Introduction
According to the latest tumor epidemiological
statistics, prostate cancer (PCa) is still the most common tumor
affecting male reproductive and urinary health, with its incidence
rate and mortality ranking fourth and eighth, respectively
(1). PCa progresses slowly in the
early stages and surgical resection, radiation therapy and androgen
deprivation therapy can effectively halt the progression of
disease. However, approximately one-third of patients with PCa
develop recurrence or metastasis after radical surgery. These
patients can be treated with androgen-deprivation therapy, also
known as castration therapy, which involves the administration of
gonadotropin-releasing hormone agonists, or with surgical
castration (2). Unfortunately, most
patients develop disease progression again ~18 months after
treatment and are no longer affected by androgen levels. Instead,
the disease evolves into a more aggressive form of
castration-resistant PCa, either metastatic castration-resistant
disease or non-metastatic castration-resistant disease (3). The first-line treatment for
castration-resistant PCa mainly includes the chemotherapy drug
docetaxel along with novel endocrine drugs, such as abiraterone or
enzalutamide (4). Although the
early treatment response is good, drug resistance is still
inevitable in the later stages and no effective treatment exists
for these patients at present. Therefore, it is important to
explore the molecular mechanisms of PCa progression (5).
Mitochondria are responsible for the synthesis of
intracellular bioenergetics. In addition to the synthesis of ATP,
mitochondria can also synthesize macromolecular metabolic
precursors, such as lipids, proteins, DNA and RNA. Mitochondria can
also remove or use metabolic waste, such as reactive oxygen species
(ROS) and gases (6). Under normal
physiological conditions, mitochondria are important cell pressure
sensors and can coordinate cell adaptation to various stressors,
such as nutrient deprivation, oxidative stress, DNA damage and
endoplasmic reticulum stress. Therefore, mitochondria can maintain
the growth and survival of tumor cells in harsh environments,
including nutrient depletion, hypoxia and tumor treatment, which is
a key factor in promoting tumor progression (7). An early study found that tumor cells
still tended to undergo glycolysis under aerobic conditions, known
as the Warburg effect, indicating that mitochondrial respiratory
defects may be the fundamental cause of tumor development. In
recent years, mitochondrial function and tumor metabolism
reprogramming have received increasing attention (8). Researchers have found that the Warburg
effect is not a characteristic of all cancer cells. Although
damaged mitochondria may drive the Warburg effect in some cases, a
number of tumor cells that exhibit Warburg metabolism also have
complete mitochondrial respiration and some tumor subtypes also
depend on mitochondrial respiration (7,9).
Biosynthesis and other functions of mitochondria are typically
upregulated in cancer and metabolic reprogramming has become an
important tumor marker (10). The
metabolic phenotype of prostate epithelial cells is unique and the
stages of tumor development and progression from prostatic
intraepithelial neoplasia to metastasis are different. Normal
prostate epithelial cells are highly dependent on glycolysis, but
during tumor progression, PCa cells gradually reactivate
mitochondrial oxidative phosphorylation while increasing glucose
metabolism and reducing citrate production (11,12).
In PCa, androgen receptors (ARs) also participate in cellular
metabolic reprogramming, which includes aerobic glycolysis,
mitochondrial respiration and de novo fat generation,
thereby meeting the metabolic and biosynthetic needs of PCa cells
(13,14).
In the present study, differentially expressed genes
(DEGs) in PCa were first identified by analyzing three datasets:
The Cancer Genome Atlas (TCGA)-prostate adenocarcinoma (PRAD),
GSE46602 and GSE55945. This initial step focused on screening
general DEGs (upregulated or downregulated) between PCa and
adjacent normal tissues across the datasets, without restricting to
mitochondrial relevance. To narrow down the identified DEGs to
mitochondria-associated candidates, the first intersection was
performed by overlapping these general PCa DEGs with the full list
of human mitochondrial protein-coding genes from the MitoCarta3.0
database. This step yielded a preliminary set of differentially
expressed mitochondrial-related genes (DeMRGs) but still included
all genes that met both ‘differential expression in PCa’ and
‘mitochondrial localization’ criteria. To further refine and
prioritize high-confidence targets, a second targeted intersection
was conducted: The upregulated subset of the initially obtained
DeMRGs was re-intersected with the MitoCarta3.0 mitochondrial gene
list. This distinct step aimed to exclude potential false positives
and focus specifically on consistently upregulated DeMRGs, a subset
with stronger relevance to PCa progression as upregulated
mitochondrial genes are more likely to drive pro-tumor metabolic
reprogramming. Subsequently, Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analyses were performed on
these DeMRGs. The transcription factors and microRNAs (miRNAs)
associated with the DEGs were also analyzed to provide a basis for
further mechanistic studies. Furthermore, to explore the role of
mitochondrial-related genes in PCa, the present study analyzed the
correlation between key mitochondrial genes and immune infiltration
through immune infiltration analysis. Additionally, it investigated
the association of these genes with mitochondrial respiration and
their relationship with mitochondrial metabolic pathways in PCa,
aiming to identify potential mitochondrial hub genes involved in
PCa progression.
Materials and methods
Cell culture
The PCa cell lines (Du145, PC-3 and LNcap) and
normal prostate cells (RWPE-1) purchased from American Type Culture
Collection) were cultured in medium containing 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C constant
temperature and 5% CO2. Du145 cells were maintained in
MEM (Biotecnómica), whereas PC-3 and LNcap were maintained in RPMI
(Biotecnómica) and RWPE-1 was maintained in K-SFM
(Biotecnómica).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the RWPE-1, PC-3, LNcap
and Du145 cell lines using RNAiso Plus reagent (Thermo Fisher
Scientific, Inc.). cDNA was synthesized using a PrimeScript™ RT
reagent kit (Takara Biotechnology Co., Ltd) at 42°C for 15 min and
85°C for 5 sec. Two-step qPCR was performed using SYBR Green
Reagent (Takara Biotechnology Co., Ltd.), C1000 Touch Thermal
Cycler and the CFX96 Real-Time System according to the
manufacturer's instructions. The primer sequences are listed in
Table I. The results were
calculated using the 2−ΔΔCq method, and the data are
expressed as a ratio of the control gene GAPDH (15).
 | Table I.Sequences of the reverse
transcription-quantitative PCR primers. |
Table I.
Sequences of the reverse
transcription-quantitative PCR primers.
| Genes | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| ACACB |
AGATCGCCTCCACCGTTGTC |
CTGTCACCTCACCTGTCTTCTACT |
| PDK4 |
TGAGTGTTCAAGGATGCTCTGTG |
GGTCTGGTTGGTTAAGTGTAGCA |
| GATM |
TCATTGGACCTGGTATTGTGCTTTC |
GTAATGAGGAGGTTGTGGTTAGT |
|
|
| AGG |
| MCCC2 |
AGGTGGCATTATTACAGGCATTGG |
CGGATGATGGGTCACTGACACTT |
| MRPL12 |
ACATCGCCAGCCTCACTCTC |
GGCTACCCACCACACTACAGA |
| FASN |
CAGCGGCAAGCGTGTGATG |
GTTGACCTGCGACCTCCTCC |
| GAPDH |
AATGGGCAGCCGTTAGGAA |
GAGACTAAACCAGCATAACCCG |
Public data acquisition and
preprocessing
RNA-sequencing (RNA-seq) data for patients with PCa
were derived from the TCGA-PRAD dataset (TCGA-PRAD; http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga),
which includes 502 cancer tissues and 52 adjacent tissues, and from
two datasets provided by the National Center for Biotechnology
Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo): GSE46602
(consisting of 36 cancer tissues and 14 adjacent tissues) and
GSE55945 (including sequencing data from 13 cancer tissues and 8
adjacent tissues) (16,17). In addition, the GSE70770 dataset was
used as an independent validation cohort, which contains RNA-seq
data for 220 PCa tissues and 73 adjacent non-cancerous tissues
(18). Using R v4.2.2 (https://cran.r-project.org/), the array datasets were
normalized using the limma package v3.58.1 and high-throughput
sequencing count data were normalized using DESeq2 (version 3.20.0)
to generate standardized matrices. Specifically, the DESeq function
was applied for normalization, which accounts for library size
differences and adjusts for gene-wise dispersion estimates. For
genes with multiple transcript IDs, the ID with the highest average
expression was selected. The original gene expression data
underwent log2 transformation and quantile normalization for
subsequent downstream analyses.
Identification and functional
enrichment analysis of DeMRGs
The DEGs found in PCa and adjacent tissues were
analyzed using the limma and DESeq2 packages, with the following
selection criteria: log2|fold change|≥1 and P<0.05.
DeMRGs were identified by intersecting these DEGs (from TCGA-PRAD,
GSE46602 and GSE55945) with 1,136 human mitochondrial
protein-coding genes from MitoCarta3.0 (https://www.broadinstitute.org/mitocarta/mitocarta30-inventory-mammalian-mitochondrial-proteins-and-pathways),
visualized via heat maps, Venn diagrams and volcano plots. GO
(http://geneontology.org) and KEGG (https://www.genome.jp/kegg/kegg1.html)
enrichment analyses of DeMRGs were performed using R's
clusterProfiler package (v4.10.1). Gene Set Enrichment Analysis
(GSEA; http://www.broadinstitute.org/gsea/) with KEGG gene
sets (c2.cp.KEGG.v7.4.symbols.gmt from MSigDB; http://software.broadinstitute.org/gsea/msigdb/)
was conducted, with significance defined as P<0.05 and false
discovery rate <0.05. The transcription factors and interacting
miRNAs of the identified DeMRGs were predicted using the online
platform NetworkAnalyst 3.0 (https://www.networkanalyst.ca/) and visualized with
Cytoscape software.
Protein-protein interaction (PPI)
analysis and hub gene extraction
In the present study, PPI networks of DeMRGs were
identified to determine the differences between PCa and its
adjacent tissues from the perspective of protein interactions.
First, the STRING online analysis tool (https://www.string-db.org/) was used to generate the
PPI network of DeMRGs. Then, the PPI network was analyzed using
Cytoscape software (https://cytoscape.org; CytoHubba plugin and DMNC;
v3.10.0) and screened out genes with a degree value ≥4 as core
genes for further research. The network type was full STRING. In
total, six core data sources were integrated to ensure interaction
reliability, including ‘experiments’, ‘databases’, ‘co-expression’,
‘neighborhood’, ‘gene fusion’ and ‘co-occurrence’. The interaction
score was set to a confidence threshold of 0.4.
Hub genes and PCa immune infiltration
analysis
The TCGA-PRAD differential gene volcano plot was
generated using R's ggplot2 (https://CRAN.R-project.org/package=ggplot2; v3.5.1).
Core DeMRGs in TCGA-PRAD were obtained by intersecting TCGA-PRAD
DEGs with core genes. For immune infiltration analysis, R was used
to convert count data to transcripts per million (TPM), followed by
the use of CIBERSORT (http://cibersort.stanford.edu; v3.15.0) to estimate 22
immune cell proportions (P<0.05 was considered reliable). The
correlation between each hub gene and the 22 immune cell types was
tested using Spearman's rank correlation analysis and visualized as
lollipop plots. GSEA with KEGG analysis were used for functional
enrichment. Using the GSE70770 dataset, the immune infiltration
scores for 22 immune cell subsets were calculated via single-sample
GSEA (ssGSEA) implemented through the R package Gene Set Variation
Analysis (GSVA). Specifically, the GSVA package was used to run
ssGSEA, which quantifies the enrichment level of each immune
cell-associated gene set within individual samples of GSE70770,
thereby generating the immune infiltration scores for each cell
type. Paired t-tests were used to compare the immune scores of PCa
and adjacent tissues, visualized as box plots with R's ggplot2.
Immune cell composition was also plotted with R's ggplot2. Spearman
correlation tests of genes and immune cells were visualized as heat
maps using R's ComplexHeatmap (v1.0.12). To ensure the reliability
of the observed correlations across different computational tools,
R's psych package (v2.4.3) was used to compute correlation
coefficients and P-values of core DeMRGs and immune cells; box
plots showed the differences in the immune scores of high/low
expression (samples with expression levels greater than or equal to
the median were defined as the high expression subgroup, while
those with expression levels below the median were designated as
the low expression subgroup.) subgroups. Cluster analysis heat maps
(using the pheatmap v1.0.12 function in R) illustrated the
relationships between hub gene expression and immune cells. The
correlations between the 6 core genes and immune cells were
calculated using the R package Hmisc (v5.1–1).
Bioinformatics evaluation of the PCa
mitochondrial respiratory chain and mitochondrial metabolism
Using MitoCarta3.0 and TCGA-PRAD data, paired
t-tests were conducted to identify genes with significant
differences (P<0.05) in oxidative respiratory chain complexes in
cancer cells and adjacent tissues, visualized via R's ggplot2.
Genes that were |statistical|>6 were correlated with the
CIBERSORT immune infiltration results, with correlation heat maps
generated using ggplot2. Spearman rank analysis was conducted to
assess the correlations between the 6 hub genes and oxidative
respiratory chain complex genes (|statistical|>4); these
correlations were visualized using ggplot2. Linear regression was
used to evaluate the pairwise relationships between the 4 selected
hub genes and oxidative phosphorylation (OXPHOS) genes. R's ggplot2
(stat_compare_means) was used to perform paired t-tests to analyze
the expression of damage associated molecular patterns DAMPs genes
(such as BCL2 and calreticulin) and immune receptor-related genes
(such as toll-like receptor 2 and cyclic GMP-AMP synthase). Mantel
tests and Pearson correlations were used to calculate the
correlations between the 4 hub genes, mitochondrial metabolism and
DAMPs.
Statistical analyses
SPSS 20.0 (IBM Corp.) was utilized to perform the
statistical analyses. Data are shown as the mean ± standard error
of the mean according to the results of three independent repeated
experiments. One-way analysis of variance (ANOVA) was employed to
analyze differences in the expression levels of the 6 core DeMRGs
[acetyl-CoA carboxylase β (ACACB), pyruvate dehydrogenase kinase 4
(PDK4), glycine amidinotransferase (GATM), methylcrotonyl-CoA
carboxylase subunit 2 (MCCC2), mitochondrial ribosomal protein L12
(MRPL12) and fatty acid synthase (FASN)] in the four different cell
lines, which were detected by RT-qPCR from three independent
repeated experiments. Following the one-way ANOVA, the Tukey's
honestly significant difference test was used as the post hoc test
to perform pairwise comparisons between groups, ensuring the
accurate identification of specific differences in gene expression
levels among different cell lines and between different genes.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of DeMRGs in PCa and
enrichment analyses
TCGA-PRAD contains RNA-seq data for 554 tissue
samples, including 502 cancer tissues and 52 adjacent tissues.
Differential analysis identified 1,681 genes that were
significantly downregulated and 1,216 genes that were significantly
upregulated in PCa. The DEGs were intersected with human
mitochondrial genes in the MitoCarta3.0 database to obtain 60
intersecting genes, including 34 significantly upregulated and 26
significantly downregulated genes. The top 9 DeMRGs are shown in
Fig. 1A. Similarly, 523 genes were
significantly downregulated and 323 genes were significantly
upregulated in PCa in GSE46606 and 337 genes were significantly
downregulated and 191 genes were significantly upregulated in PCa
in GSE55945. After taking the intersection of these DEGs with the
MitoCarta3.0 database, 23 significantly upregulated and 22
significantly downregulated genes were identified in GSE46602,
while 4 significantly upregulated and 23 significantly
downregulated genes were identified in GSE55945 (Fig. 1A). After conducting a differential
analysis using the training set (TCGA-PRAD, GSE 46606 and
GSE55945), To specifically screen for high-confidence upregulated
mitochondria-related genes in PCa, the upregulated subset of the
initially identified 60 DeMRGs (from intersecting general PCa DEGs
with MitoCarta3.0 mitochondrial genes) was further intersected with
the human mitochondrial genes in the MitoCarta3.0 database. This
targeted refinement yielded 6 core upregulated mitochondrial genes,
which were selected for subsequent functional and correlation
analyses. The 6 mitochondrial genes in GSE55945 and GSE46602 are
presented as heat maps (Fig.
1B).
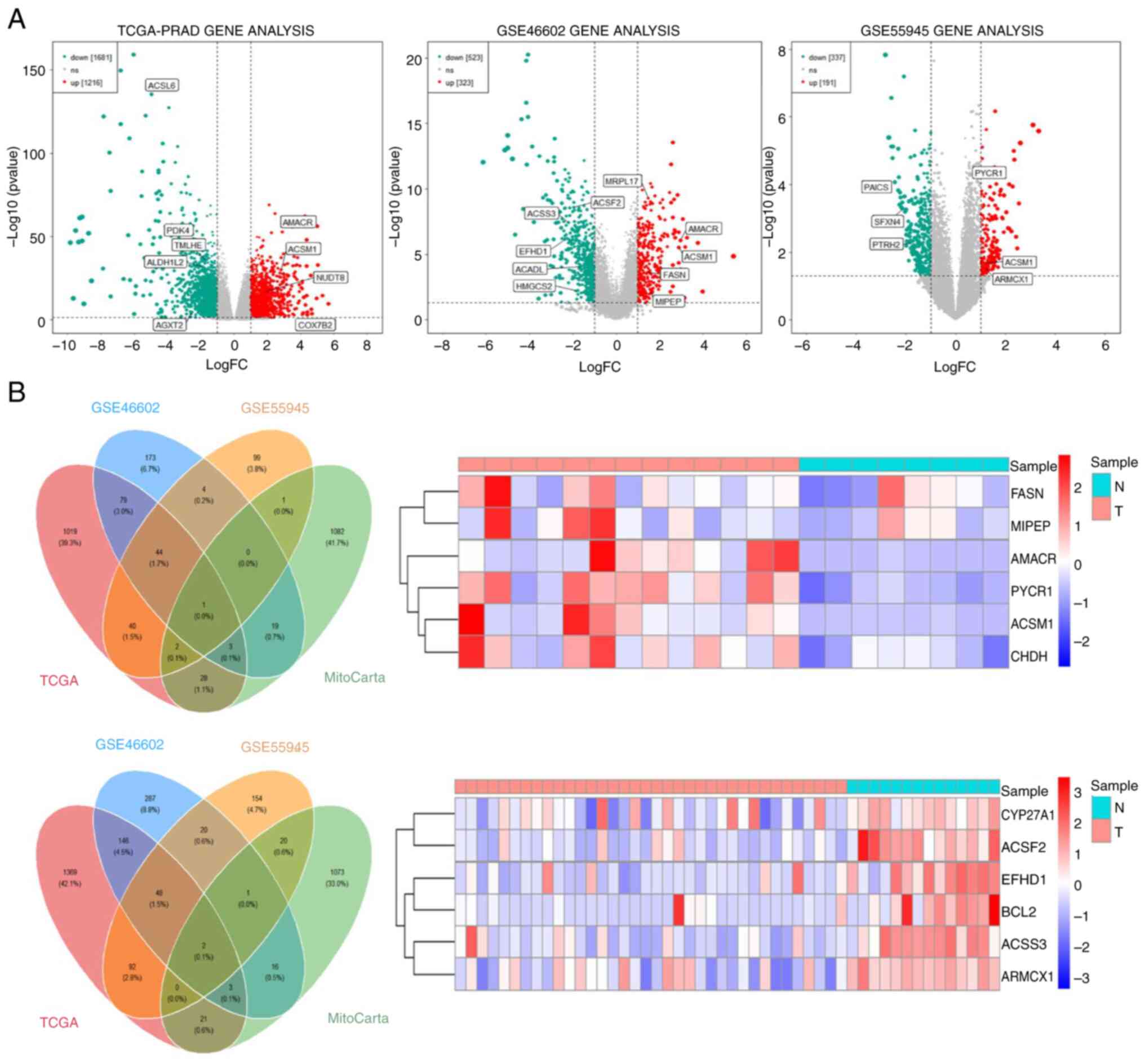 | Figure 1.Differential expression of mRNA in
mitochondria between prostate cancer and normal prostate tissue.
(A) Differential analysis was conducted on three training sets
(TCGA-PRAD, GSE46602 and GSE55945). (B) The left sides are the Venn
diagram, and the right side is the expression heatmap. The top
images are for GSE55945 and the bottom images are for GSE46602;
TCGA, The Cancer Genome Atlas; PRAD, prostate adenocarcinoma; ns,
no significance; FASN, fatty acid synthase; MIPEP, mitochondrial
intermediate peptidase; AMACR, α-methylacyl-CoA racemase; PYCR1,
pyrroline-5-carboxylate reductase 1; ACSM1, acyl-CoA synthetase
medium chain family member 1; CHDH, choline dehydrogenase; N,
normal; T, tumor; FC, fold change. |
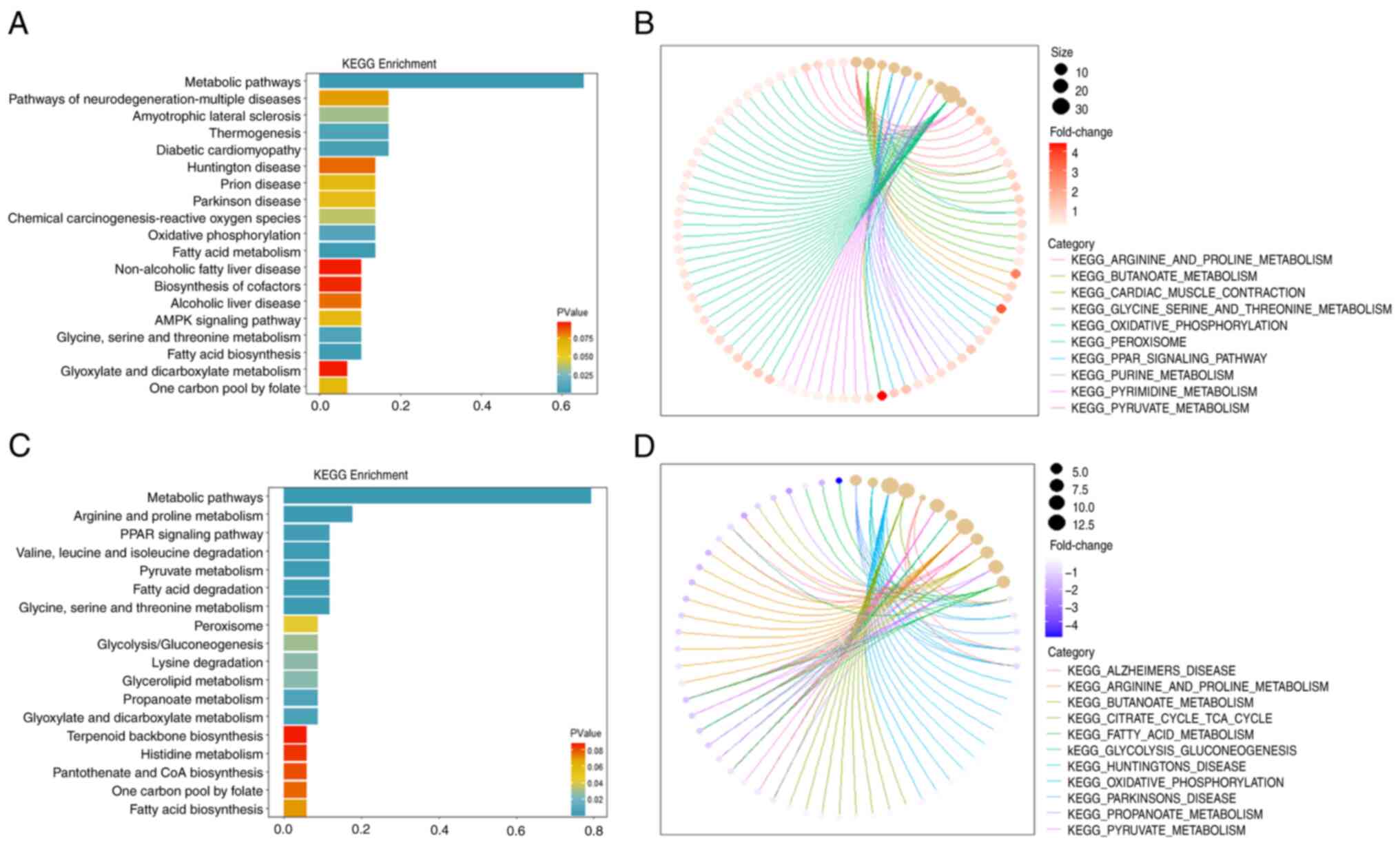
GO analysis was performed on all significantly
upregulated DeMRGs (54 genes) in the three datasets, including in
terms of molecular function, biological process and cell component.
This analysis showed that the DeMRGs significantly upregulated in
PCa were related to ‘nucleotide phosphate metabolic process’,
‘oxidative phosphorylation’, ‘mitochondrial transport’,
‘mitochondrial transmembrane transport’ and ‘ribosomes’ (Fig. S1A). By contrast, the 63
significantly downregulated DeMRGs in PCa were associated with
‘small molecule catabolic process’, ‘organic acid biosynthetic
process’ and ‘mitochondrial gene expression’ (Fig. S1B).
Correlation analysis of core
genes
KEGG analysis and GSEA showed that the upregulated
DeMRGs were enriched in multiple metabolic pathways and were
associated with multiple diseases, including ‘Huntington’ disease’,
‘Parkinson disease’ and ‘prion diseases’ (Fig. 2A and B). Similarly, the KEGG and
GSEA results showed that the downregulated DeMRGs were enriched in
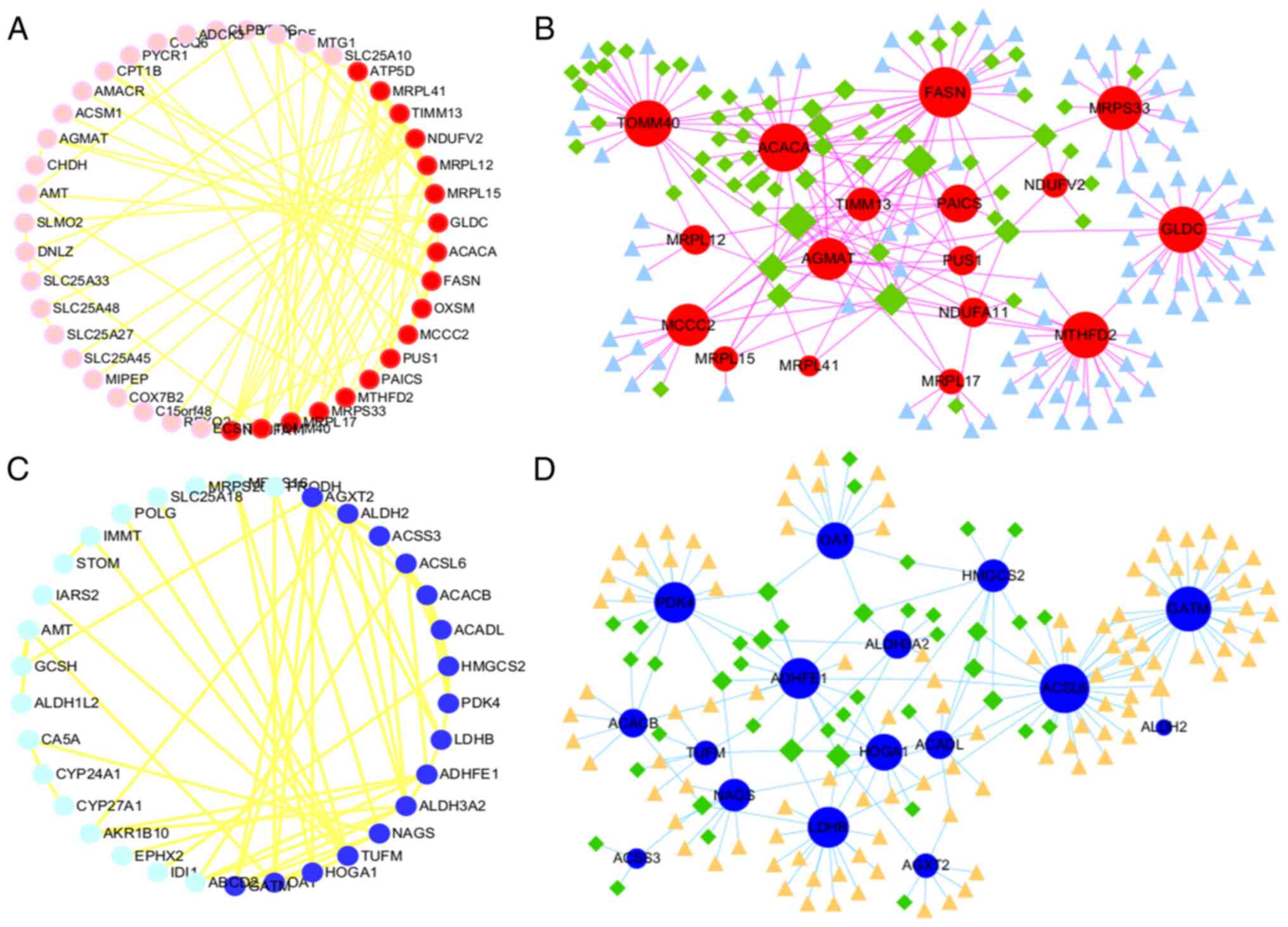
multiple metabolic pathways and ‘PPAR signaling pathway’ (Fig. 2C and D). A PPI network was
constructed using the STRING website to analyze the 54 upregulated
genes and Cytoscape analysis was used to screen out 18 core genes
with a degree value >4 (Fig.
3A). Subsequently, NetworkAnalyst 3.0 was used to predict the
transcription factors and interacting miRNAs of these 18 genes
(Fig. 3B). Similarly, the 63
downregulated genes were used to construct a PPI network through
the STRING website and 16 genes were screened out based on a degree
value >4 (Fig. 3C). The
predicted transcription factors and interacting miRNAs of these 16
genes were also screened out (Fig.
3D). The results collectively suggest that the core genes
selected in the present study are related to multiple transcription
factors and miRNAs.
Correlation between core DeMRGs and
immune infiltration in PCa
The TPM expression matrix of PCa tissue (502 cases)
was used to calculate its immune score using the CIBERSORT
deconvolution algorithm. The genes examined are the intersection of
the 60 DeMRGs in TCGA-PRAD and the 34 core upregulated genes
selected earlier, resulting in 15 intersection genes, which were
the core DeMRGs of TCGA-PRAD. Subsequently, using the TPM
expression matrix of PCa tissues (502 cases) from TCGA-PRAD
dataset, the proportions of 22 immune cell subsets were first
estimated via the CIBERSORT deconvolution algorithm to generate
immune infiltration profiles. On this basis, a correlation analysis
was further performed between these 15 intersection genes and the
aforementioned immune infiltration results (Fig. 4A). It can be observed that there are
significant differences in the positive and negative correlations
between different immune cells and specific genes. These
differences help to reveal the association between immune cell
functions and gene expression, and provide references for research
on tumor immune regulation, gene functions and other related
fields. The 6 genes most closely related to immunity were screened
out, as shown in Fig. 4B. The
differences in the expression of the 6 genes in PCa and adjacent
tissues were significant. ACACB, PDK4 and GATM were downregulated
in PCa, while MCCC2, MRPL12 and FASN were upregulated in PCa.
Lollipop plots of the 6 genes were constructed and the results
showed a significant negative correlation between MRPL12 and
quiescent CD4 memory T cells, memory B cells and M1 macrophage
infiltration, while the trend was the opposite for Tregs and
NK-activated cells (P<0.05; Fig
4C). MCCC2 was significantly negatively correlated with the
infiltration of memory B cells, activated dendritic cells and Tregs
cells, while the trend was the opposite for plasma cells and
stationary mast cells (P<0.05; Fig.
4D). A significant negative correlation was found between FASN
and γ-δ T cells, quiescent CD4 memory T cells and memory B cells,
while the opposite was found for plasma cells and M0 macrophages
(P<0.05; Fig. 4E). PDK4 was
significantly negatively correlated with Tregs, memory B cells and
follicular helper T cells, while it was positively correlated with
activated mast cells, resting CD4+ memory T cells and
CD8 T cells (Fig. 4F). GATM was
significantly negatively correlated with plasma cells and Tregs,
while it was positively correlated with resting CD4+
memory T cells, resting dendritic cells and γ-δ T cells (Fig. 4G). ACACB was significantly
negatively correlated with Tregs, plasma cells and M2 macrophages,
while it was positively correlated with resting CD4+
memory T cells, resting dendritic cells and activated dendritic
cells (Fig. 4H). Subsequently, a
single-gene GSEA was conducted; the results showed that MCCC2,
MRPL12 and FASN were negatively correlated with ‘KEGG_APOPTOSIS’,
‘KEGG_B_CELL_RECEPTOR_SIGNALING_PATHWAY’ and
‘KEGG_INTESINAL_IMMUNE_NETWORK_FOR_IGA_PRODUCTION’. However, ACACB,
PDK4 and GATM showed the opposite results (Fig. S2). These results imply a close
association between the core DeMRGs and infiltrating immune
cells.
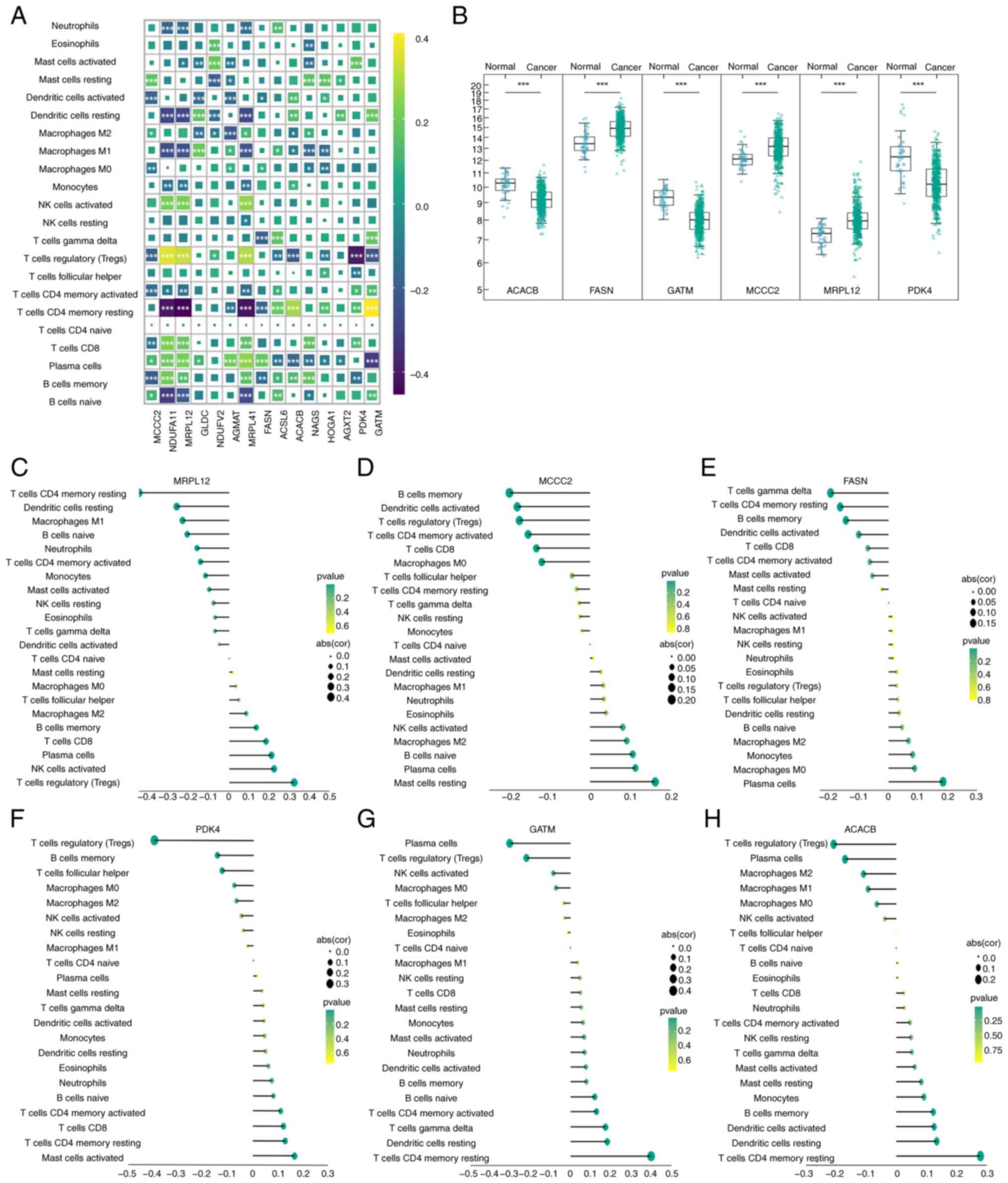 | Figure 4.Relationship between core
differential mitochondrial mRNA and tumor immune infiltration in
PCa and normal prostate tissue. (A) TCGA-PRAD dataset was used to
calculate the immune score of the transcripts per million
expression matrix of PCa tissue (502 cases), which is shown as a
heatmap. The genes on the x-axis are the core genes selected from
TCGA-PRAD. The color of the heatmap represents the correlation
coefficient. The larger the box is, the more statistically
significant the correlation. (B) The box plot shows the 6 genes
most closely related to immunity, with P-values determined by
paired t-test. The R package ggplot2 was used to examine the
correlation between hub genes and immune cells, with the results
presented as lollipop plots. The relationship between (C) MRPL12,
(D) MCCC2, (E) FASN, (F) PDK4, (G) GTAM and (H) ACACB with immune
cell infiltration. The size of the points represents the absolute
value of the correlation coefficient and the larger the points, the
more correlated they are. The greener the point, the smaller the
P-value. *P<0.05, **P<0.01, ***P<0.001. PCa, prostate
cancer; TCGA, The Cancer Genome Atlas; PRAD, prostate
adenocarcinoma; ACACB, acetyl-CoA carboxylase β; PDK4, pyruvate
dehydrogenase kinase 4; GATM, glycine amidinotransferase; MCCC2,
methylcrotonyl-CoA carboxylase subunit 2; MRPL12, mitochondrial
ribosomal protein L12; FASN, fatty acid synthase. |
Validation of the correlation between
the 6 characteristic genes and immune cell infiltration
GSE70770 was used for validation, which included a
total of 293 tissue samples: 220 PCa tissues and 73 adjacent cancer
samples. The results showed that in PCa, the 6 genes were
significantly downregulated in activated B cells, effector CD8
memory T cells, immature dendritic cells, mast cells and NK cells
of PCa tissues compared with the adjacent cancer tissues (Fig. 5A). Next, the proportion of 26 immune
cells in PCa and adjacent tissues were analyzed using the GSE70770
dataset (Fig. 5B). There were
significant stratified differences in the proportions of different
types of immune cells (such as activated B cells, activated CD4/CD8
T cells, macrophages and neutrophils) between the two types of
tissues. These observed differences are suggestive of immune cell
remodeling within the tumor microenvironment, thereby providing
critical insights that hold value for investigating the molecular
mechanisms underlying tumor immune escape and identifying candidate
targets for immune-based therapeutic interventions. The correlation
between the 6 core genes and immune cells were calculated using the
Hmisc R package and displayed it in the form of a heat map
(Fig. 5C). The results showed that
these genes were correlated (P<0.001) with NK cells, NK-T cells,
B cells, dendritic cells, mast cells and CD8 memory T cells. The
correlation coefficients and P-values for these 6 genes and the
immune cells were calculated separately. The results showed that
high expression of MCCC2, MRPL12 and FASN was associated with low
expression of activated B cells, CD8 memory T cells, NK cells and
NK-T cells. In addition, low expression of PDK4, ACACB and GATM was
associated with low expression of activated B cells, CD8 memory T
cells, NK cells and NK-T cells; the strongest immune correlation
was with GATM (Fig. 6A). A heat map
was used to visually display the differences between cancer and the
adjacent immune cells, as well as the relationship between the
expression levels of these 6 core genes and immune cells. The
results showed that with low expression of PDK4 and GATM, memory B
cells and CD4 memory T cells were highly expressed, while CD8
memory T cells showed lower expression levels (Fig. 6B).
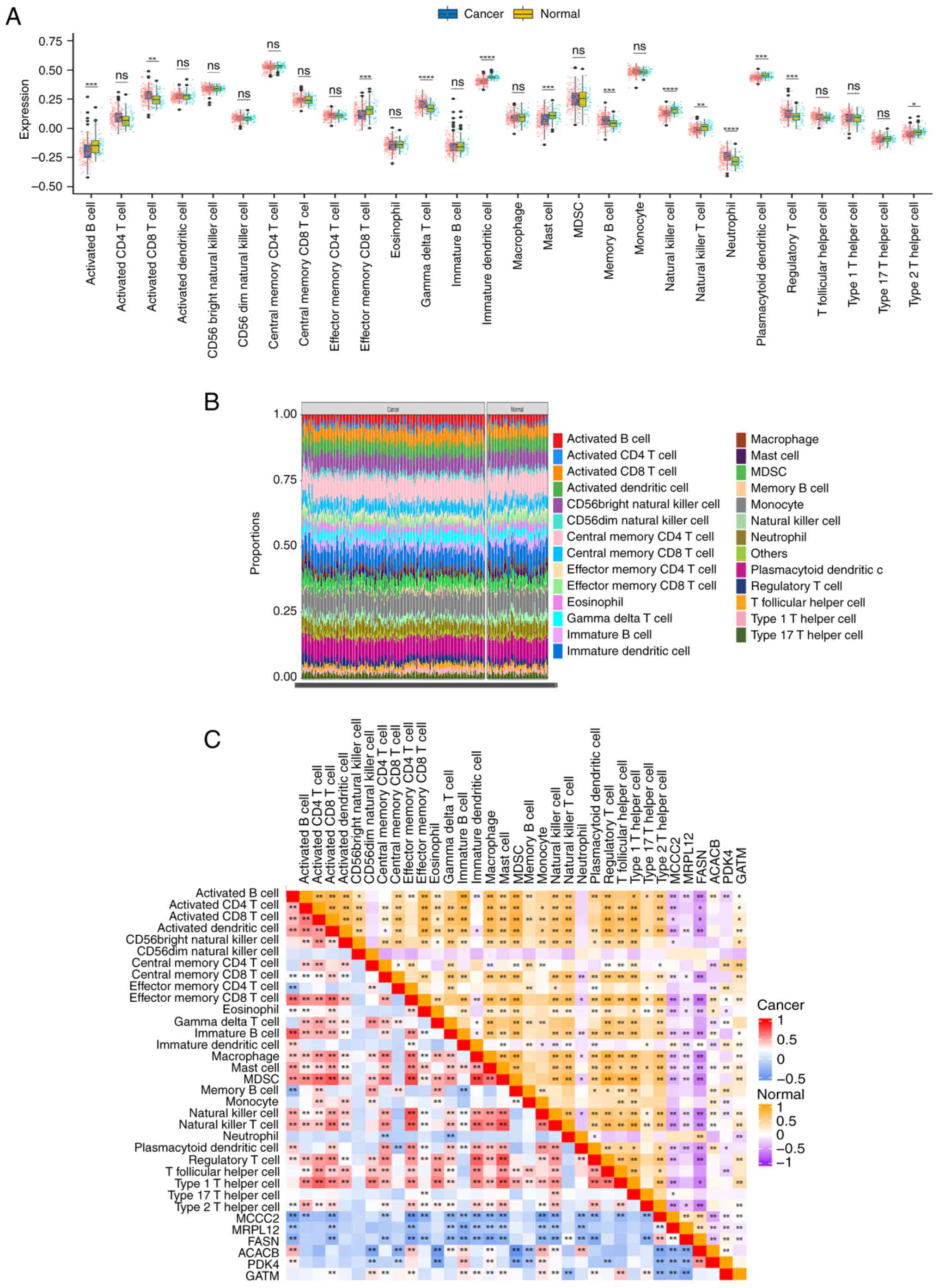 | Figure 5.Validation of the relationship
between MCCC2, MRPL12, FASN, PDK4, ACACB and GATM genes and tumor
immunity in prostate cancer and normal prostate tissue. (A)
Validation of the relationship between the MCCC2, MRPL12, FASN,
PDK4, ACACB and GATM genes and tumor immunity in PCa and normal
prostate tissue. The data is sourced from the validation dataset
GSE70770. (B) The proportion of 26 immune cells in PCa and adjacent
tissues in the validation dataset GSE70770. (C) The correlation
between 6 core genes and immune cells in the validation dataset
GSE70770. The darker the color, the stronger the correlation. The
upper right corner shows the correlation between these genes and
immune cells, while the lower left corner shows the correlation
between these genes and immune cells in PCa. *P<0.05,
**P<0.01, ***P<0.001; ****P<0.0001. ACACB, acetyl-CoA
carboxylase β; PDK4, pyruvate dehydrogenase kinase 4; GATM, glycine
amidinotransferase; MCCC2, methylcrotonyl-CoA carboxylase subunit
2; MRPL12, mitochondrial ribosomal protein L12; FASN, fatty acid
synthase; PCa, prostate cancer. |
 | Figure 6.Validation of the relationship
between the MCCC2, MRPL12, FASN, PDK4, ACACB and GATMM genes and
immune cells in prostate cancer and normal prostate tissue. (A) The
relationship between the core MCCC2, MRPL12, FASN, PDK4, ACACB and
GATMM genes and immune cells, using the validation dataset
GSE70770. (B) The differences between cancer and adjacent immune
cells, as well as the relationship between the expression levels of
the 6 core genes and immune cells, using the validation dataset
GSE70770. *P<0.05, **P<0.01, ***P<0.001; ****P<0.0001.
ns, no significance; ACACB, acetyl-CoA carboxylase β; PDK4,
pyruvate dehydrogenase kinase 4; GATM, glycine amidinotransferase;
MCCC2, methylcrotonyl-CoA carboxylase subunit 2; MRPL12,
mitochondrial ribosomal protein L12; FASN, fatty acid synthase. |
Relationship between the 6 core genes,
mitochondrial respiration and PCa progression
Next, the TCGA-PRAD dataset was used to study the
relationship between mitochondrial respiration and PCa as well as
its relationship with the 6 core genes and selected statistically
significant genes (P<0.05; Fig.
7A). Genes such as COX10, COX11 and COX14 were upregulated,
while genes such as FOXRED1, AGOX3 and FMC1 showed a downward
trend. Genes with |statistical|>6 were selected and their
correlation with immune infiltrating cells was calculated using
CIBERSORT. The results showed that these mitochondrial respiratory
chain genes were negatively correlated with resting CD4 memory T
cells and positively correlated with Tregs (Fig. 7B). Using the TCGA-PRAD dataset, the
correlation between the 6 core genes and mitochondrial respiratory
chain genes (|statistical|>4) was calculated. The results showed
that MRPL12, PDK4, ACACB and GATM had the strongest correlation
with mitochondrial respiration, and these 4 genes were selected as
hub genes (Fig. 7C). The 6
mitochondrial respiratory chain genes most closely related to these
4 hub genes were then selected and a linear regression scatter plot
was created. The results showed that MRPL12 was significantly
positively correlated with TMEM186, UQCC3, COX7A1 and ATPAF1
(Fig. S3A), while PDK4, ACACB and
GATM were positively correlated with COX7A1 and negatively
correlated with TMEMI186, SDHAF3, UQCC3, COA7 and ATPAF1 (Fig. S3B-D). These results suggest that
the core differential genes related to mitochondrial respiration
are closely related to immune infiltrating cells in PCa.
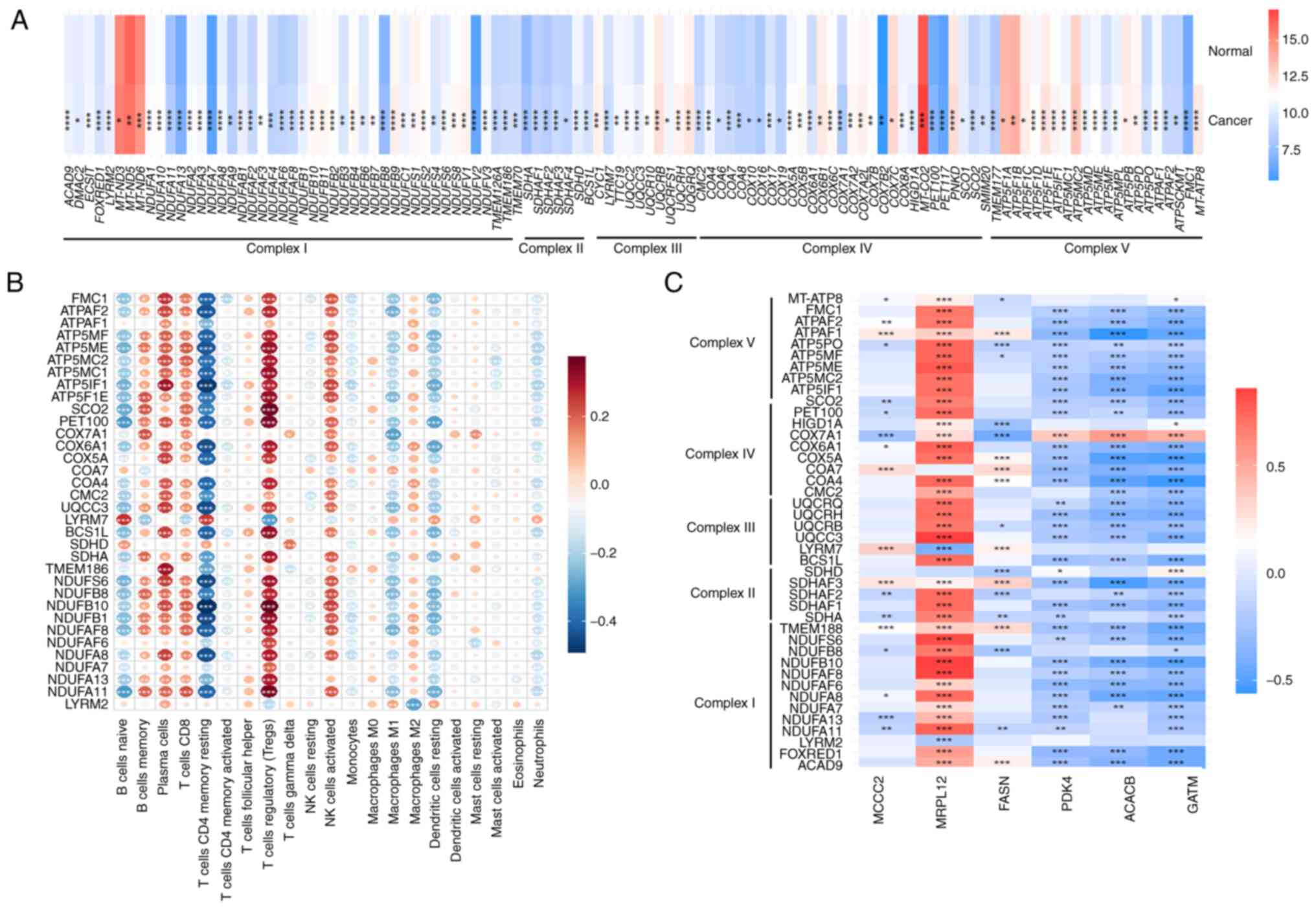 | Figure 7.Relationship between mitochondrial
respiration and prostate cancer. (A) Relationship between
mitochondrial respiration and prostate cancer, as well as the
relationship with 6 core genes. The dataset used is TCGA-PRAD. The
genes of the five stages of mitochondrial respiration were provided
by the MitoCarta 3.0 database, and statistically significant genes
were screened out; (B) Using the TCGA-PRAD dataset, genes with
|statistical|>6 from (A) were selected and the correlation
between these genes and CIBERSORT immune infiltrating cells was
calculated. (C) Using the TCGA-PRAD dataset, the correlation
between the 6 core genes and mitochondrial respiratory chain genes
(|statistical|>4) was calculated. *P<0.05, **P<0.01,
***P<0.001; ****P<0.0001. TCGA, The Cancer Genome Atlas;
PRAD, prostate adenocarcinoma; ACACB, acetyl-CoA carboxylase β;
PDK4, pyruvate dehydrogenase kinase 4; GATM, glycine
amidinotransferase; MCCC2, methylcrotonyl-CoA carboxylase subunit
2; MRPL12, mitochondrial ribosomal protein L12; FASN, fatty acid
synthase; |
Potential association of hub genes
with DAMPs and mitochondrial metabolism
The TCGA-PRAD dataset was then used to create violin
plots to determine the differences between DAMP-related genes (11
genes) and immune receptor-related genes (21 genes) in PCa and
adjacent tissues. Genes with significant differences were selected
for further research (P<0.001; Fig.
8A). An analysis of the correlation between the 4 hub genes and
significantly different DAMP-related genes and immune receptors
showed that they have a strong correlation (Fig. 8B). To further investigate the
potential correlation between the 4 hub genes (MRPL12, PDK4, ACACB
and GATM) and DAMPs and mitochondrial metabolism, the Mantel test
was used to analyze statistical significance. The results showed
that they were closely related to the mitochondrial metabolic
pathways in gluconeogenesis, the tricarboxylic acid (TCA) cycle and
pyruvate/ketone/lipid/amino acid metabolism in PCa (Fig. 8C).
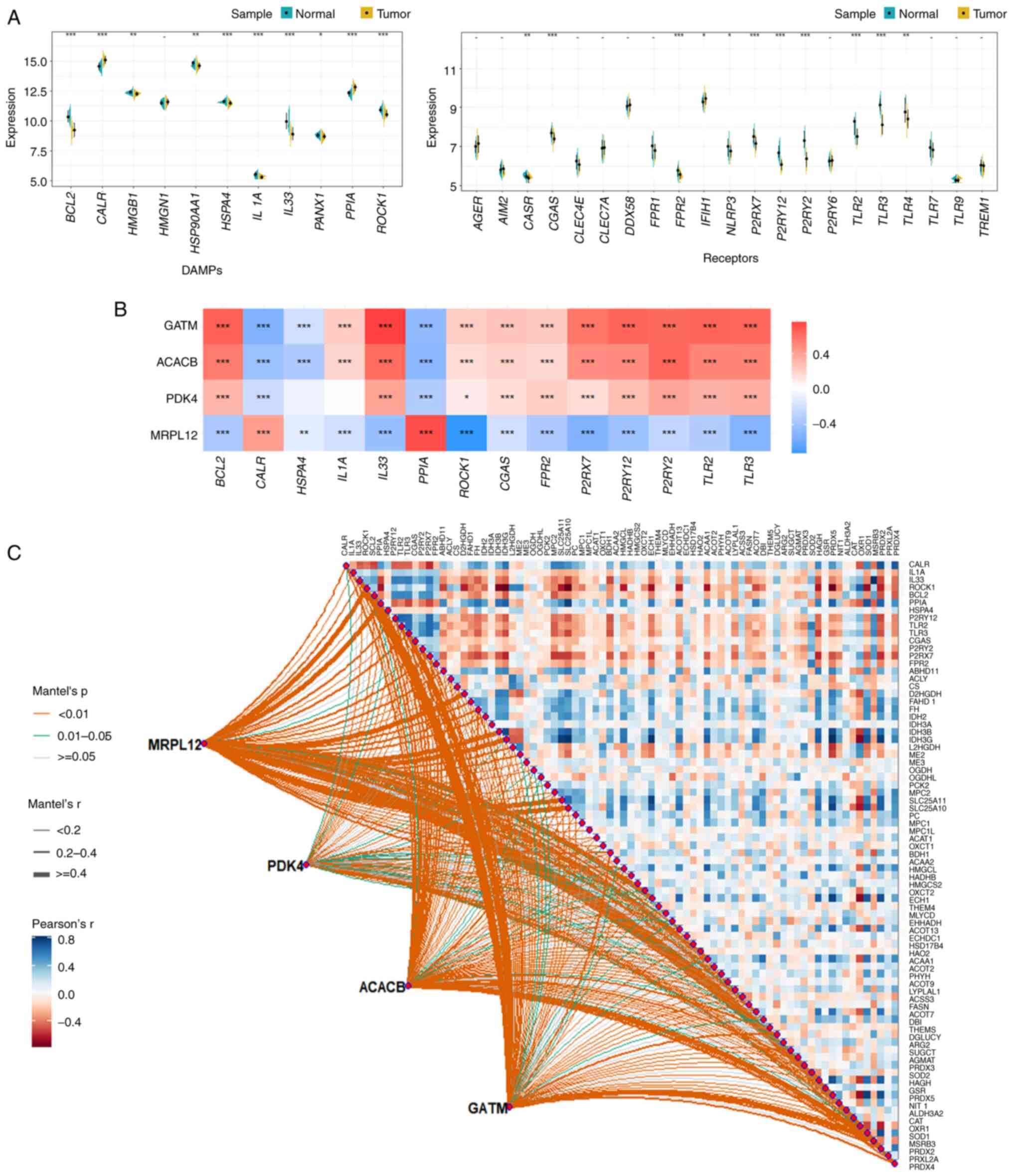 | Figure 8.Potential correlation between 4 hub
genes (MRPL12, PDK4, ACACB and GATM) and immune receptors and
mitochondrial metabolism. (A) Using the TCGA-PRAD dataset, the
differences between DAMP-related genes (11) and immune receptor-related genes
(21) in PCa and adjacent tissues
were detected; the genes with significant differences
(**P<0.001) were selected for further study. (B) The correlation
between the 4 hub genes and the significant differences in
DAMP-related genes and immune receptors selected in (a). (C)
Further investigation of the potential correlation between the 4
hub genes and DAMPs. *P<0.05, **P<0.01, ***P<0.001. PRAD,
prostate adenocarcinoma; ACACB, acetyl-CoA carboxylase β; PDK4,
pyruvate dehydrogenase kinase 4; GATM, glycine amidinotransferase;
MCCC2, methylcrotonyl-CoA carboxylase subunit 2; MRPL12,
mitochondrial ribosomal protein L12; FASN, fatty acid synthase;
TCGA, The Cancer Genome Atlas. |
Validating expression levels of
DeMRGs
Finally, expression of the core genes was validated
using the RWPE-1 (the non-PCa cell line used for comparison), PC-3,
LNcap and Du145 cell lines. The results showed that ACACB, PDK4 and
GATM were downregulated in PCa, while MCCC2, MRPL12 and FASN were
upregulated in PCa, consistent with the bioinformatics analysis
(Fig. 9).
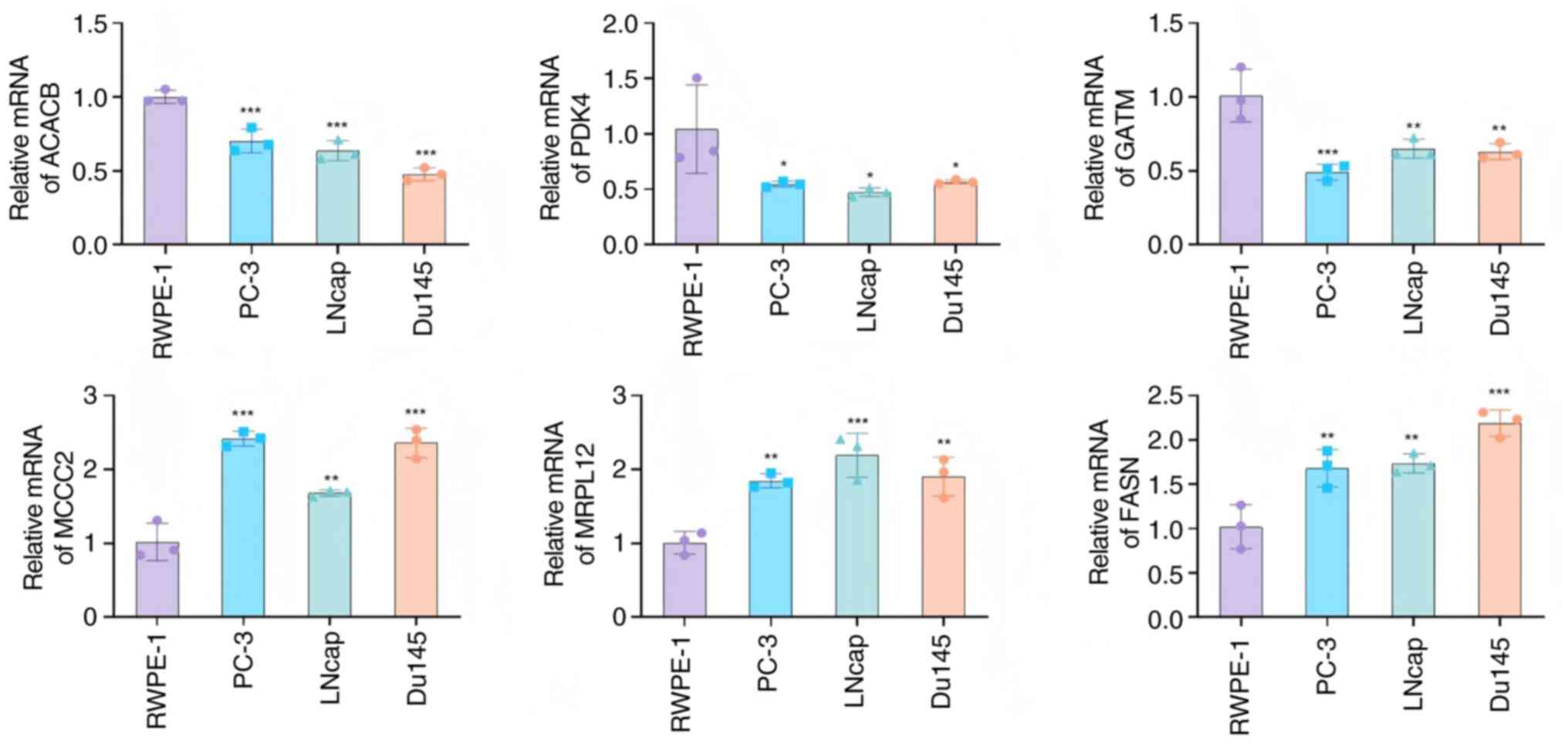 | Figure 9.RT-qPCR validation of the core genes.
Relative RNA levels of ACACB, PDK4, GATM, MCCC2, MRPL12 and FASN
were determined by the RT-qPCR for the RWPE-1, PC-3, LNcap and
Du145 cell lines. *P<0.05, **P<0.01, ***P<0.001; n=3.
RT-qPCR, reverse transcription-quantitative PCR; ACACB, acetyl-CoA
carboxylase β; PDK4, pyruvate dehydrogenase kinase 4; GATM, glycine
amidinotransferase; MCCC2, methylcrotonyl-CoA carboxylase subunit
2; MRPL12, mitochondrial ribosomal protein L12; FASN, fatty acid
synthase. |
Discussion
Mitochondria are the ‘power factories’ in cells,
providing 80% of the energy required for cellular life activities.
Mitochondria play an important role in maintaining normal
physiological metabolism and body development and are involved in
the occurrence and development of various diseases (such as breast
cancer and renal cell carcinoma) (19). Variations in mitochondrial DNA
(mtDNA) sequences are common in certain tumors (such as ovarian
cancer, oncocytoma and prostate cancer). Two types of mtDNA cancer
variants can be identified: Novel mutations as oncogenic inducers
and functional variants as adaptors, allowing cancer cells to grow
in different environments. These mtDNA variants come from three
sources: Familial variants, somatic mutations produced within each
cell or individual and variants associated with ancient mtDNA
lineages (haplotypes) that are considered to adapt to constantly
changing tissues or geographical environments (20,21).
In addition to mtDNA sequence mutations, mtDNA copy number and
mtDNA sequence transfer to the nucleus may also lead to certain
cancer types [such as breast epithelial cell lines, patient-derived
xenograft models of triple-negative breast cancer and high-grade
serous ovarian cancer (HGSC) and primary HGSC tumors] (22). There is a strong functional
correlation between mtDNA mutations in eosinophilic tumors and PCa
(21). In PCa, significant changes
occur in the mitochondrial membrane, leading to the increased
uptake of integrated membrane proteins. This process leads to
increased rigidity and higher enzyme activity of cancer cell
mitochondria (23). NKX3.1 is
expressed in the prostate epithelium and its function is to protect
the prostate from damage and inflammation as well as maintain the
luminal prostate stem cells. In addition, it can regulate the
expression of mitochondrial genes to promote mitochondrial
homeostasis and prevent the occurrence of PCa (24).
Mitochondrial changes have a significant impact on
the phenotype of prostate tumor cells. First, the progression of
PCa is accompanied by an increase in ROS, which promotes its
invasiveness. During the transformation and later stages of PCa
development, PCa cells increase their mitochondrial respiration and
glycolytic rates to meet energy needs. Due to enhanced
mitochondrial respiration, ROS levels increase, inducing signaling
pathways related to PCa growth and survival (25–27).
Mitochondrial respiration and metabolism are related to PCa, while
normal prostate epithelial cells are highly dependent on glycolysis
and produce citrate from glucose. TCA cycle cannot effectively
produce a large amount of citrate. During the transformation
process, PCa cells gradually reactivate OXPHOS, increase glucose
metabolism and reduce citrate production. In addition, the late
stages of PCa are marked by an increase in TCA cycle and citrate
levels, which are used by cancer cells for biomolecular synthesis
to support their growth (28).
Epithelial-mesenchymal transition (EMT) provides enhanced migration
and invasion capabilities for cancer cells, promoting tumor
dissemination and metastasis. A study has shown that the
downregulation of mitochondrial proteins involved in OXPHOS is
associated with increased EMT and invasive disease characteristics
(29). The changes in mitochondrial
gene expression are also related to the growth, survival and drug
resistance of PCa. Previous research has found that mitochondrial
fission factor and dynamic related protein-1 are amplified in
castration-resistant PCa, leading to low patient survival rates
(29). In addition to serving as a
power source for cells, mitochondria also play an important role in
cell death pathways, such as apoptosis. Mitochondria are crucial in
the activation of cell apoptosis through intrinsic pathways in
response to excessive oxidative stress and DNA damage (30,31).
Higher expression of Bcl-x is associated with higher-grade PCa
tumors, as well as the presence of lymph node metastasis and
distant metastasis. Given that Bcl-x is a key regulator of
mitochondrial-mediated apoptosis, these findings further indicate
that mitochondria play an important role in PCa immune escape
(32,33).
In PCa, ARs reprogram the entire cellular metabolic
pathway, including aerobic glycolysis and mitochondrial
respiration, as well as de novo fat generation, to support
the metabolic and biosynthetic needs of PCa cells (34,35).
The m6A-mediated circular RNA (circ)RBM33-FMR1 complex can activate
mitochondrial metabolism by stabilizing pyruvate dehydrogenase E1
subunit α1 mRNA, thereby promoting the progression of PCa and
reducing the arylsulfatase family member I efficacy of circRBM33 in
PCa treatment (36). Treatment
targeting mitochondria may help with PCa as the naturally occurring
amino acid 5-aminolevulinic acid (5-ALA) immediately enhances
mitochondrial ROS production after exposure to IR and reduces
mitochondrial membrane potential by increasing intracellular PpIX
(the metabolite of 5-ALA) in PC-3 and DU-145 PCA cell lines. IR is
accompanied by mitochondrial dysfunction induced by 5-ALA and an
increase in ATP production, which switches energy metabolism to a
quiescent state. Under hypoxic conditions, IR induces ROS burst and
mitochondrial dysfunction with 5-ALA to reduce cancer stemness and
radiation resistance (37). RNA
polymerase mitochondria (POLRMT) are crucial for mitochondrial
transcription mechanisms and other mitochondrial functions.
Upregulated POLRMT is important for PCa cell growth and
mitochondrial POLRMT may serve as a target for PCa treatment
(38).
In the present study, MRPL12, PDK4, ACACB and GATM
were identified as hub genes. MRPL12 is a core component of the
mitochondrial ribosome (mitoribosome), which is essential for
synthesizing proteins encoded by mtDNA, including key subunits of
the OXPHOS complex (39). In
hepatocellular carcinoma, MRPL12 is upregulated via the
PI3K/mTOR/YY1 pathway, which enhances OXPHOS and mitochondrial DNA
content, thereby driving malignant phenotypes (39). PDK4 is a gene involved in fatty acid
metabolism. Together with ACACB, FABP3 and other genes, it
constitutes a metabolic gene signature for prostate cancer. By
inhibiting the activity of the pyruvate dehydrogenase complex, PDK4
promotes glycolysis and suppresses OXPHOS, thereby supporting the
Warburg effect in tumors (40).
Following androgen receptor inhibition, PCa cells switch to a
dependence on mitochondrial oxidative metabolism, and PDK4 may play
a regulatory role in this process (41). ACACB is a rate-limiting enzyme in
fatty acid synthesis and is involved in lipid metabolic
reprogramming in prostate cancer. As a member of the same 5-gene
metabolic signature as PDK4, its low expression is associated with
high-risk PCa (40). Dysregulated
lipid metabolism may promote immune evasion by altering the
function of immune cells (such as CD8+ T cells) in the
tumor microenvironment (42). GATM
is involved in creatine synthesis, and its expression is associated
with the MYC pathway and OXPHOS. In PCa, high GATM expression may
lead to mitochondrial dysfunction (such as reduced aspartate
levels), driving metabolic shifts toward glycolysis (43).
Despite the novel findings regarding mitochondrial
hub genes in PCa, the present study has several limitations that
should be acknowledged. First, the identification and validation of
DeMRGs and hub genes relied heavily on publicly available datasets
(TCGA-PRAD, GSE46602, GSE55945 and GSE70770) and in vitro
experiments using established PCa cell lines (Du145, PC-3 and
LNcap) and a normal prostate epithelial cell line (RWPE-1). The
lack of primary PCa tissue samples from a patient cohort (such as
clinical specimens with detailed pathological staging, treatment
history and long-term follow-up data) limits the generalizability
of the results to diverse clinical populations, as public datasets
may have inherent biases in sample selection and data
standardization. Second, the functional validation of the four hub
genes (MRPL12, PDK4, ACACB and GATM) was limited to correlative
analyses (such as associations with immune cell infiltration,
mitochondrial respiration and metabolic pathways) and gene
expression verification via RT-qPCR. No in-depth mechanistic
experiments were performed to elucidate the direct roles of these
hub genes in PCa progression, for instance,
gain-of-function/loss-of-function assays (such as overexpression
plasmids, small interfering RNA or CRISPR-Cas9) to assess changes
in PCa cell proliferation, migration, invasion or mitochondrial
function (such as mitochondrial membrane potential, ROS production
and ATP levels) were not conducted. Additionally, in vivo
validation using PCa xenograft models or genetically engineered
mouse models was absent, which would be critical to confirm the
in vivo relevance of these hub genes. In summary, while the
present study provides a foundation for understanding the role of
mitochondrial hub genes in PCa, future studies addressing these
limitations, including large-scale clinical validation, mechanistic
experiments and subtype-specific analyses, will be necessary to
advance the translational potential of the findings.
In summary, the present study found that ACACB, PDK4
and GATM were downregulated, while MCCC2, MRPL12 and FASN are
upregulated in PCa; among these genes, MCCC2 and FASN were linked
to adverse PCa phenotypes and MRPL12, PDK4, ACACB and GATM showed
the strongest associations with mitochondrial metabolic pathways in
PCa, providing potential therapeutic targets for PCa treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by Hunan Provincial Natural Science
Foundation of China (grant nos. 2025JJ50553 and 2025JJ80574) and
Changsha Municipal Natural Science Foundation (grant no.
kq2502330).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SL and HS conducted most experiments and wrote the
manuscript. LH, LL, YW conceived, designed and interpreted data.
All authors edited the manuscript. All authors read and approved
the final version of the manuscript. SL and HS confirm the
authenticity of all the raw data
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Moris L, Cumberbatch MG, Van den Broeck T,
Gandaglia G, Fossati N, Kelly B, Pal R, Briers E, Cornford P, De
Santis M, et al: Benefits and risks of primary treatments for
High-risk localized and locally advanced prostate cancer: An
international multidisciplinary systematic review. Eur Urol.
77:614–627. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rebello RJ, Oing C, Knudsen KE, Loeb S,
Johnson DC, Reiter RE, Gillessen S, Van der Kwast T and Bristow RG:
Prostate cancer. Nat Rev Dis Primers. 7:92021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schaeffer E, Srinivas S, Antonarakis ES,
Armstrong AJ, Bekelman JE, Cheng H, D'Amico AV, Davis BJ, Desai N,
Dorff T, et al: NCCN guidelines insights: Prostate cancer, version
1.2021. J Natl Compr Canc Netw. 19:134–143. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Linder S, van der Poel HG, Bergman AM,
Zwart W and Prekovic S: Enzalutamide therapy for advanced prostate
cancer: Efficacy, resistance and beyond. Endocr Relat Cancer.
26:R31–R52. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parida S, Pal I, Parekh A, Thakur B,
Bharti R, Das S and Mandal M: GW627368X inhibits proliferation and
induces apoptosis in cervical cancer by interfering with EP4/EGFR
interactive signaling. Cell Death Dis. 7:e21542016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sathya S, Sudhagar S, Sarathkumar B and
Lakshmi BS: EGFR inhibition by pentacyclic triterpenes exhibit cell
cycle and growth arrest in breast cancer cells. Life Sci. 95:53–62.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang H, Kim B, Park J, Youn H and Youn B:
The Warburg effect on radioresistance: Survival beyond growth.
Biochim Biophys Acta Rev Cancer. 1878:1889882023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen H, Zhou L, Wu X, Li R, Wen J, Sha J
and Wen X: The PI3K/AKT pathway in the pathogenesis of prostate
cancer. Front Biosci (Landmark Ed). 21:1084–1091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumar R, Srinivasan S, Pahari P, Rohr J
and Damodaran C: Activating stress-activated protein
kinase-mediated cell death and inhibiting epidermal growth factor
receptor signaling: A promising therapeutic strategy for prostate
cancer. Mol Cancer Ther. 9:2488–2496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu W, Yang Q, Fung KM, Humphreys MR, Brame
LS, Cao A, Fang YT, Shih PT, Kropp BP and Lin HK: Linking
γ-aminobutyric acid A receptor to epidermal growth factor receptor
pathways activation in human prostate cancer. Mol Cell Endocrinol.
383:69–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hirokawa N, Noda Y, Tanaka Y and Niwa S:
Kinesin superfamily motor proteins and intracellular transport. Nat
Rev Mol Cell Biol. 10:682–696. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Drechsler H and McAinsh AD: Kinesin-12
motors cooperate to suppress microtubule catastrophes and drive the
formation of parallel microtubule bundles. Proc Natl Acad Sci USA.
113:E1635–E1644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao H, Bo Q, Wu Z, Liu Q, Li Y, Zhang N,
Guo H and Shi B: KIF15 promotes bladder cancer proliferation via
the MEK-ERK signaling pathway. Cancer Manag Res. 11:1857–1868.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mortensen MM, Høyer S, Lynnerup AS,
Ørntoft TF, Sørensen KD, Borre M and Dyrskjøt L: Expression
profiling of prostate cancer tissue delineates genes associated
with recurrence after prostatectomy. Sci Rep. 5:160182015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arredouani MS, Lu B, Bhasin M, Eljanne M,
Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA and
Sanda MG: Identification of the transcription factor single-minded
homologue 2 as a potential biomarker and immunotherapy target in
prostate cancer. Clin Cancer Res. 15:5794–5802. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ross-Adams H, Lamb AD, Dunning MJ, Halim
S, Lindberg J, Massie CM, Egevad LA, Russell R, Ramos-Montoya A,
Vowler SL, et al: Integration of copy number and transcriptomics
provides risk stratification in prostate cancer: A discovery and
validation cohort study. EBioMedicine. 2:1133–1144. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hocaoglu H and Sieber M: Mitochondrial
respiratory quiescence: A new model for examining the role of
mitochondrial metabolism in development. Semin Cell Dev Biol.
138:94–103. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boso D, Piga I, Trento C, Minuzzo S, Angi
E, Iommarini L, Lazzarini E, Caporali L, Fiorini C, D'Angelo L, et
al: Pathogenic mitochondrial DNA variants are associated with
response to anti-VEGF therapy in ovarian cancer PDX models. J Exp
Clin Cancer Res. 43:3252024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kopinski PK, Singh LN, Zhang S, Lott MT
and Wallace DC: Mitochondrial DNA variation and cancer. Nat Rev
Cancer. 21:431–445. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim M, Gorelick AN, Vàzquez-García I,
Williams MJ, Salehi S, Shi H, Weiner AC, Ceglia N, Funnell T, Park
T, et al: Single-cell mtDNA dynamics in tumors is driven by
coregulation of nuclear and mitochondrial genomes. Nat Genet.
56:889–899. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zichri SB, Kolusheva S, Shames AI,
Schneiderman EA, Poggio JL, Stein DE, Doubijensky E, Levy D,
Orynbayeva Z and Jelinek R: Mitochondria membrane transformations
in colon and prostate cancer and their biological implications.
Biochim Biophys Acta Biomembr. 1863:1834712021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Papachristodoulou A, Rodriguez-Calero A,
Panja S, Margolskee E, Virk RK, Milner TA, Martina LP, Kim JY, Di
Bernardo M, Williams AB, et al: NKX3.1 Localization to mitochondria
suppresses prostate cancer initiation. Cancer Discov. 11:2316–2333.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mamouni K, Kallifatidis G and Lokeshwar
BL: Targeting mitochondrial metabolism in prostate cancer with
triterpenoids. Int J Mol Sci. 22:24662021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen CL, Lin CY and Kung HJ: Targeting
mitochondrial OXPHOS and their regulatory signals in prostate
cancers. Int J Mol Sci. 22:134352021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han C, Wang Z, Xu Y, Chen S, Han Y, Li L,
Wang M and Jin X: Roles of reactive oxygen species in biological
behaviors of prostate cancer. Biomed Res Int. 2020:12696242020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Li N, Liu Q, Guo J, Pan Q, Cheng
B, Xu J, Dong B, Yang G, Yang B, et al: Antiandrogen treatment
induces stromal cell reprogramming to promote castration resistance
in prostate cancer. Cancer Cell. 41:1345–1362.e9. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guerra F, Guaragnella N, Arbini AA, Bucci
C, Giannattasio S and Moro L: Mitochondrial dysfunction: A novel
potential driver of Epithelial-to-mesenchymal transition in cancer.
Front Oncol. 7:2952017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Civenni G, Bosotti R, Timpanaro A, Vàzquez
R, Merulla J, Pandit S, Rossi S, Albino D, Allegrini S, Mitra A, et
al: Epigenetic control of mitochondrial fission enables
Self-renewal of stem-like tumor cells in human prostate cancer.
Cell Metab. 30:303–318.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jeong SY and Seol DW: The role of
mitochondria in apoptosis. BMB Rep. 41:11–22. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Castilla C, Congregado B, Chinchón D,
Torrubia FJ, Japón MA and Sáez C: Bcl-xL is overexpressed in
hormone-resistant prostate cancer and promotes survival of LNCaP
cells via interaction with proapoptotic Bak. Endocrinology.
147:4960–4967. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krajewska M, Krajewski S, Epstein JI,
Shabaik A, Sauvageot J, Song K, Kitada S and Reed JC:
Immunohistochemical analysis of bcl-2, bax, bcl-X, and mcl-1
expression in prostate cancers. Am J Pathol. 148:1567–1576.
1996.PubMed/NCBI
|
|
34
|
Massie CE, Lynch A, Ramos-Montoya A, Boren
J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, et al:
The androgen receptor fuels prostate cancer by regulating central
metabolism and biosynthesis. EMBO J. 30:2719–2733. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Audet-Walsh É, Dufour CR, Yee T, Zouanat
FZ, Yan M, Kalloghlian G, Vernier M, Caron M, Bourque G, Scarlata
E, et al: Nuclear mTOR acts as a transcriptional integrator of the
androgen signaling pathway in prostate cancer. Genes Dev.
31:1228–1242. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhong C, Long Z, Yang T, Wang S, Zhong W,
Hu F, Teoh JY, Lu J and Mao X: M6A-modified circRBM33 promotes
prostate cancer progression via PDHA1-mediated mitochondrial
respiration regulation and presents a potential target for ARSI
therapy. Int J Biol Sci. 19:1543–1563. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Owari T, Tanaka N, Nakai Y, Miyake M, Anai
S, Kishi S, Mori S, Fujiwara-Tani R, Hojo Y, Mori T, et al:
5-Aminolevulinic acid overcomes hypoxia-induced radiation
resistance by enhancing mitochondrial reactive oxygen species
production in prostate cancer cells. Br J Cancer. 127:350–363.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, Yao L, Wang T, Gu X, Wu Y and Jiang
T: Identification of the mitochondrial protein POLRMT as a
potential therapeutic target of prostate cancer. Cell Death Dis.
14:6652023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ji X, Yang Z, Li C, Zhu S, Zhang Y, Xue F,
Sun S, Fu T, Ding C, Liu Y, et al: Mitochondrial ribosomal protein
L12 potentiates hepatocellular carcinoma by regulating
mitochondrial biogenesis and metabolic reprogramming. Metabolism.
152:1557612023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fan Y, Wang J, Wang Y, Li Y, Wang S, Weng
Y, Yang Q, Chen C, Lin L, Qiu Y, et al: Development and clinical
validation of a novel 5 gene signature based on fatty acid
Metabolism-related genes in oral squamous cell carcinoma. Oxid Med
Cell Longev. 2022:32853932022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Crowell PD, Giafaglione JM, Jones AE,
Nunley NM, Hashimoto T, Delcourt AML, Petcherski A, Agrawal R,
Bernard MJ, Diaz JA, et al: MYC is a regulator of androgen receptor
inhibition-induced metabolic requirements in prostate cancer. Cell
Rep. 42:1132212023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li L, Chao Z, Peng H, Hu Z, Wang Z and
Zeng X: Tumor ABCC4-mediated release of PGE2 induces CD8+ T cell
dysfunction and impairs PD-1 blockade in prostate cancer. Int J
Biol Sci. 20:4424–4437. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Löf C, Sultana N, Goel N, Heron S,
Wahlström G, House A, Holopainen M, Käkelä R and Schleutker J: ANO7
expression in the prostate modulates mitochondrial function and
lipid metabolism. Cell Commun Signal. 23:712025. View Article : Google Scholar : PubMed/NCBI
|