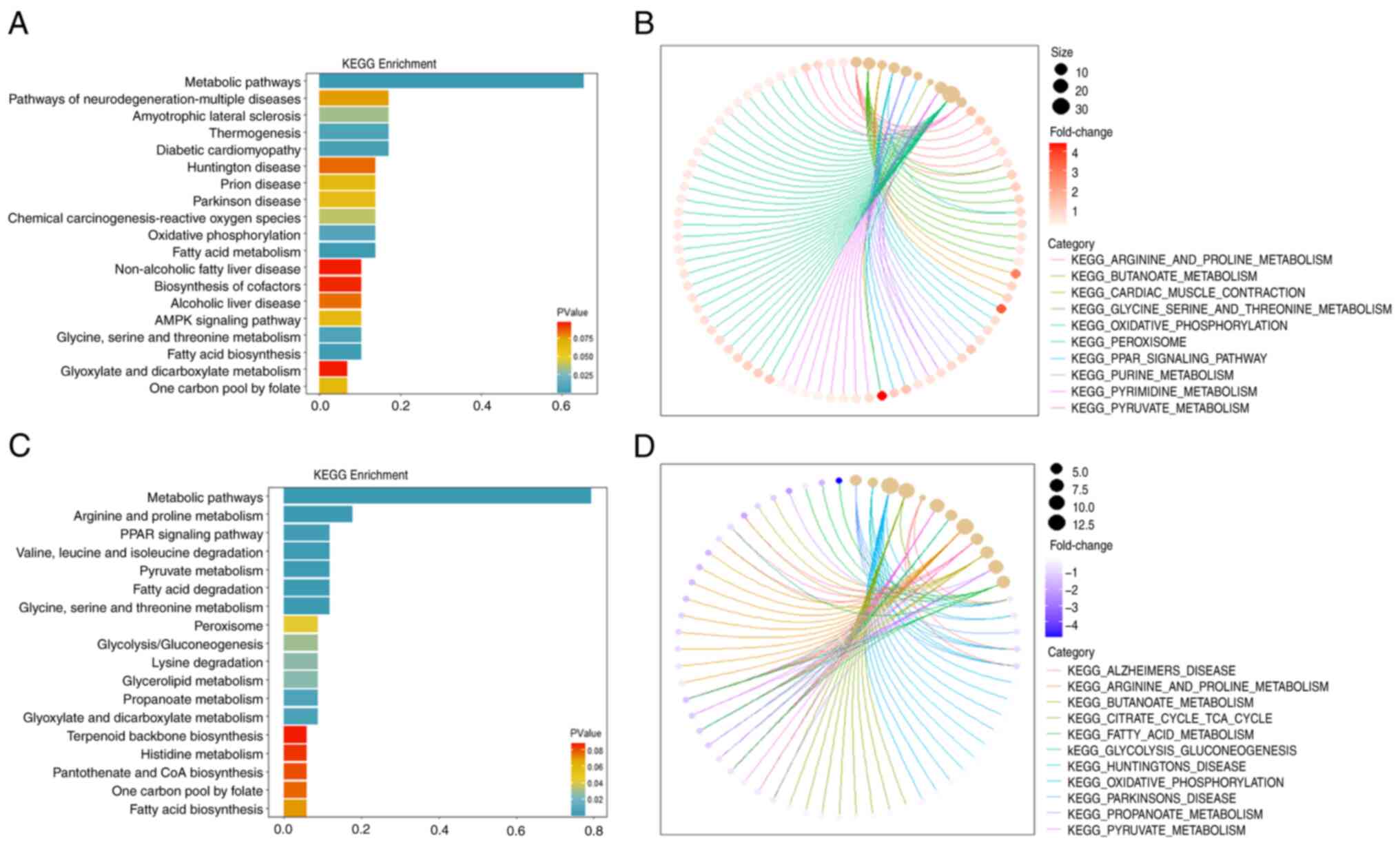
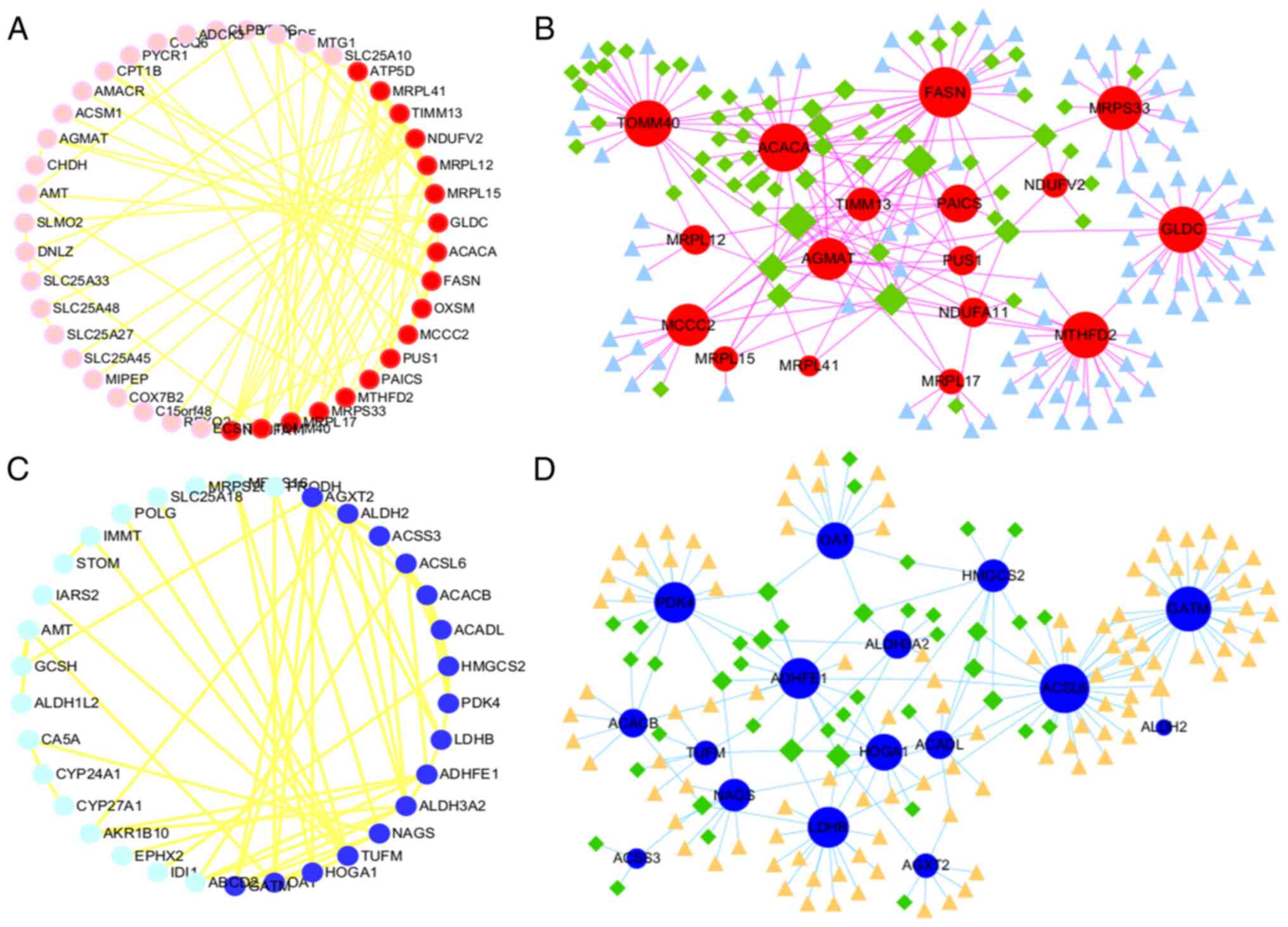
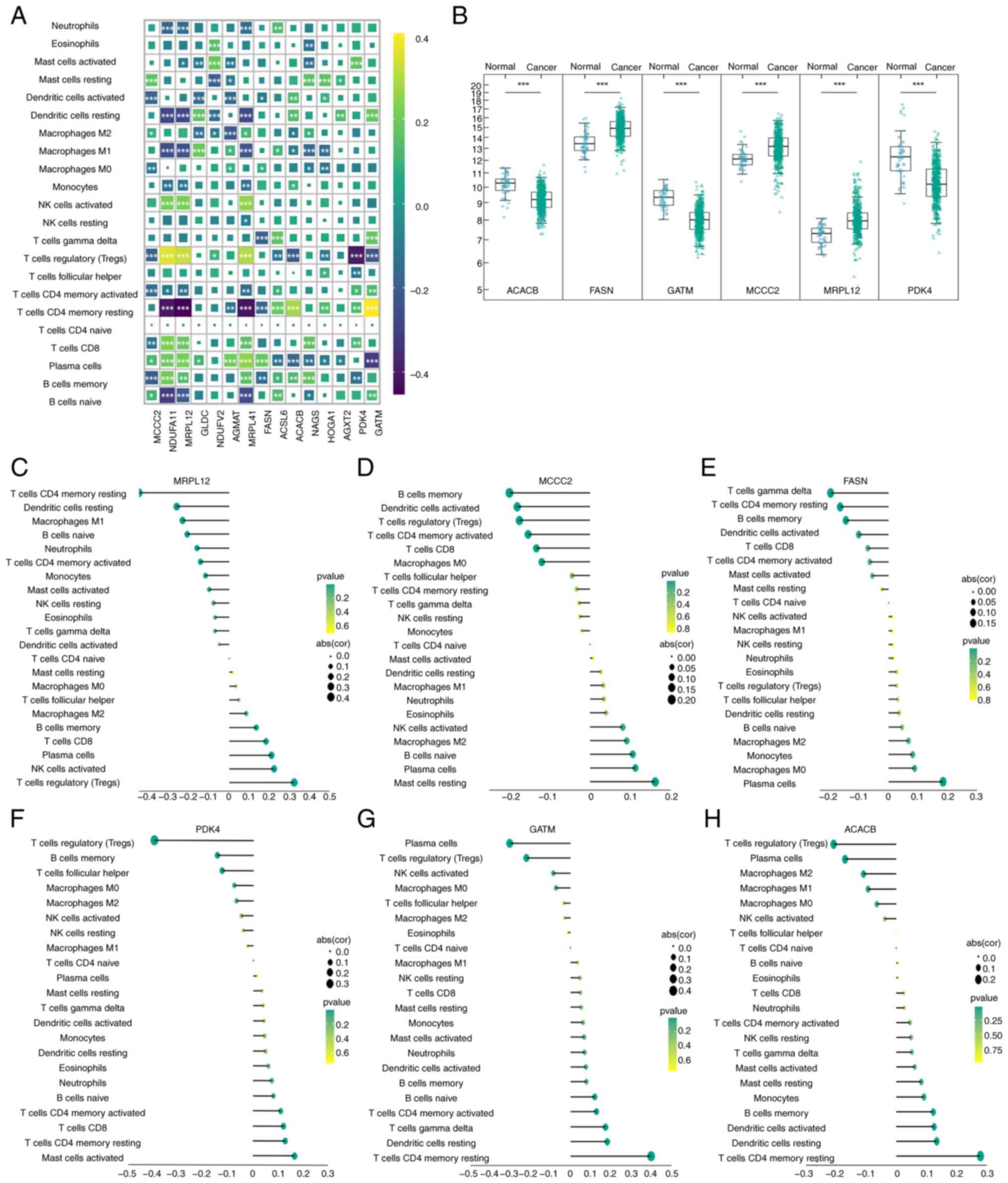
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Moris L, Cumberbatch MG, Van den Broeck T,
Gandaglia G, Fossati N, Kelly B, Pal R, Briers E, Cornford P, De
Santis M, et al: Benefits and risks of primary treatments for
High-risk localized and locally advanced prostate cancer: An
international multidisciplinary systematic review. Eur Urol.
77:614–627. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rebello RJ, Oing C, Knudsen KE, Loeb S,
Johnson DC, Reiter RE, Gillessen S, Van der Kwast T and Bristow RG:
Prostate cancer. Nat Rev Dis Primers. 7:92021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schaeffer E, Srinivas S, Antonarakis ES,
Armstrong AJ, Bekelman JE, Cheng H, D'Amico AV, Davis BJ, Desai N,
Dorff T, et al: NCCN guidelines insights: Prostate cancer, version
1.2021. J Natl Compr Canc Netw. 19:134–143. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Linder S, van der Poel HG, Bergman AM,
Zwart W and Prekovic S: Enzalutamide therapy for advanced prostate
cancer: Efficacy, resistance and beyond. Endocr Relat Cancer.
26:R31–R52. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parida S, Pal I, Parekh A, Thakur B,
Bharti R, Das S and Mandal M: GW627368X inhibits proliferation and
induces apoptosis in cervical cancer by interfering with EP4/EGFR
interactive signaling. Cell Death Dis. 7:e21542016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sathya S, Sudhagar S, Sarathkumar B and
Lakshmi BS: EGFR inhibition by pentacyclic triterpenes exhibit cell
cycle and growth arrest in breast cancer cells. Life Sci. 95:53–62.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang H, Kim B, Park J, Youn H and Youn B:
The Warburg effect on radioresistance: Survival beyond growth.
Biochim Biophys Acta Rev Cancer. 1878:1889882023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen H, Zhou L, Wu X, Li R, Wen J, Sha J
and Wen X: The PI3K/AKT pathway in the pathogenesis of prostate
cancer. Front Biosci (Landmark Ed). 21:1084–1091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumar R, Srinivasan S, Pahari P, Rohr J
and Damodaran C: Activating stress-activated protein
kinase-mediated cell death and inhibiting epidermal growth factor
receptor signaling: A promising therapeutic strategy for prostate
cancer. Mol Cancer Ther. 9:2488–2496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu W, Yang Q, Fung KM, Humphreys MR, Brame
LS, Cao A, Fang YT, Shih PT, Kropp BP and Lin HK: Linking
γ-aminobutyric acid A receptor to epidermal growth factor receptor
pathways activation in human prostate cancer. Mol Cell Endocrinol.
383:69–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hirokawa N, Noda Y, Tanaka Y and Niwa S:
Kinesin superfamily motor proteins and intracellular transport. Nat
Rev Mol Cell Biol. 10:682–696. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Drechsler H and McAinsh AD: Kinesin-12
motors cooperate to suppress microtubule catastrophes and drive the
formation of parallel microtubule bundles. Proc Natl Acad Sci USA.
113:E1635–E1644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao H, Bo Q, Wu Z, Liu Q, Li Y, Zhang N,
Guo H and Shi B: KIF15 promotes bladder cancer proliferation via
the MEK-ERK signaling pathway. Cancer Manag Res. 11:1857–1868.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mortensen MM, Høyer S, Lynnerup AS,
Ørntoft TF, Sørensen KD, Borre M and Dyrskjøt L: Expression
profiling of prostate cancer tissue delineates genes associated
with recurrence after prostatectomy. Sci Rep. 5:160182015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arredouani MS, Lu B, Bhasin M, Eljanne M,
Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA and
Sanda MG: Identification of the transcription factor single-minded
homologue 2 as a potential biomarker and immunotherapy target in
prostate cancer. Clin Cancer Res. 15:5794–5802. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ross-Adams H, Lamb AD, Dunning MJ, Halim
S, Lindberg J, Massie CM, Egevad LA, Russell R, Ramos-Montoya A,
Vowler SL, et al: Integration of copy number and transcriptomics
provides risk stratification in prostate cancer: A discovery and
validation cohort study. EBioMedicine. 2:1133–1144. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hocaoglu H and Sieber M: Mitochondrial
respiratory quiescence: A new model for examining the role of
mitochondrial metabolism in development. Semin Cell Dev Biol.
138:94–103. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boso D, Piga I, Trento C, Minuzzo S, Angi
E, Iommarini L, Lazzarini E, Caporali L, Fiorini C, D'Angelo L, et
al: Pathogenic mitochondrial DNA variants are associated with
response to anti-VEGF therapy in ovarian cancer PDX models. J Exp
Clin Cancer Res. 43:3252024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kopinski PK, Singh LN, Zhang S, Lott MT
and Wallace DC: Mitochondrial DNA variation and cancer. Nat Rev
Cancer. 21:431–445. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim M, Gorelick AN, Vàzquez-García I,
Williams MJ, Salehi S, Shi H, Weiner AC, Ceglia N, Funnell T, Park
T, et al: Single-cell mtDNA dynamics in tumors is driven by
coregulation of nuclear and mitochondrial genomes. Nat Genet.
56:889–899. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zichri SB, Kolusheva S, Shames AI,
Schneiderman EA, Poggio JL, Stein DE, Doubijensky E, Levy D,
Orynbayeva Z and Jelinek R: Mitochondria membrane transformations
in colon and prostate cancer and their biological implications.
Biochim Biophys Acta Biomembr. 1863:1834712021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Papachristodoulou A, Rodriguez-Calero A,
Panja S, Margolskee E, Virk RK, Milner TA, Martina LP, Kim JY, Di
Bernardo M, Williams AB, et al: NKX3.1 Localization to mitochondria
suppresses prostate cancer initiation. Cancer Discov. 11:2316–2333.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mamouni K, Kallifatidis G and Lokeshwar
BL: Targeting mitochondrial metabolism in prostate cancer with
triterpenoids. Int J Mol Sci. 22:24662021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen CL, Lin CY and Kung HJ: Targeting
mitochondrial OXPHOS and their regulatory signals in prostate
cancers. Int J Mol Sci. 22:134352021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han C, Wang Z, Xu Y, Chen S, Han Y, Li L,
Wang M and Jin X: Roles of reactive oxygen species in biological
behaviors of prostate cancer. Biomed Res Int. 2020:12696242020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Li N, Liu Q, Guo J, Pan Q, Cheng
B, Xu J, Dong B, Yang G, Yang B, et al: Antiandrogen treatment
induces stromal cell reprogramming to promote castration resistance
in prostate cancer. Cancer Cell. 41:1345–1362.e9. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guerra F, Guaragnella N, Arbini AA, Bucci
C, Giannattasio S and Moro L: Mitochondrial dysfunction: A novel
potential driver of Epithelial-to-mesenchymal transition in cancer.
Front Oncol. 7:2952017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Civenni G, Bosotti R, Timpanaro A, Vàzquez
R, Merulla J, Pandit S, Rossi S, Albino D, Allegrini S, Mitra A, et
al: Epigenetic control of mitochondrial fission enables
Self-renewal of stem-like tumor cells in human prostate cancer.
Cell Metab. 30:303–318.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jeong SY and Seol DW: The role of
mitochondria in apoptosis. BMB Rep. 41:11–22. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Castilla C, Congregado B, Chinchón D,
Torrubia FJ, Japón MA and Sáez C: Bcl-xL is overexpressed in
hormone-resistant prostate cancer and promotes survival of LNCaP
cells via interaction with proapoptotic Bak. Endocrinology.
147:4960–4967. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krajewska M, Krajewski S, Epstein JI,
Shabaik A, Sauvageot J, Song K, Kitada S and Reed JC:
Immunohistochemical analysis of bcl-2, bax, bcl-X, and mcl-1
expression in prostate cancers. Am J Pathol. 148:1567–1576.
1996.PubMed/NCBI
|
|
34
|
Massie CE, Lynch A, Ramos-Montoya A, Boren
J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, et al:
The androgen receptor fuels prostate cancer by regulating central
metabolism and biosynthesis. EMBO J. 30:2719–2733. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Audet-Walsh É, Dufour CR, Yee T, Zouanat
FZ, Yan M, Kalloghlian G, Vernier M, Caron M, Bourque G, Scarlata
E, et al: Nuclear mTOR acts as a transcriptional integrator of the
androgen signaling pathway in prostate cancer. Genes Dev.
31:1228–1242. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhong C, Long Z, Yang T, Wang S, Zhong W,
Hu F, Teoh JY, Lu J and Mao X: M6A-modified circRBM33 promotes
prostate cancer progression via PDHA1-mediated mitochondrial
respiration regulation and presents a potential target for ARSI
therapy. Int J Biol Sci. 19:1543–1563. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Owari T, Tanaka N, Nakai Y, Miyake M, Anai
S, Kishi S, Mori S, Fujiwara-Tani R, Hojo Y, Mori T, et al:
5-Aminolevulinic acid overcomes hypoxia-induced radiation
resistance by enhancing mitochondrial reactive oxygen species
production in prostate cancer cells. Br J Cancer. 127:350–363.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, Yao L, Wang T, Gu X, Wu Y and Jiang
T: Identification of the mitochondrial protein POLRMT as a
potential therapeutic target of prostate cancer. Cell Death Dis.
14:6652023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ji X, Yang Z, Li C, Zhu S, Zhang Y, Xue F,
Sun S, Fu T, Ding C, Liu Y, et al: Mitochondrial ribosomal protein
L12 potentiates hepatocellular carcinoma by regulating
mitochondrial biogenesis and metabolic reprogramming. Metabolism.
152:1557612023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fan Y, Wang J, Wang Y, Li Y, Wang S, Weng
Y, Yang Q, Chen C, Lin L, Qiu Y, et al: Development and clinical
validation of a novel 5 gene signature based on fatty acid
Metabolism-related genes in oral squamous cell carcinoma. Oxid Med
Cell Longev. 2022:32853932022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Crowell PD, Giafaglione JM, Jones AE,
Nunley NM, Hashimoto T, Delcourt AML, Petcherski A, Agrawal R,
Bernard MJ, Diaz JA, et al: MYC is a regulator of androgen receptor
inhibition-induced metabolic requirements in prostate cancer. Cell
Rep. 42:1132212023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li L, Chao Z, Peng H, Hu Z, Wang Z and
Zeng X: Tumor ABCC4-mediated release of PGE2 induces CD8+ T cell
dysfunction and impairs PD-1 blockade in prostate cancer. Int J
Biol Sci. 20:4424–4437. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Löf C, Sultana N, Goel N, Heron S,
Wahlström G, House A, Holopainen M, Käkelä R and Schleutker J: ANO7
expression in the prostate modulates mitochondrial function and
lipid metabolism. Cell Commun Signal. 23:712025. View Article : Google Scholar : PubMed/NCBI
|