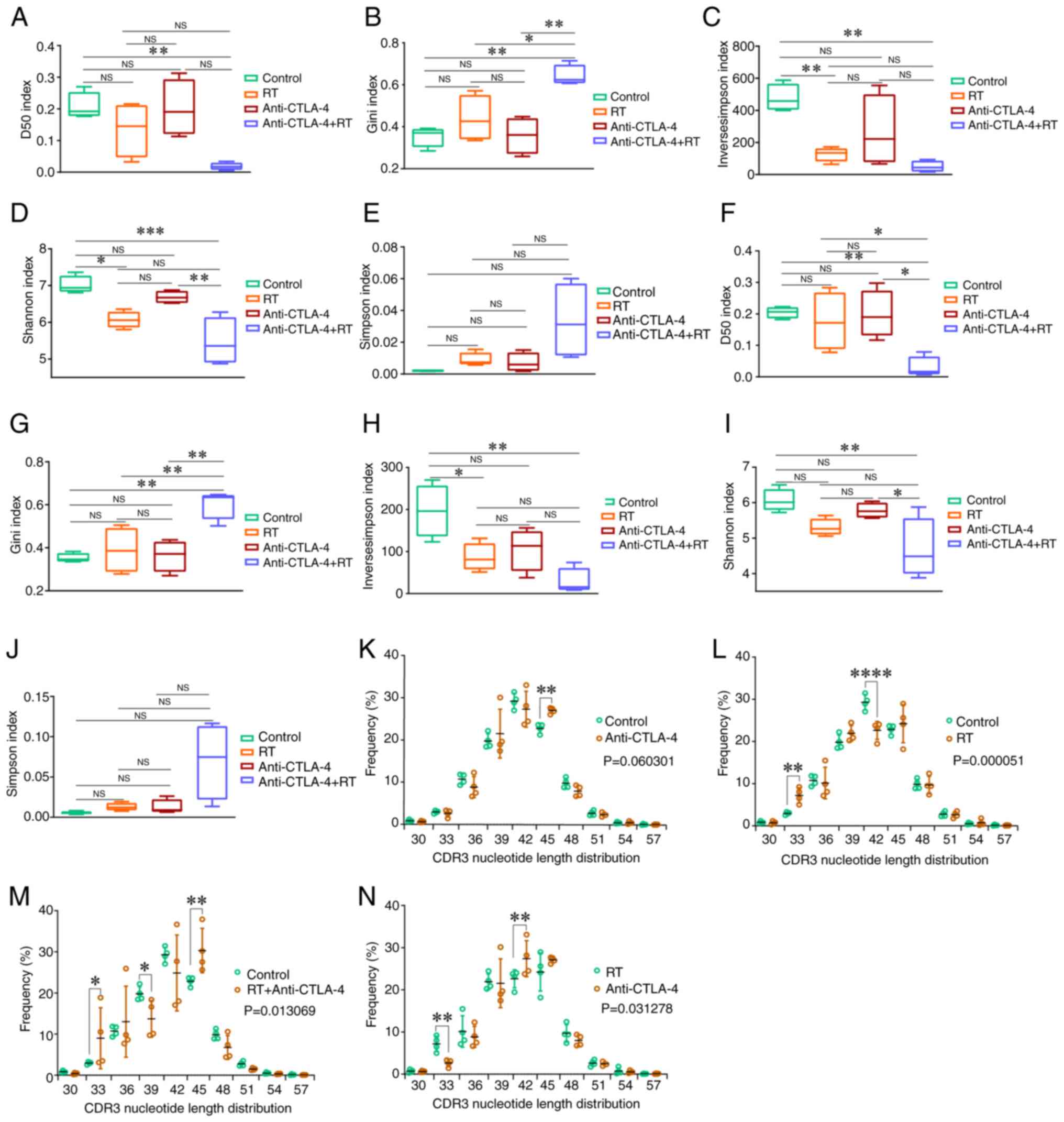
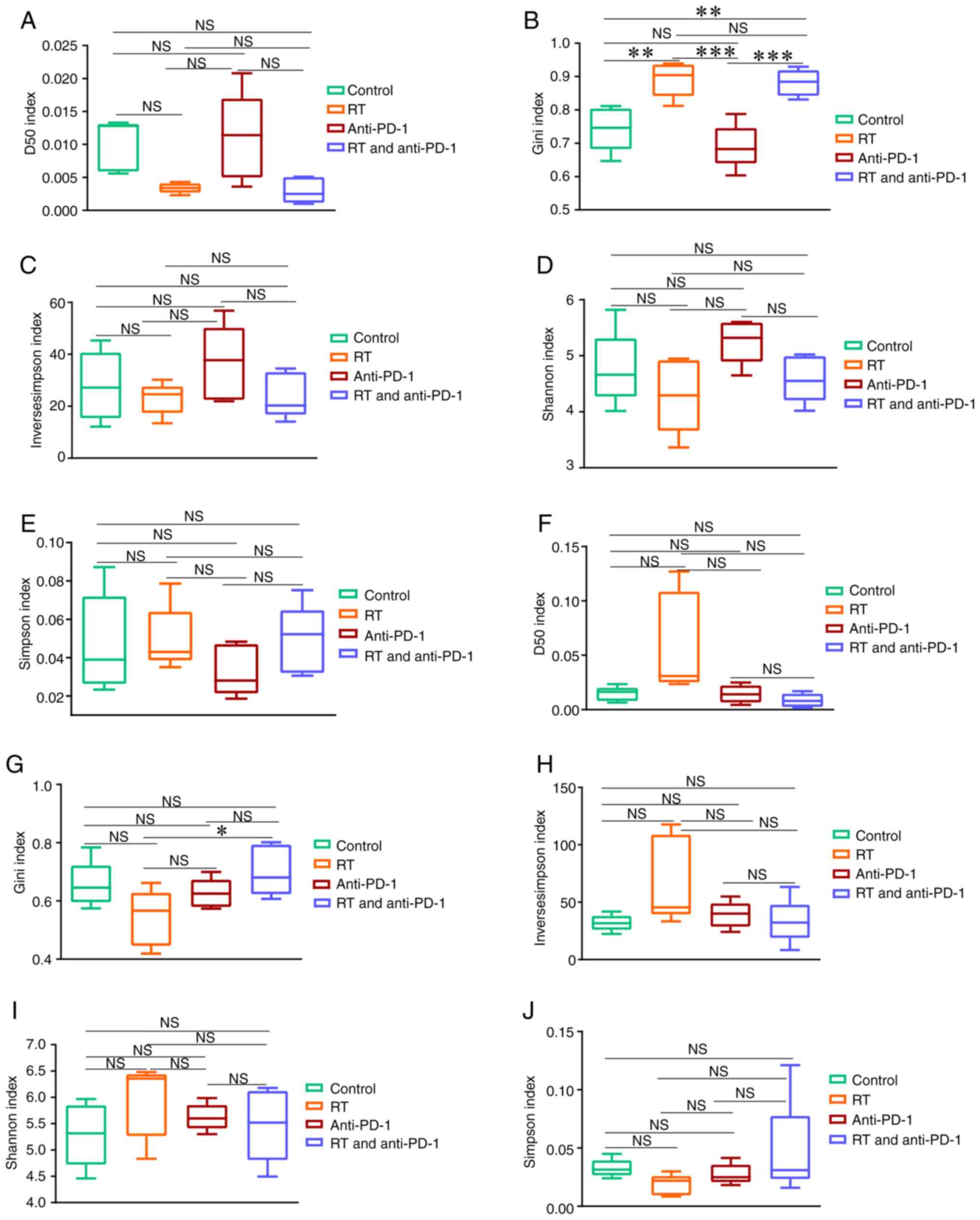
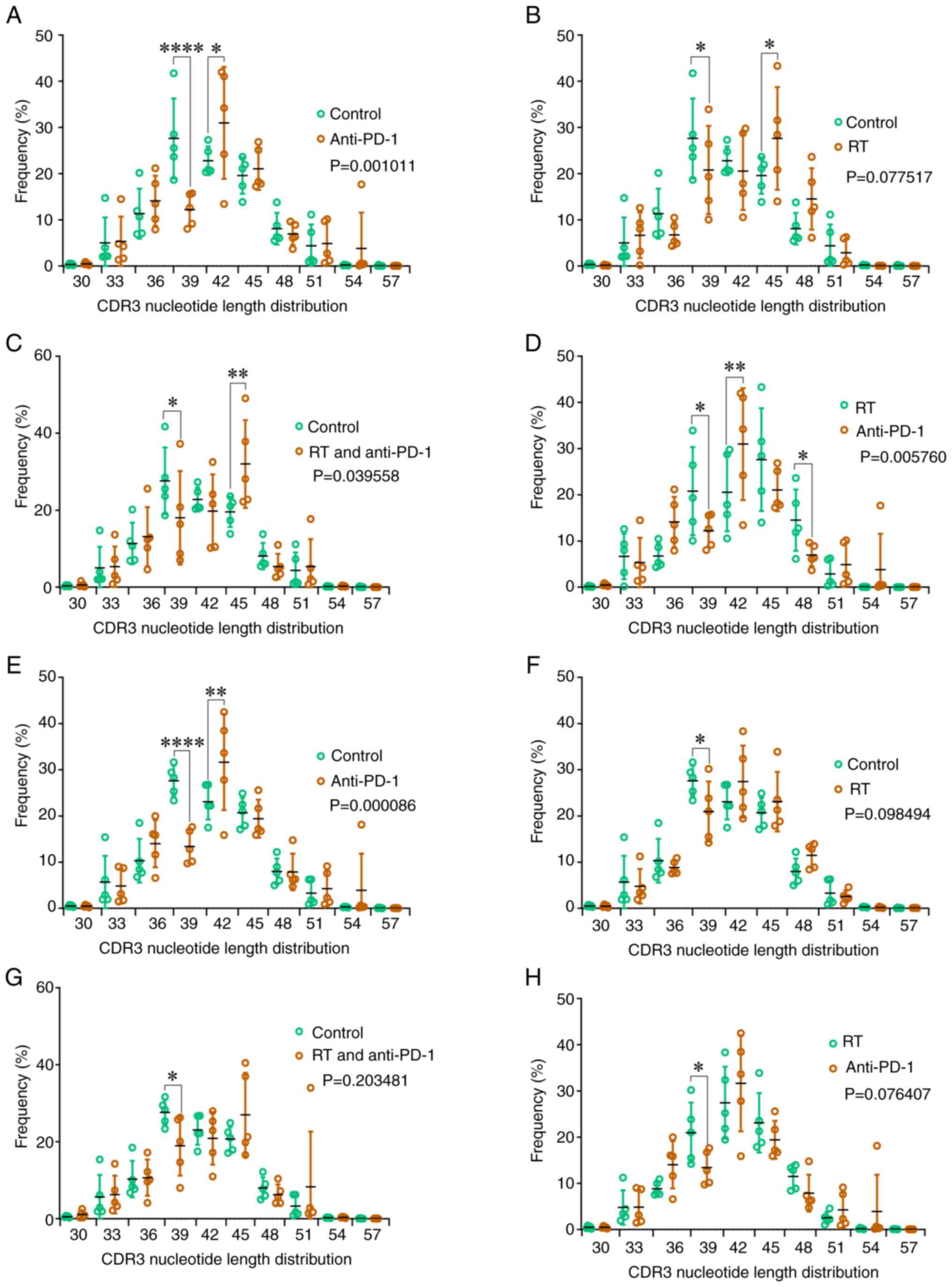
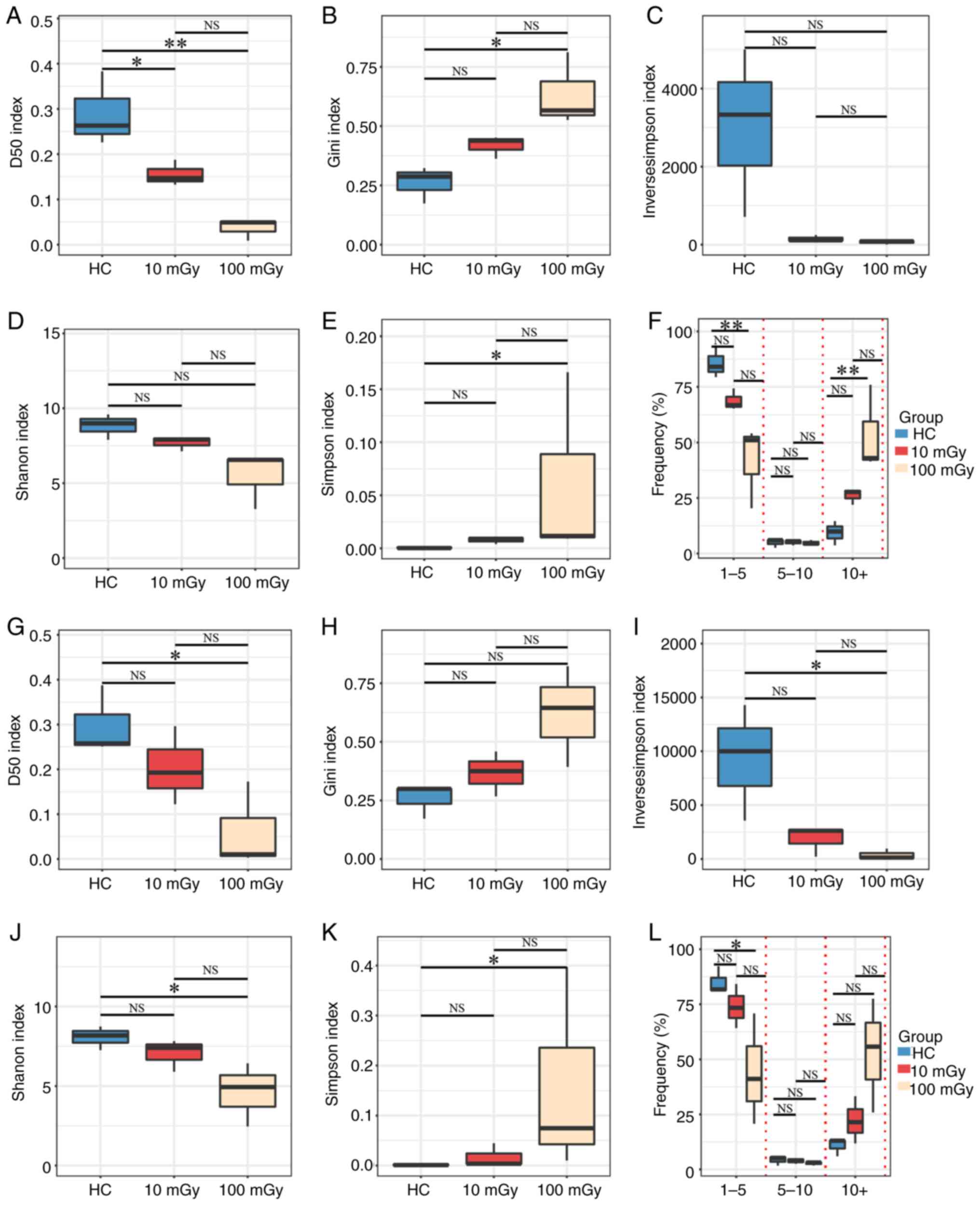
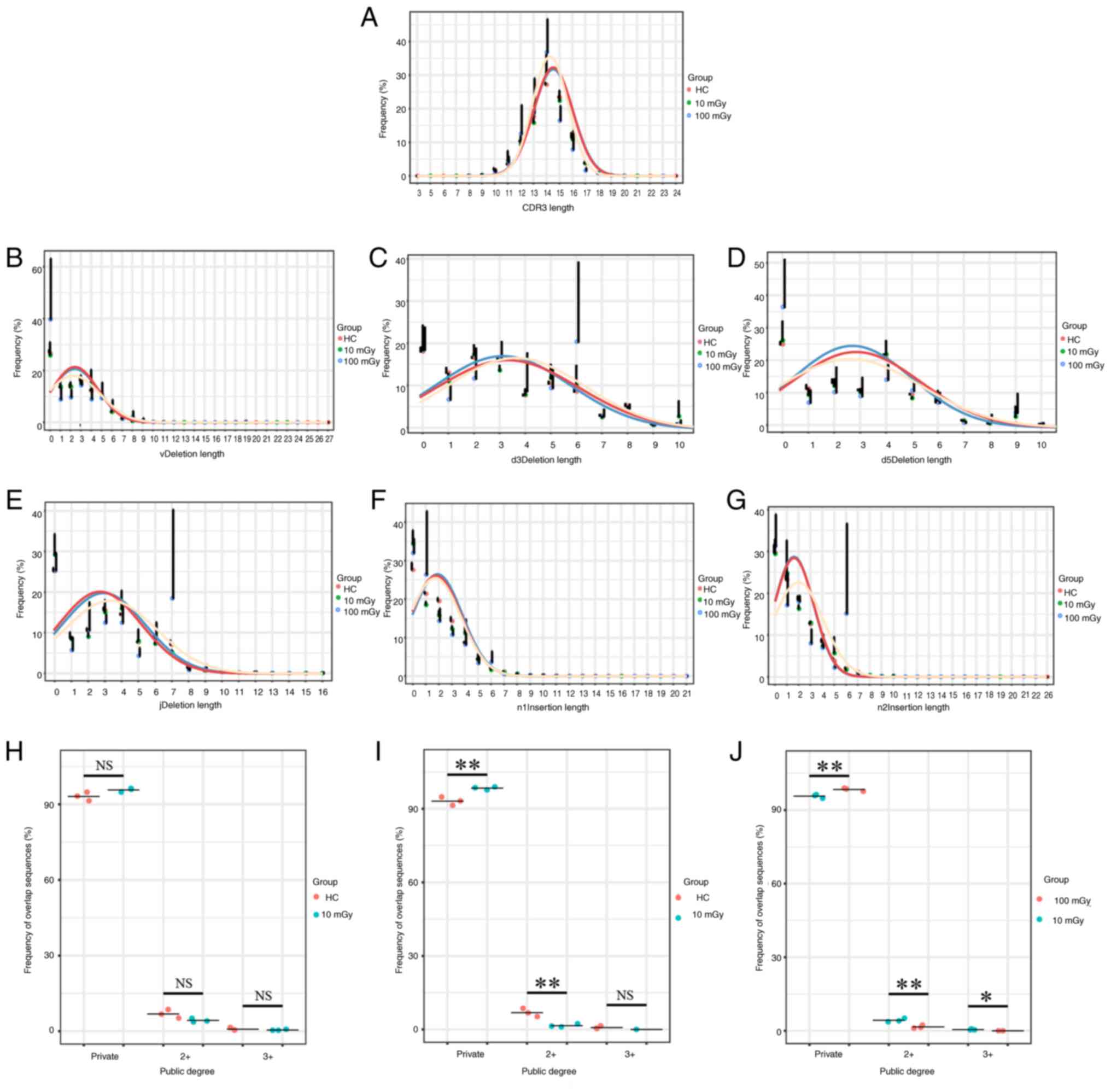
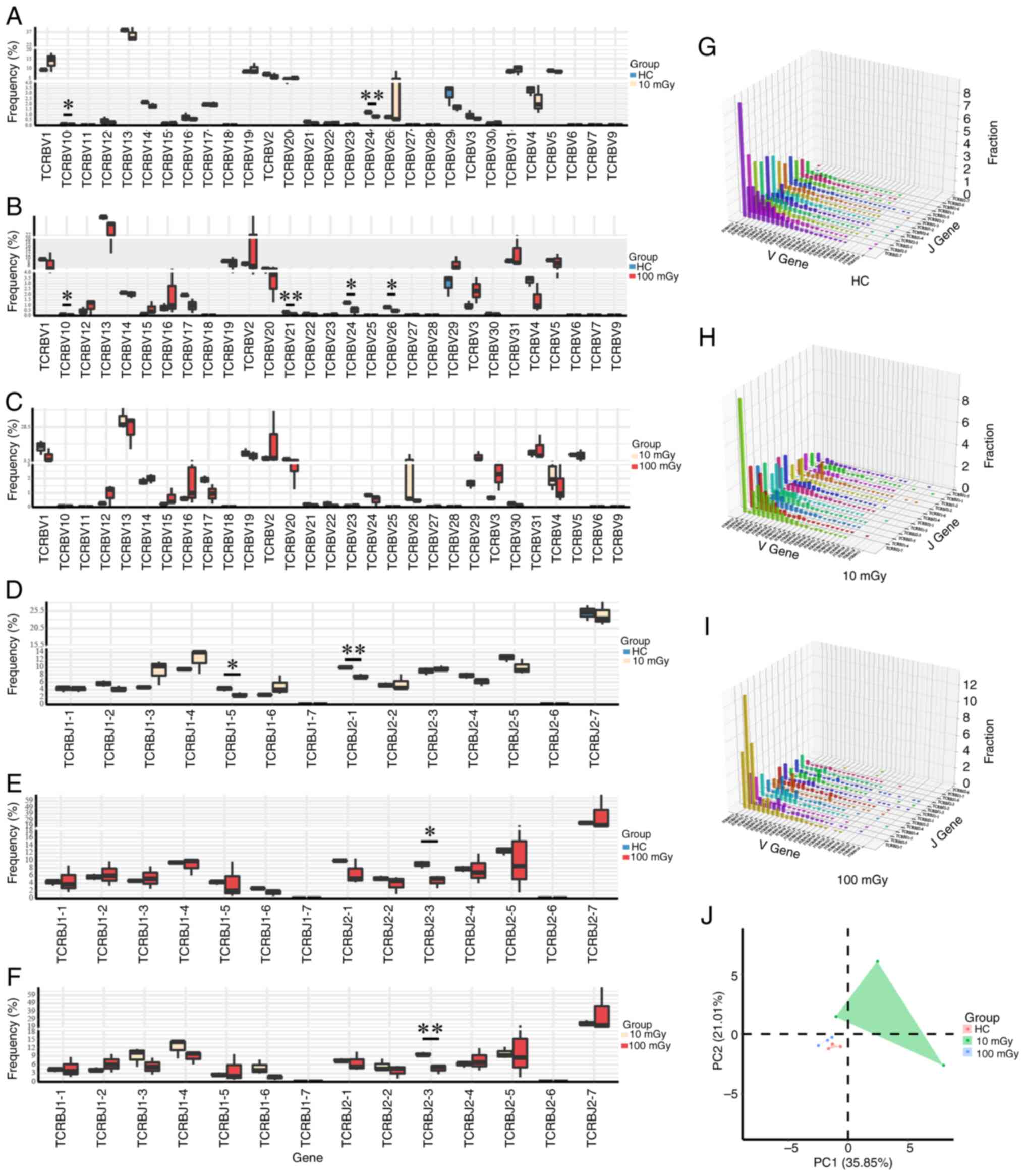
|
1
|
Chen Z, Ma Y, Hua J, Wang Y and Guo H:
Impacts from economic development and environmental factors on life
expectancy: A comparative study based on data from both developed
and developing countries from 2004 to 2016. Int J Environ Res
Public Health. 18:85592021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tchkonia T, Palmer AK and Kirkland JL: New
horizons: Novel approaches to enhance healthspan through targeting
cellular senescence and related aging mechanisms. J Clin Endocrinol
Metab. 106:e1481–e1487. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao W, Qin K, Li F and Chen W:
Socioeconomic inequalities in cancer incidence and mortality: An
analysis of GLOBOCAN 2022. Chin Med J (Engl). 137:1407–1413. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng R, Zhang S, Zeng H, Wang S, Sun K,
Chen R, Li L, Wei W and He J: Cancer incidence and mortality in
China, 2016. J Natl Cancer Cent. 2:1–9. 2022.PubMed/NCBI
|
|
6
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhattacharya S and Asaithamby A:
Repurposing DNA repair factors to eradicate tumor cells upon
radiotherapy. Transl Cancer Res. 6 (Suppl 5):S822–S839. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mortezaee K and Najafi M: Immune system in
cancer radiotherapy: Resistance mechanisms and therapy
perspectives. Crit Rev Oncol Hematol. 157:1031802021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watanabe T, Sato GE, Yoshimura M, Suzuki M
and Mizowaki T: The mutual relationship between the host immune
system and radiotherapy: Stimulating the action of immune cells by
irradiation. Int J Clin Oncol. 28:201–208. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hoffman RM: Back to the future: Are
tumor-targeting bacteria the next-generation cancer therapy?
Methods Mol Biol. 1317:239–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dewan MZ, Galloway AE, Kawashima N,
Dewyngaert JK, Babb JS, Formenti SC and Demaria S: Fractionated but
not single-dose radiotherapy induces an immune-mediated abscopal
effect when combined with anti-CTLA-4 antibody. Clin Cancer Res.
15:5379–5388. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Verbrugge I, Hagekyriakou J, Sharp LL,
Galli M, West A, McLaughlin NM, Duret H, Yagita H, Johnstone RW,
Smyth MJ and Haynes NM: Radiotherapy increases the permissiveness
of established mammary tumors to rejection by immunomodulatory
antibodies. Cancer Res. 72:3163–3174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hou X, Yang Y, Chen J, Jia H, Zeng P, Lv
L, Lu Y, Liu X and Diao H: TCRβ repertoire of memory T cell reveals
potential role for Escherichia coli in the pathogenesis of primary
biliary cholangitis. Liver Int. 39:956–966. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou XL, Wang L, Ding YL, Xie Q and Diao
HY: Current status and recent advances of next generation
sequencing techniques in immunological repertoire. Genes Immun.
17:153–164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou X, Lu C, Chen S, Xie Q, Cui G, Chen J,
Chen Z, Wu Z, Ding Y, Ye P, et al: High throughput sequencing of T
cell antigen receptors reveals a conserved TCR repertoire. Medicine
(Baltimore). 95:e28392016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hou X, Wang M, Lu C, Xie Q, Cui G, Chen J,
Du Y, Dai Y and Diao H: Analysis of the repertoire features of TCR
beta chain CDR3 in human by high-throughput sequencing. Cell
Physiol Biochem. 39:651–667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zvyagin IV, Pogorelyy MV, Ivanova ME,
Komech EA, Shugay M, Bolotin DA, Shelenkov AA, Kurnosov AA,
Staroverov DB, Chudakov DM, et al: Distinctive properties of
identical twins' TCR repertoires revealed by high-throughput
sequencing. Proc Natl Acad Sci USA. 111:5980–5985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Larimore K, McCormick MW, Robins HS and
Greenberg PD: Shaping of human germline IgH repertoires revealed by
deep sequencing. J Immunol. 189:3221–3230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robins HS, Campregher PV, Srivastava SK,
Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH and Carlson
CS: Comprehensive assessment of T-cell receptor beta-chain
diversity in alphabeta T cells. Blood. 114:4099–4107. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robins HS, Srivastava SK, Campregher PV,
Turtle CJ, Andriesen J, Riddell SR, Carlson CS and Warren EH:
Overlap and effective size of the human CD8+ T cell receptor
repertoire. Sci Transl Med. 2:47ra642010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Ji Z and Smith KN: Analysis of
TCR β CDR3 sequencing data for tracking anti-tumor immunity.
Methods Enzymol. 629:443–464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li S, Sun J, Allesøe R, Datta K, Bao Y,
Oliveira G, Forman J, Jin R, Olsen LR, Keskin DB, et al: RNase
H-dependent PCR-enabled T-cell receptor sequencing for highly
specific and efficient targeted sequencing of T-cell receptor mRNA
for single-cell and repertoire analysis. Nat Protoc. 14:2571–2594.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baumeister SH, Freeman GJ, Dranoff G and
Sharpe AH: Coinhibitory pathways in immunotherapy for cancer. Annu
Rev Immunol. 34:539–573. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Candéias SM, Mika J, Finnon P, Verbiest T,
Finnon R, Brown N, Bouffler S, Polanska J and Badie C: Low-dose
radiation accelerates aging of the T-cell receptor repertoire in
CBA/Ca mice. Cell Mol Life Sci. 74:4339–4351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rudqvist NP, Pilones KA, Lhuillier C,
Wennerberg E, Sidhom JW, Emerson RO, Robins HS, Schneck J, Formenti
SC and Demaria S: Radiotherapy and CTLA-4 blockade shape the TCR
repertoire of tumor-infiltrating T cells. Cancer Immunol Res.
6:139–150. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dovedi SJ, Cheadle EJ, Popple AL, Poon E,
Morrow M, Stewart R, Yusko EC, Sanders CM, Vignali M, Emerson RO,
et al: Fractionated radiation therapy stimulates antitumor immunity
mediated by both resident and infiltrating polyclonal T-cell
populations when combined with PD-1 blockade. Clin Cancer Res.
23:5514–5526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Robins H, Desmarais C, Matthis J,
Livingston R, Andriesen J, Reijonen H, Carlson C, Nepom G, Yee C
and Cerosaletti K: Ultra-sensitive detection of rare T cell clones.
J Immunol Methods. 375:14–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carlson CS, Emerson RO, Sherwood AM,
Desmarais C, Chung MW, Parsons JM, Steen MS,
LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, et al:
Using synthetic templates to design an unbiased multiplex PCR
assay. Nat Commun. 4:26802013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bolotin DA, Shugay M, Mamedov IZ,
Putintseva EV, Turchaninova MA, Zvyagin IV, Britanova OV and
Chudakov DM: MiTCR: Software for T-cell receptor sequencing data
analysis. Nat Methods. 10:813–814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Elhanati Y, Sethna Z, Callan CG Jr, Mora T
and Walczak AM: Predicting the spectrum of TCR repertoire sharing
with a data-driven model of recombination. Immunol Rev.
284:167–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pogorelyy MV, Minervina AA, Chudakov DM,
Mamedov IZ, Lebedev YB, Mora T and Walczak AM: Method for
identification of condition-associated public antigen receptor
sequences. Elife. 7:e330502018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Murugan A, Mora T, Walczak AM and Callan
CG Jr: Statistical inference of the generation probability of
T-cell receptors from sequence repertoires. Proc Natl Acad Sci USA.
109:16161–16166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gomez-Tourino I, Kamra Y, Baptista R,
Lorenc A and Peakman M: T cell receptor β-chains display abnormal
shortening and repertoire sharing in type 1 diabetes. Nat Commun.
8:17922017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hou X, Zeng P, Chen J and Diao H: No
difference in TCRβ repertoire of CD4+ naive T cell between patients
with primary biliary cholangitis and healthy control subjects. Mol
Immunol. 116:167–173. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hou X, Zeng P, Zhang X, Chen J, Liang Y,
Yang J, Yang Y, Liu X and Diao H: Shorter TCR β-chains are highly
enriched during thymic selection and antigen-driven selection.
Front Immunol. 10:2992019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang C, Li X, Wu J, Zhang W, Sun S, Lin
L, Wang X, Li H, Wu X, Zhang P, et al: The landscape and diagnostic
potential of T and B cell repertoire in immunoglobulin A
nephropathy. J Autoimmun. 97:100–107. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Zhu H, Zhang Q and Zhao Y: Clonal
analysis of peripheral blood T cells in patients with hashimoto's
thyroiditis at different stages using TCR sequencing.
Immunobiology. 230:1528902025. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Venturi V, Kedzierska K, Turner SJ,
Doherty PC and Davenport MP: Methods for comparing the diversity of
samples of the T cell receptor repertoire. J Immunol Methods.
321:182–195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Z, Tang Y, Tan Y, Wei Q and Yu W:
Cancer-associated fibroblasts in radiotherapy: Challenges and new
opportunities. Cell Commun Signal. 17:472019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Van Allen EM, Miao D, Schilling B, Shukla
SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger
SM, et al: Genomic correlates of response to CTLA-4 blockade in
metastatic melanoma. Science. 9:207–211. 2015. View Article : Google Scholar
|
|
42
|
Chan TA, Wolchok JD and Snyder A: Genetic
basis for clinical response to CTLA-4 blockade in melanoma. N Engl
J Med. 12:19842015. View Article : Google Scholar
|
|
43
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 3:124–128. 2015.
View Article : Google Scholar
|
|
44
|
Ruocco MG, Pilones KA, Kawashima N, Cammer
M, Huang J, Babb JS, Liu M, Formenti SC, Dustin ML and Demaria S:
Suppressing T cell motility induced by anti-CTLA-4 monotherapy
improves antitumor effects. J Clin Invest. 122:3718–3730. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Krishna C, Chowell D, Gönen M, Elhanati Y
and Chan TA: Genetic and environmental determinants of human TCR
repertoire diversity. Immun Ageing. 17:262020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Riaz N, Havel JJ, Makarov V, Desrichard A,
Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH,
et al: Tumor and microenvironment evolution during immunotherapy
with nivolumab. Cell. 171:934–949.e916. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hebeisen M, Allard M, Gannon PO, Schmidt
J, Speiser DE and Rufer N: Identifying individual T cell receptors
of optimal avidity for tumor antigens. Front Immunol. 6:5822015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yan C, Ma X, Guo Z, Wei X, Han D, Zhang T,
Chen X, Cao F, Dong J, Zhao G, et al: Time-spatial analysis of T
cell receptor repertoire in esophageal squamous cell carcinoma
patients treated with combined radiotherapy and PD-1 blockade.
Oncoimmunology. 11:20256682022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pickman Y, Dunn-Walters D and Mehr R: BCR
CDR3 length distributions differ between blood and spleen and
between old and young patients, and TCR distributions can be used
to detect myelodysplastic syndrome. Phys Biol. 10:0560012013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu S, Hou XL, Sui WG, Lu QJ, Hu YL and
Dai Y: Direct measurement of B-cell receptor repertoire's
composition and variation in systemic lupus erythematosus. Genes
Immun. 18:22–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Weng NP: Numbers and odds: TCR repertoire
size and its age changes impacting on T cell functions. Semin
Immunol. 69:1018102023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hollingsworth BA, Aldrich JT, Case CM Jr,
DiCarlo AL, Hoffman CM, Jakubowski AA, Liu Q, Loelius SG, PrabhuDas
M, Winters TA and Cassatt DR: Immune dysfunction from radiation
exposure. Radiat Res. 200:396–416. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Price DA, Asher TE, Wilson NA, Nason MC,
Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK,
Roederer M, et al: Public clonotype usage identifies protective
Gag-specific CD8+ T cell responses in SIV infection. J Exp Med.
206:923–936. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Venturi V, Quigley MF, Greenaway HY, Ng
PC, Ende ZS, McIntosh T, Asher TE, Almeida JR, Levy S, Price DA, et
al: A mechanism for TCR sharing between T cell subsets and
individuals revealed by pyrosequencing. J Immunol. 186:4285–4294.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhao Y, Nguyen P, Ma J, Wu T, Jones LL,
Pei D, Cheng C and Geiger TL: Preferential use of public TCR during
autoimmune encephalomyelitis. J Immunol. 196:4905–4914. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hou X, Wang G, Fan W, Chen X, Mo C, Wang
Y, Gong W, Wen X, Chen H, He D, et al: T-cell receptor repertoires
as potential diagnostic markers for patients with COVID-19. Int J
Infect Dis. 113:308–317. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Forrester HB and Radford IR: Ionizing
radiation-induced chromosomal rearrangements occur in
transcriptionally active regions of the genome. Int J Radiat Biol.
80:757–767. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sui W, Hou X, Zou G, Che W, Yang M, Zheng
C, Liu F, Chen P, Wei X, Lai L and Dai Y: Composition and variation
analysis of the TCR β-chain CDR3 repertoire in systemic lupus
erythematosus using high-throughput sequencing. Mol Immunol.
67:455–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kuehm LM, Wolf K, Zahour J, DiPaolo RJ and
Teague RM: Checkpoint blockade immunotherapy enhances the frequency
and effector function of murine tumor-infiltrating T cells but does
not alter TCRβ diversity. Cancer Immunol Immunother. 68:1095–1106.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Valença-Pereira F, Sheridan RM, Riemondy
KA, Thornton T, Fang Q, Barret B, Paludo G, Thompson C, Collins P,
Santiago M, et al: Inactivation of GSK3β by Ser(389)
phosphorylation prevents thymocyte necroptosis and impacts Tcr
repertoire diversity. Cell Death Differ. 32:880–898. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kershaw MH, Devaud C, John LB, Westwood JA
and Darcy PK: Enhancing immunotherapy using chemotherapy and
radiation to modify the tumor microenvironment. Oncoimmunology.
2:e259622013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Huang J and Li JJ: Multiple dynamics in
tumor microenvironment under radiotherapy. Adv Exp Med Biol.
1263:175–202. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
D'Alonzo RA, Keam S, Gill S, Rowshanfarzad
P, Nowak AK, Ebert MA and Cook AM: Fractionated low-dose
radiotherapy primes the tumor microenvironment for immunotherapy in
a murine mesothelioma model. Cancer Immunol Immunother. 74:442025.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Pituch KC, Miska J, Krenciute G, Li G,
Panek WK, Gottschalk S and Balyasnikova IV: P06.01 Functional
analysis of IL13Rα2-specific chimeric antigen receptor T cells in
immunocompetent mouse of glioblastoma. Neuro Oncol. 19:iii48–iii49.
2017. View Article : Google Scholar
|
|
65
|
Hsieh J, Ng S, Bosinger S, Wu JH, Tharp
GK, Garcia A, Hossain MS, Yuan S, Waller EK and Galipeau J: A GMCSF
and IL7 fusion cytokine leads to functional thymic-dependent T-cell
regeneration in age-associated immune deficiency. Clin Transl
Immunology. 4:e372015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Savino W, Mendes-da-Cruz DA, Lepletier A
and Dardenne M: Hormonal control of T-cell development in health
and disease. Nat Rev Endocrinol. 12:77–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Besman M, Zambrowicz A and Matwiejczyk M:
Review of thymic peptides and hormones: From their properties to
clinical application. International Journal of Peptide Research and
Therapeutics. 31:102024. View Article : Google Scholar
|
|
68
|
Kim KS: Regulation of T cell repertoires
by commensal microbiota. Front Cell Infect Microbiol.
12:10043392022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bessell CA, Isser A, Havel JJ, Lee S, Bell
DR, Hickey JW, Chaisawangwong W, Glick Bieler J, Srivastava R, Kuo
F, et al: Commensal bacteria stimulate antitumor responses via T
cell cross-reactivity. JCI Insight. 5:e1355972020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fang M, Miao Y, Zhu L, Mei Y, Zeng H, Luo
L, Ding Y, Zhou L, Quan X, Zhao Q, et al: Age-related dynamics and
spectral characteristics of the tcrβ repertoire in healthy
children: Implications for immune aging. Aging Cell. 24:e144602025.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen M, Su Z and Xue J: Targeting T-cell
aging to remodel the aging immune system and revitalize geriatric
immunotherapy. Aging Dis. 15:10.14336/AD.2025.0061. 2025.
|
|
72
|
Han J, Duan J, Bai H, Wang Y, Wan R, Wang
X, Chen S, Tian Y, Wang D, Fei K, et al: TCR repertoire diversity
of peripheral PD-1(+)CD8(+) T cells predicts clinical outcomes
after immunotherapy in patients with non-small cell lung cancer.
Cancer Immunol Res. 8:146–154. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang J, Bie Z, Zhang Y, Li L, Zhu Y, Zhang
Y, Nie X, Zhang P, Cheng G, Di X, et al: Prognostic value of the
baseline circulating T cell receptor β chain diversity in advanced
lung cancer. Oncoimmunology. 10:18996092021. View Article : Google Scholar : PubMed/NCBI
|