Introduction
Improvements in socioeconomic conditions have
enhanced the average life expectancy of humans (1). However, an aging population has been
associated with some public health concerns (2). In particular, the cancer incidence and
mortality rates have been increasing (3). According to published epidemiological
studies, global cancer incidence in 2012 reached 14.1 million new
cases with 8.2 million associated deaths (4). By 2022, however, these figures had
risen to ~19.98 million newly diagnosed cases and 9.74 million
cancer-attributable deaths worldwide (3). Globally, cancer, a chronic disease, is
a major cause of death (5,6).
Chemotherapy, radiotherapy (RT) and surgical
resection were the main therapeutic strategies for cancer until the
21st century. RT was traditionally assumed to function by
irreversibly damaging tumor cell DNA, which led to cell death or
loss of replicative ability (7).
Preclinical and clinical studies have reported that the immune
system also determines the response to RT (8,9). More
than a century ago, clinicians observed complete remission in
advanced cancer cases after acute bacterial infections (10). This observation established the
concept of immunological approaches for cancer treatment.
Immunotherapy has since transformed cancer treatment and improved
the current understanding of tumor biology (11). It has been highlighted that cancer
treatment should not only target the cancer cells but also consider
the entire tumor microenvironment (TME). The cellular mediators of
the anticancer effects of immunotherapy are immune cells (11). Previous studies have reported that
the combination of RT and immunotherapy significantly improves
cancer treatment outcomes (12,13).
However, the effect of the combination of RT and immunotherapy on
the T-cell receptor β chain (TCRβ) repertoire is unclear.
The immune system determines the anticancer efficacy
of both RT and immunotherapy. T cells are involved in adaptive
immune responses and complementarity-determining region 3 (CDR3)
determines the specificity of T cells (14). CDR3 is generated through the
recombination of germline V, D and J genes, as well as the deletion
and insertion of nucleotides at the V(D)J junctions (15–17).
As CDR3 closely interacts with antigen peptides, its sequence
diversity is a key indicator of T cell diversity. Recombination
events at the TCR loci can lead to the production of non-functional
(out-of-frame) TCRs with frameshift mutations or premature stop
codons (18,19). In this situation, the T cells
attempt to rearrange the second allele. If a functional (in-frame)
TCR is successfully formed, the T cells carry both functional and
non-functional TCR genes (18).
There is a certain level of mRNA in the T cells, although
non-functional TCR genes cannot be translated into functional TCRβ
chains. Non-functional TCRs can represent the pre-selection TCR
repertoire as they are not influenced by the functional selection
processes (both positive and negative selection) (19–21).
Functional TCRs can be used to investigate the TCR repertoire after
selection. Certain studies have demonstrated that deep sequencing
analysis of TCRβ CDR3 can provide useful insights into the effect
of treatment on the T cell response (22,23).
Cytotoxic T lymphocyte-associated antigen-4 (CTLA-4)
and programmed cell death-1 (PD-1) are immunosuppressive molecules
expressed on the immune cell surface and can regulate immune
activation (24). Furthermore,
CTLA-4 and PD-1 prevent the occurrence of autoimmune effects. The
present study analyzed a publicly available sequencing dataset of
the immune repertoire of a mouse cancer model treated with a
combination of RT and anti-CTLA-4/anti-PD-1 therapy. Furthermore, a
public dataset of T cell repertoire sequencing derived from the
hematopoietic stem cells (HSCs) of mice exposed to various
radiation doses was also analyzed. The present study focused on
examining the effects of RT and immunotherapy on the TCRβ
repertoire in cancer treatment based on various parameters, such as
TCRβ CDR3 diversity, frequency distribution of CDR3, length
distribution of CDR3, V/J gene usage, V-J pairing and overlap
indices. The findings from the present study will enhance the
current understanding of radiation-induced adaptive immune
responses and the TCRβ repertoire after RT and
anti-CTLA-4/anti-PD-1 therapy, which can potentially provide
valuable insights into the complex dynamics driving global cancer
patterns.
Materials and methods
TCR sequencing data from public
databases
The TCR sequencing data of T cells derived from
CBA/HmCherry mouse HSCs exposed to radiation doses of 0,
10 and 100 mGy were obtained online from the Adaptive
Biotechnologies immuneACCESS database (https://clients.adaptivebiotech.com/pub/candeias-2019-mouse;
immuneACCESS DOI: http://doi.org/10.21417/SC012019) (25). Meanwhile, the TCR data of
tumor-bearing mice treated with the combination of RT and
anti-CTLA-4/anti-PD-1 therapy were downloaded from different
sources
[https://clients.adaptivebiotech.com/pub/rudqvist-2017cancerimmunology-research,
immune ACCESS DOI: https://doi.org/10.21417/B7H34S (26) and
https://clients.adaptivebiotech.com/pub/dovedi-2017-clincancerres,
immune ACCESS DOI: https://doi.org/10.21417/B7TS67 (27)]. The T cell repertoires in 45 samples
(9, 16 and 20 samples from mice exposed to different radiation
doses, mice treated with RT and anti-CTLA-4 therapy for breast
cancer and mice treated with RT and anti-PD-1 therapy for colon
cancer, respectively) were analyzed. TCRβ sequencing was performed
on peripheral blood samples exhibiting the highest levels of
donor-derived T lymphocyte reconstitution. Based on the percentage
of T lymphocytes, the top samples were chosen from the following
groups: 11–15% (HSCs exposed to 0 mGy), 4–13% (HSCs exposed to 10
mGy) and 3.5–10% (HSCs exposed to 100 mGy). The yield of unique
TCRβ rearrangements was extremely low in 1 NSG mouse reconstituted
with 100 mGy-irradiated HSCs, which exhibited a low T-cell
frequency of merely 3.5% among leukocytes. Thus, these mice were
classified as outliers based on the Hubert and Vandervieren test
and excluded from subsequent analyses (25). The original research had obtained
ethical approval.
DNA extraction, TCRβ immunosequencing
and data analysis
The experimental procedures, sample collection, DNA
extraction and TCRβ immunosequencing were previously described in
the original study (26), while
data analysis based on process data was conducted as part of the
present study. Briefly, blood samples were collected from mice 6
months after receiving HSC transplants with radiation doses of 0,
10 and 100 mGy. Additionally, breast cancer and colon cancer
tissues were obtained from mice treated with RT combined with anti
CTLA-4, RT combined with anti PD-1 and untreated mice. The mouse
blood samples and the untreated and treated tumor samples had been
subjected to TCRβ CDR3 immunosequencing using the ImmunoSEQ™ assay
(28,29). A synthetic immune receptor library
was used to identify and reduce PCR biases and computational
techniques were applied to remove any remaining biases after
sequencing and ensure the quantitative accuracy of the ImmunoSEQ™
detection kit (29). PCR bias
assessed the total amplification bias by computing the following
two metrics for the amplification bias of each template compared
with the mean: The dynamic range (maximum bias divided by minimum
bias) and the sum of squared logarithmic bias values. The process
of altering primer concentrations was repeated until improvements
ceased (29). In the
biased-controlled multiplex PCR experiment, the extracted DNA was
amplified using 54 forward primers targeting V genes and 13 reverse
primers targeting J genes. High-throughput sequencing was then
performed. The raw data were processed and analyzed with an
ImmunoSEQ™ analyzer (http://www.adaptivebiotech.com/immunoseq). Then, a
reanalysis of the data from the original study was performed in the
present study. Subsequently, correction of sequencing errors and
PCR amplification bias was performed with MiTCR (http://mitcr.milaboratory.com/) (30). The international ImMunoGeneTics
database (www.imgt.org) provided gene definitions
for the TCRβ V, D and J genes. As the CDR3 sequence can be
generated by various pathways, it cannot directly provide the
probability distribution of hidden recombination events (31,32).
Consequently, TCR nucleotide sequence probabilities were calculated
using the existing recombination model (33). According to the probability model,
specific sequences of CDR3 may be generated based on the original
recombination process. This enables the annotation of the V(N)D(N)J
genes for each distinctive CDR3 and the determination of the amino
acid sequence for each gene. Furthermore, the combat function in
the sva package (R version 4.4.1) was used to address batch effects
across different datasets. Next, TCR data were subjected to various
bioinformatics analyses based on the R statistical programming
language (R version 4.4.1), which encompassed the diversity of TCRβ
repertoire, usage of V/J gene, V-J gene pairing, length
distribution of CDR3 and vDeletion, d3Deletion, d5Deletion,
jDeletion, n1Insertion and n2Insertion distribution (14,34–37).
These analyses were evaluated using previously published research
(14,34,37).
The D50 index, Gini index, inverse Simpson index, Shannon index and
Simpson index were used to calculate the TCR repertoire diversity
(38,39). Shared TCR repertoires between
species were quantified using overlap coefficients of |X and Y|/min
(|X|, |Y|). The TCRβ overlap was calculated based on the number of
common nucleotide sequences in each sharing category (summed across
all subjects in which this sequence was found)-[Percentage
(n)=number of common nucleotide sequences appeared in ≥n
subjects/total number of nucleotide sequences in sample]. In
addition, principal coordinate analysis (PCA) was used to perform
cluster analysis on the V/J genes of mice in the radiation exposure
group and healthy control (HC) group.
Statistical analysis
Means between two groups were compared using the
unpaired t-test or the Mann-Whitney U test. One-way ANOVA or
Kruskal-Wallis test was used to compare group means across three or
more groups and Bonferroni was used to correct for multiple
comparisons. Additionally, two-way ANOVA followed by Tukey's test
was used. Data are represented as mean ± SD or percentages (%). All
statistical analyses were performed using SPSS (version 20; IBM
Corp.). P<0.05 was considered to indicate a statistically
significant difference.
Results
RT and anti-CTLA-4 therapy alters the
TCR repertoire
Alterations in TCRβ repertoire diversity
The TCRβ repertoires of T cells sorted from tumors
of the untreated (control), RT, anti-CTLA-4 and anti-CTLA-4 + RT
groups were analyzed. The diversity indices were determined based
on D50, Gini, inverse Simpson, Shannon and Simpson indices
(Fig. 1). When considering the Gini
or Simpson indexes, a smaller value is associated with a higher
CDR3 diversity. The greater the index, the more diverse the CDR3,
when it comes to the D50, Shannon and inverse Simpson diversity
indices. In the post-selection repertoire (in-frame sequences;
Fig. 1A-E), the D50 index in the
anti-CTLA-4 + RT group was lower than that in the control
(P<0.01; Fig. 1A). The Gini
index in the anti-CTLA-4 + RT group was higher than that in the
control (P<0.01), RT (P<0.05) and anti-CTLA-4 (P<0.01)
groups (Fig. 1B). The inverse
Simpson index in the RT and the anti-CTLA-4 + RT groups was lower
than that in the control group (both P<0.01; Fig. 1C). The Shannon index in the RT group
(P<0.05) and anti-CTLA-4 + RT (P<0.001) groups was lower than
that in the control group, furthermore, it was also significantly
lower in the anti-CTLA-4 + RT group compared with that in the
anti-CTLA-4 group (Fig. 1D). Other
parameters were not significantly different between the control and
treatment groups. Furthermore, similar trends were observed in the
pre-selection repertoire (out-of-frame TCRβ sequences; Fig. 1F-J). In summary, the diversity of
the TCRβ CDR3 amino acid sequence in the treatment group
(especially in the RT + anti-CTLA-4 group) was lower than that in
the control group.
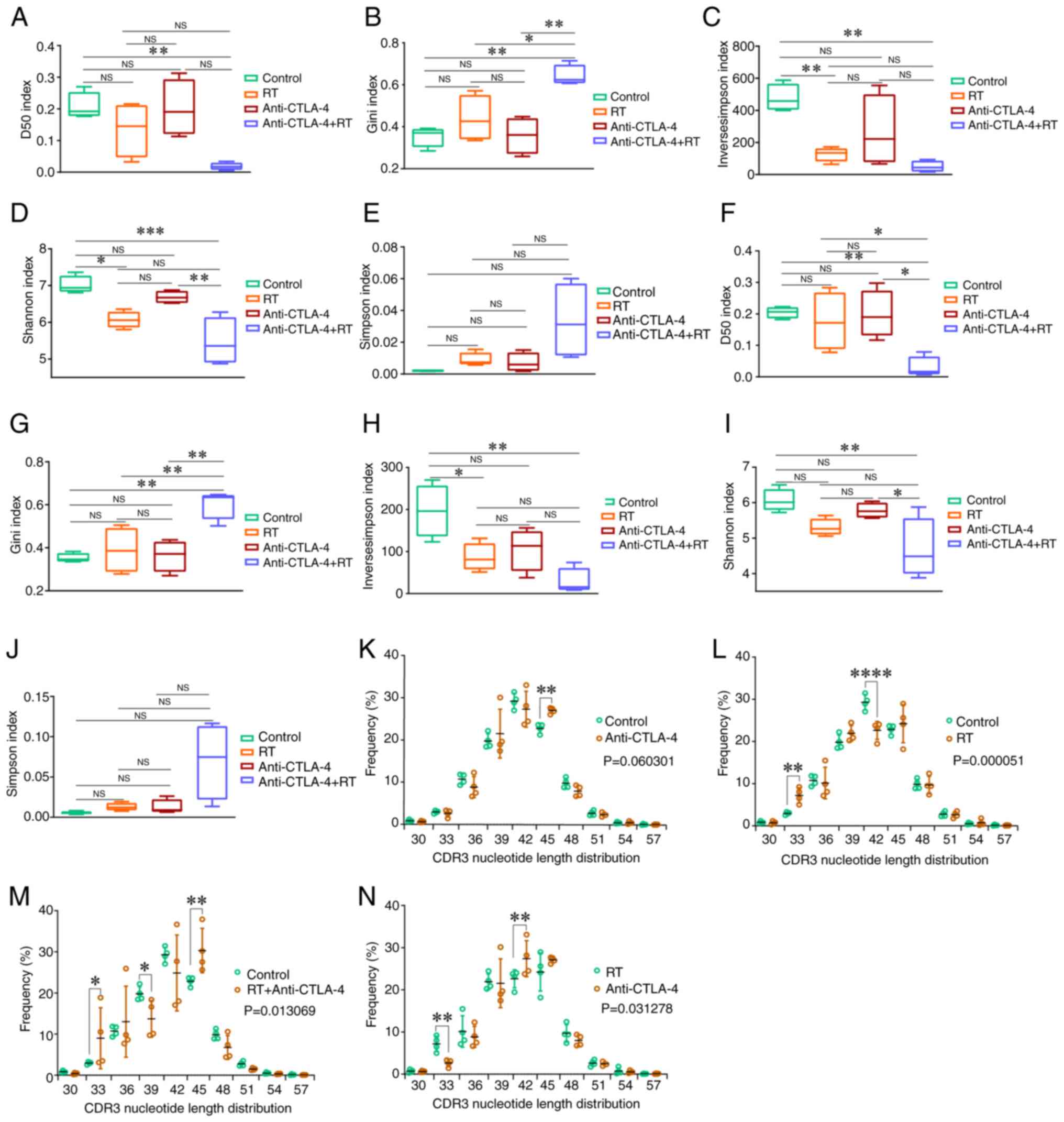 | Figure 1.CDR3 nucleotide length distribution
and diversity analysis of the TCRβ repertoire in mice with RT and
anti-CTLA-4 therapy, and the control group. The TCRβ CDR3 diversity
in the post-selection repertoire (in-frame sequences) was estimated
by (A) the D50 index, (B) Gini index, (C) Inverse Simpson index,
(D) Shannon index and (E) Simpson index. (F-J) The TCRβ CDR3
diversity in the pre-selection repertoire (out-of-frame sequences)
was estimated by the aforementioned five diversity indices. Data
are presented as the mean ± SD; comparisons were made using one-way
ANOVA or Kruskal-Wallis test and corrected for multiple comparisons
using Bonferroni. *P<0.05, **P<0.01 and ***P<0.001. (K-N)
The comparison of CDR3 length between the different groups. All of
the CDR3 lengths that differed significantly (P<0.05) between
treatment group mice and controls are presented. Data are presented
as the mean ± SD; the overall differences of the CDR3 length
distribution (ten length categories) between groups were compared
using two-way ANOVA, and Tukey's test was performed after the
two-way ANOVA to compare the differences in each CDR3 length
between groups. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. CDR3, complementarity-determining region 3; TCRβ,
T-cell receptor β-chain; CTLA-4, cytotoxic T lymphocyte-associated
antigen-4; RT, radiotherapy; NS, not significant. |
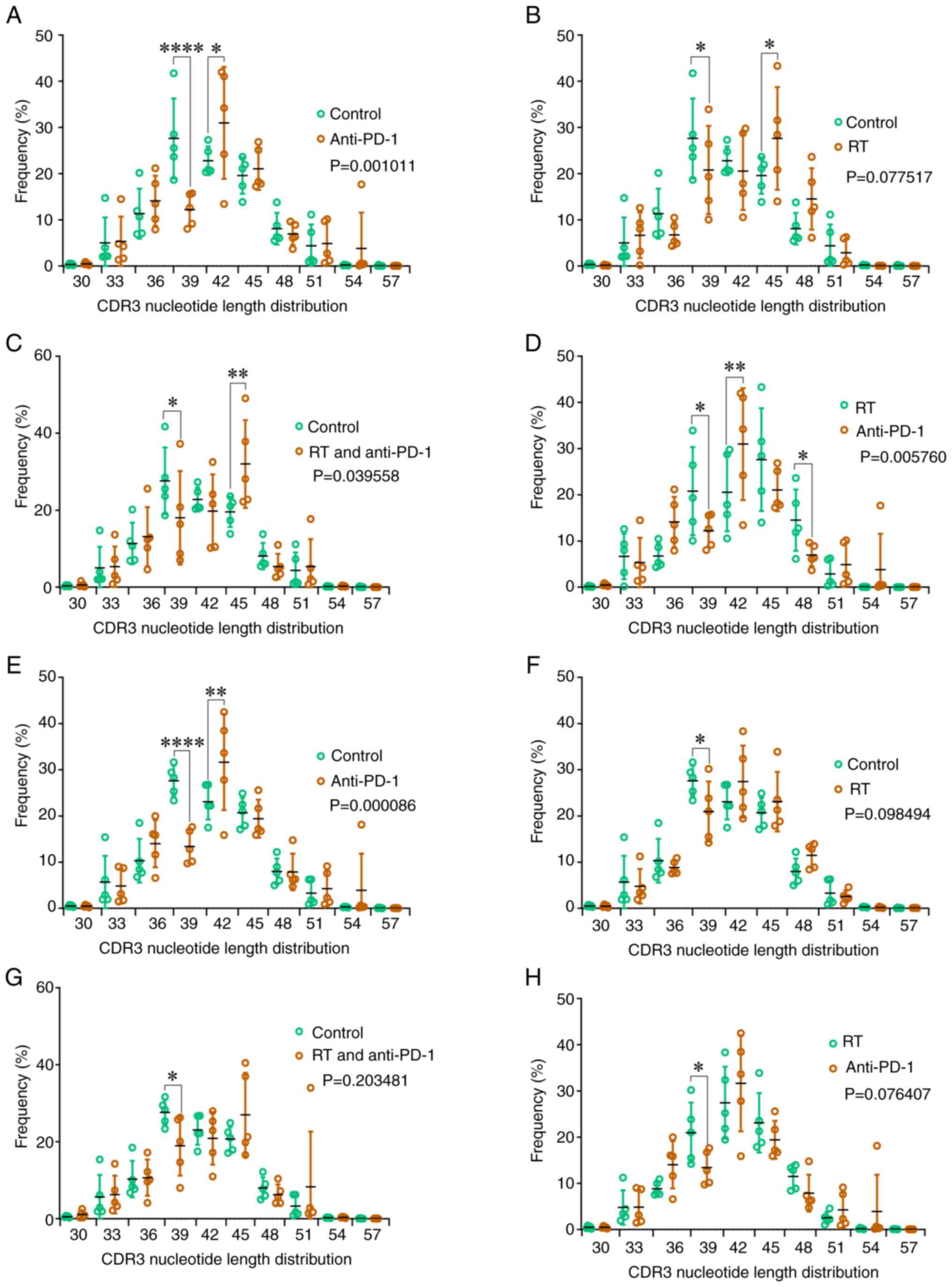
Alterations in CDR3 length
distribution
To further understand the changes in the TCRβ
repertoire induced by radiation and anti-CTLA-4 treatment, the
length distribution of the CDR3 nucleotide sequences was examined.
The overall difference in CDR3 length between the anti-CTLA-4 group
and the control group did not reach statistical significance
(P>0.05), but the CDR3 length was longer at 45-nucleotides
compared with the control group (P<0.01) (Fig. 1K). The CDR3 length in the RT group
was lower than that in the control group (P<0.001; Fig. 1L). Additionally, the CDR3 length
distribution was significantly different between the RT +
anti-CTLA-4 and control groups (P<0.05, Fig. 1M). Furthermore, the CDR3 length in
the anti-CTLA-4 group was higher than that in the RT group
(P<0.05; Fig. 1N).
RT and anti-PD-1 therapy alters the
TCR repertoire
Alterations in TCRβ repertoire diversity
Next, the present study examined the TCRβ diversity
and complexity after RT and/or anti-PD-1 treatment. The D50, Gini,
inverse Simpson, Shannon and Simpson indices were used to evaluate
the TCRβ amino acid sequence diversity in the control, RT,
anti-PD-1 and RT + anti-PD-1 groups. The diversity of the in-frame
TCRβ sequences was first examined (Fig.
2). As shown in Fig. 2B, within
the radiation field (radiation irradiated tumors), the Gini index
in the RT and RT + anti-PD-1 groups was higher than that in the
control group (both P<0.01). Additionally, the Gini index in the
RT and RT + anti-PD-1 groups was higher than that in the anti-PD-1
group (both P<0.01). Additionally, although changes in other
parameters were observed between the treatment group and the
control group, these differences did not reach statistical
significance (P>0.05; Fig. 2A and
C-E). Next, the diversity of TCRβ outside the radiation field
(tumors that have not been irradiated with radiation) was examined.
The Gini index in the RT + anti-PD-1 group was higher than that in
the RT group (P<0.05; Fig. 2G).
Similar to within the radiation field, although changes in other
parameters between the treatment group and the control group could
be observed outside the radiation field, these differences did not
reach statistical significance (P>0.05; Fig. 2F and H-J). The differences in TCRβ
diversity in the post-selection repertoire were similar to those in
the pre-selection repertoire (Fig.
S1).
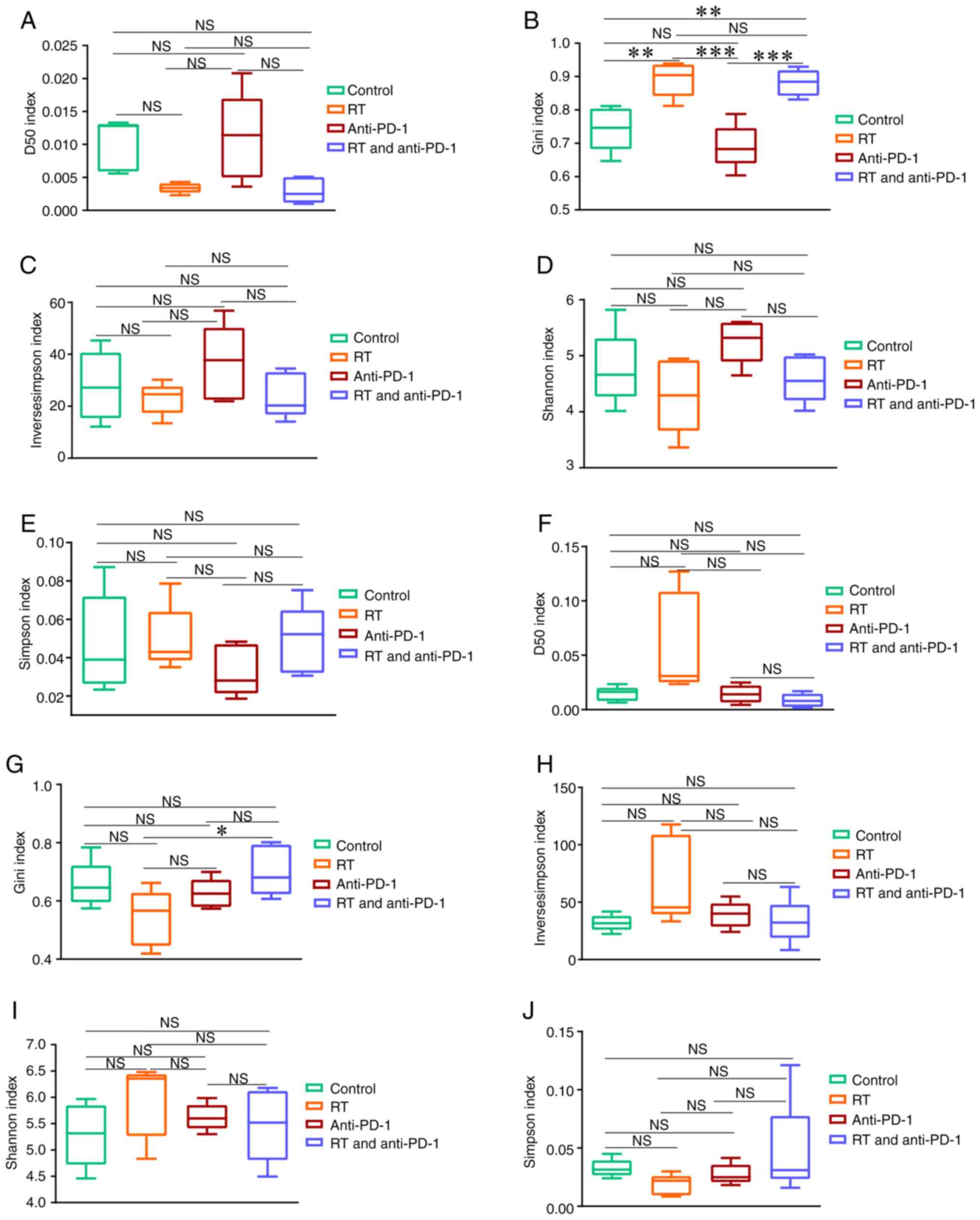 | Figure 2.Diversity indexes of the
post-selection repertoire in mice with RT and anti-PD-1 therapy,
and the control group. The diversity of the treatment area was
measured by (A) the D50 index, (B) Gini index, (C) Inverse Simpson
index, (D) Shannon index and (E) Simpson index. (F) The D50 index,
(G) Gini index, (H) Inverse Simpson index, (I) Shannon index and
(J) Simpson index were all used to evaluate the diversity of the
outside the RT field. Data are presented as the mean ± SD;
comparisons were made using one-way ANOVA corrected for multiple
comparisons using Bonferroni. *P<0.05, **P<0.01 and
***P<0.001 (two-tailed). PD-1, programmed cell death-1; NS, not
significant; RT, radiotherapy. |
Alterations in CDR3 length
distribution
The CDR3 length distribution within and outside the
radiation area was examined. The TCRβ CDR3 length within the
radiation area in the anti-PD-1 and RT + anti-PD-1 groups was
significantly longer than that in the control group (P<0.05;
Fig. 3A and C). Additionally, the
CDR3 length of the anti-PD-1 group was shorter than that of the RT
group (P<0.01; Fig. 3D). The
CDR3 length outside the radiation area in the anti-PD-1 group was
longer than that in the control group (P<0.001; Fig. 3E). In addition, whether within or
outside the radiation field, the difference in CDR3 length between
the RT group and the control group did not reach statistical
significance (P>0.05; Fig. 3 and
F). Furthermore, it was also observed that the CDR3 length was
similar outside the radiation field between the RT + anti-PD-1
group and the control group, as well as between the RT group and
the anti-PD-1 group (P>0.05; Fig. 3G
and H).
Analysis of the TCR profile of T cells
differentiated from the HSCs of mice exposed to various radiation
doses
The findings of the present study indicated that the
RT-immunotherapy combination decreased CDR3 diversity and altered
the CDR3 length. However, the potential reasons for these
alterations were not elucidated. Hence, the changes in the TCR
repertoire in the peripheral blood samples of NSG mice 6 months
after the transplantation of HSCs derived from
CBA/HmCherry mice exposed to different radiation doses
for 7 days were next investigated.
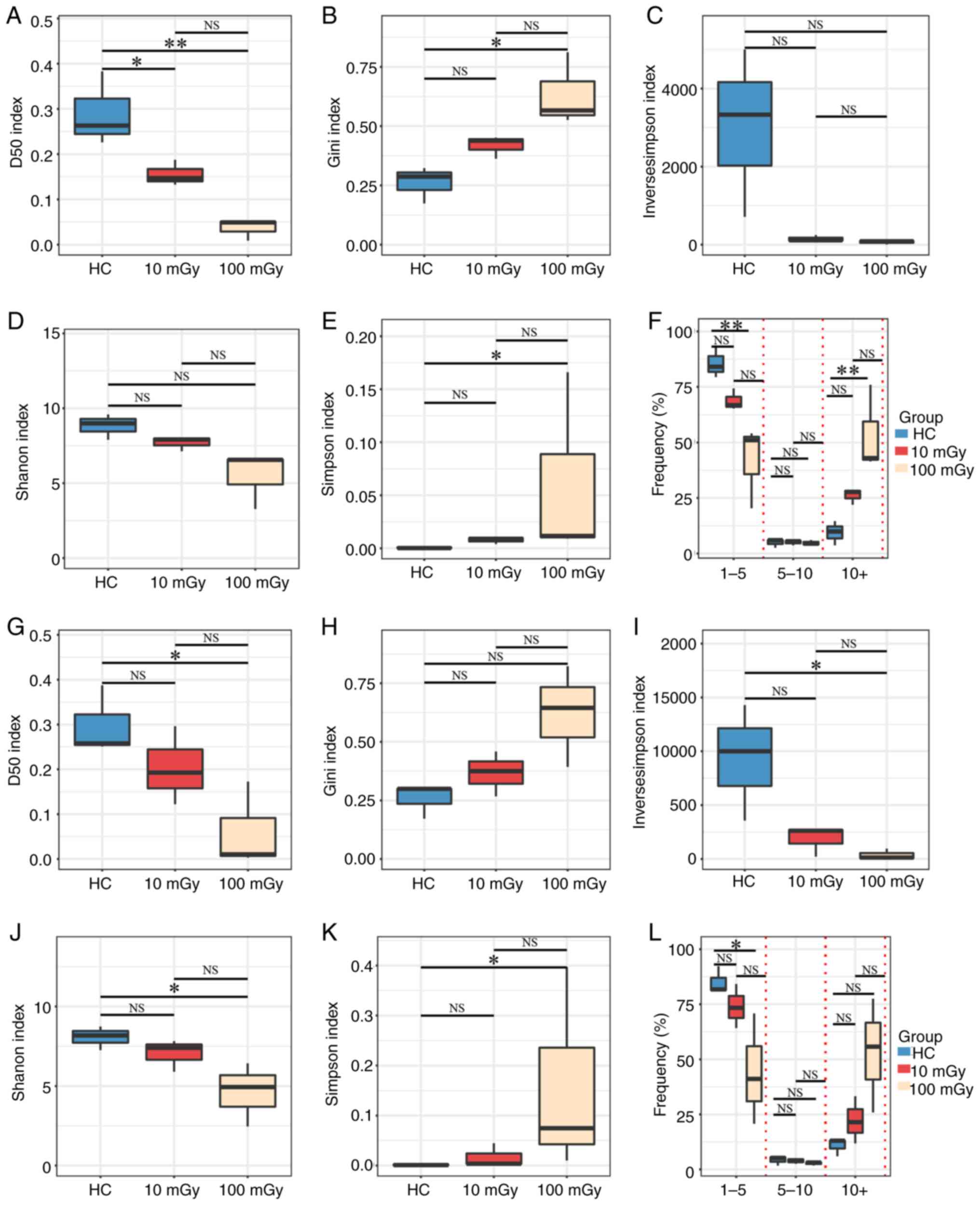
Diversity of the TCRβ repertoire
varies under different radiation doses
The D50 index, Gini index, inverse Simpson index,
Shannon index and Simpson index were examined to assess the impact
of different radiation doses on TCRβ diversity and nucleotide
sequence clonal expansion in the peripheral blood of NSG mice
following the aforementioned treatment. As shown in Fig. 4, the highest diversity was observed
in the HC group. The TCR diversity in the radiation (10 or 100
mGy)-exposed groups was lower than that in the HC group in both
post-selection (Fig. 4A-E) and
pre-selection repertoires (Fig.
4G-K). Additionally, a trend towards decreased TCR diversity
with increasing radiation dose was observed, although this trend
did not reach statistical significance.
Based on the read frequencies of unique TCRβ
nucleotide sequences (after correction), the TCRβ sequences were
categorized into the following three groups: Low-abundance (1–5
reads), medium-abundance (5–10 reads) and high-abundance (>10
reads) sequences, represented as percentages of the total sequences
for each TCRβ [Fig. 4F (in-frame)
and Fig. 4L (out-of-frame)]. The
average percentages of low-abundance and medium-abundance TCRβ
sequences were the highest in the HC group, followed by the 10
mGy-exposed and 100 mGy-exposed groups. Meanwhile, the average
percentage of high-abundance TCRβ sequences was the highest in the
100 mGy-exposed group, followed by the 10 mGy-exposed and HC
groups. However, the differences were not always significant
(P>0.05), particularly between the HC group and the 10
mGy-exposed group, and between the 10 mGy-exposed and 100
mGy-exposed groups. These results indicate that the HC group
exhibits the highest TCRβ diversity and the lowest clonality,
whereas the 100 mGy-exposed group exhibits the lowest TCRβ
diversity and the highest clonality.
Length distribution and overlap degree
of TCRβ CDR3
In the present study, the CDR3 length distribution
in the 10 mGy-exposed and 100 mGy-exposed groups was not
significantly different from that in the HC group, although some
changes were observed (Fig. 5A). To
understand the molecular mechanisms underlying the effect of
radiation on TCRs, six recombination positions (vDeletion,
d3Deletion, d5Deletion, jDeletion, n1Insertion and n2Insertion) for
both in-frame and out-of-frame insertion/deletion (InDels) events
were analyzed. The length distributions for the six recombination
events (indels) were similar between the 100 mGy-exposed and HC/10
mGy-exposed groups in the post-selection (Fig. 5B-G) and pre-selection repertoires
(Fig. S2). A previous study has
demonstrated that the TCRβ CDR3 length has been associated with the
degree of sequence sharing (34).
Thus, the overlap indices of TCRβ nucleotide sequences were
calculated for each group. Based on the shared degree of amino acid
sequences between samples, the samples were divided into the
following three groups: Private, co-owned by ≥2 and co-owned by ≥3
samples. The results indicated that, as the radiation dose
increased, the percentage of private sequences was significantly
increased and the degree of overlap was significantly decreased
(Fig. 5H-J).
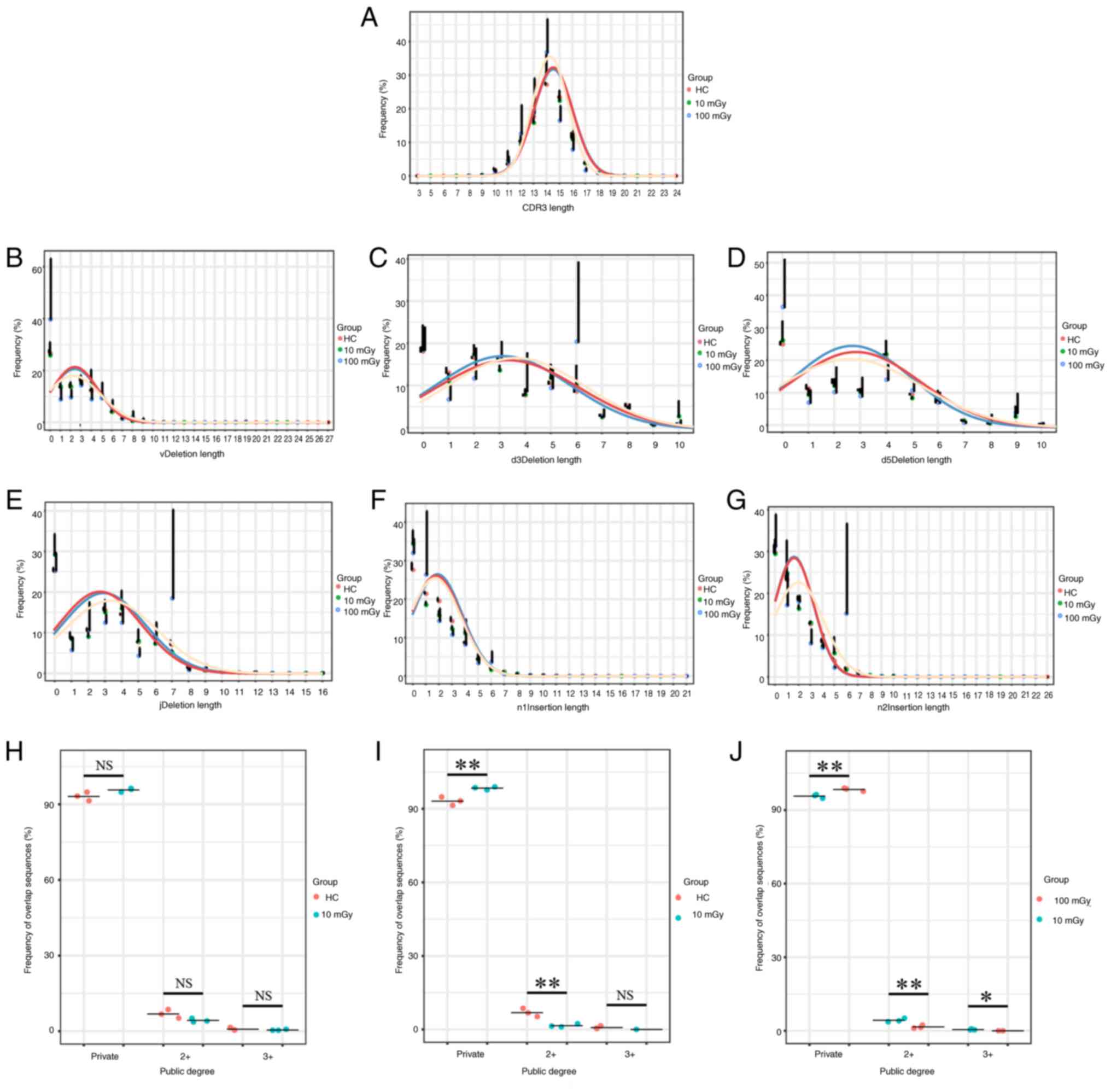 | Figure 5.Altered CDR3 length distribution,
InDel patterns and public degree in the RT group. (A) The changes
in the CDR3 length in the radiation group mice from the 10 and 100
mGy groups were compared with those of the HC group. (B-G) TCRβ
CDR3 sequences from the radiotherapy and HC groups in the
post-selection repertoire were analyzed for the frequency of
clonotypes with a specific number of InDels at each of the six
rearrangement sites: (B) vDdel, (C) d3Del, (D) d5Del, (E) jDel, (F)
n1Ins and (G) n2Ins. (H-J) Overlap indices were calculated from 9
HC, 9 mice with 10 mGy and 9 mice with 100 mGy, where (H) is the
comparison between HC group mice and mice receiving 10 mGy
radiation, (I) is the comparison between HC group mice and mice
receiving 100 mGy radiation and (J) is the comparison between mice
receiving 10 mGy radiation and mice receiving 100 mGy radiation.
Data are presented as the mean ± SD and the means between the two
groups were compared using the unpaired t-test or the Mann-Whitney
U test. One-way ANOVA or Kruskal-Wallis test was used to compare
group means across three or more groups and corrected for multiple
comparisons using Bonferroni. *P<0.05 and **P<0.01. TCRβ,
T-cell receptor β-chain; CDR3, complementarity-determining region
3; InDel, insertions and deletions; NS, not significant; HC,
healthy control. |
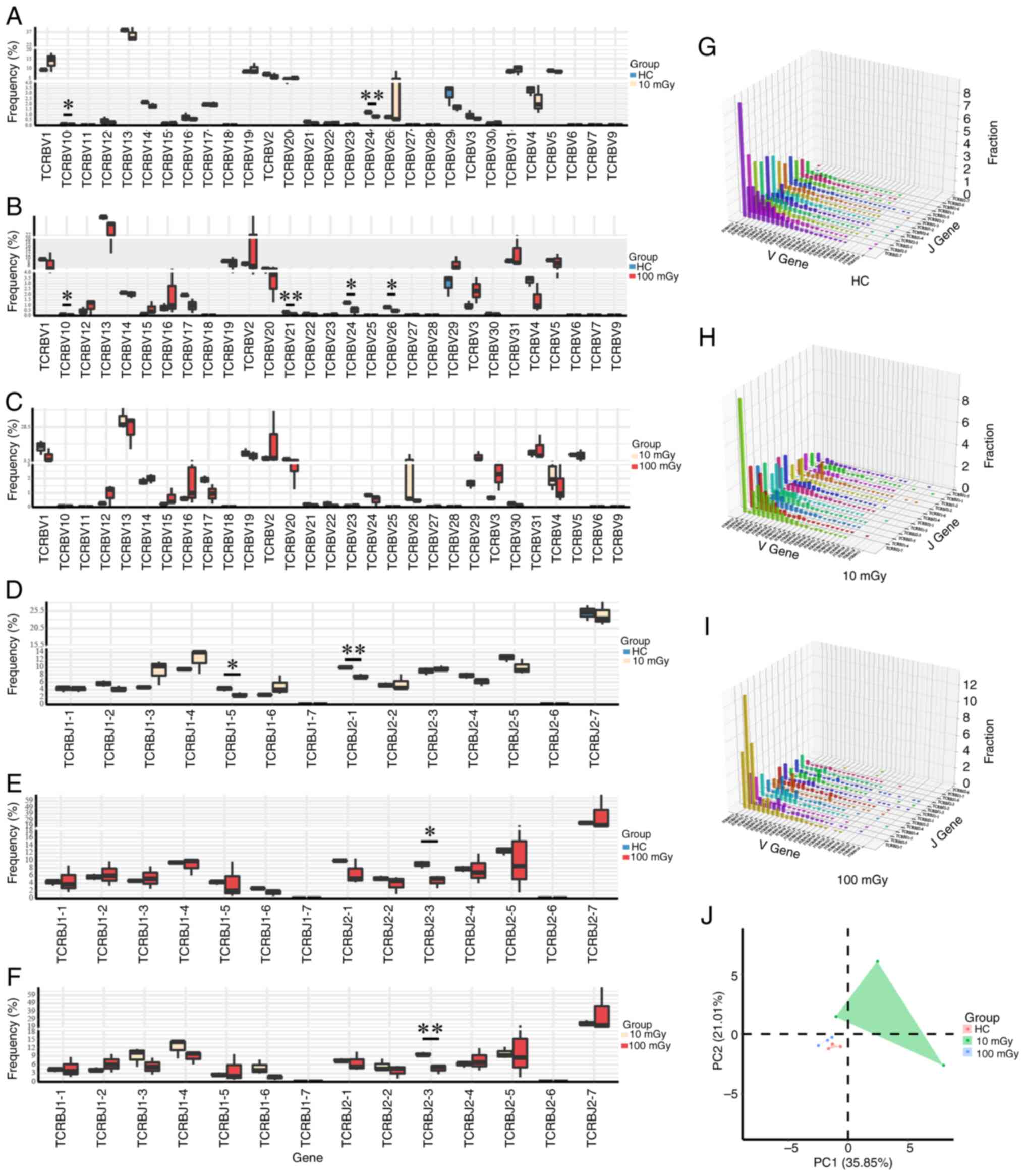
Radiation alters the usage frequency
of the TCRβ chain variable (TRBV) and TCRβ chain joining (TRBJ)
gene segments
To identify whether there was a skewed and
restricted VDJ segment usage following radiation, the usage
frequency of the TRBV and TRBJ genes in the radiation-exposed group
was compared with that in the HC group in both the post-selection
(Fig. 6A-F) and pre-selection
(Fig. S3A-F) repertoires. For the
post-selection repertoire, the usage frequency of TRBV10 and TRBV24
in the radiation-exposed groups was significantly lower than that
in the HC group. For the pre-selection repertoires, the usage
frequencies of TRBJ1-2, TRBJ1-5 and TRBJ1-6 in the
radiation-exposed groups were lower than that in the HC group, but
not all showed statistical differences. Furthermore, through PCA,
the radiation-exposed mice can be distinguished from the HC mice
based on the usage of TRBV genes, in the post-selection (Fig. 6J) and pre-selection (Fig. S3J) repertoires. Furthermore, the
three-dimensional landscapes of TRBV and TRBJ gene usage were
plotted. In the post-selection (Fig.
6G-I) repertoires, the top three V-J combinations in the 10
mGy-exposed group were TRBV13-TRBJ2-7, TRBV13-TRBJ1-4 and
TRBV13-TRBJ2-5, while those in the 100 mGy-exposed group were
TRBV13-TRBJ2-7, TRBV2-TRBJ2-7 and TRBV31-TRBJ2-7. The top three V-J
combinations in the HC group were TRBV13-TRBJ2-7, TRBV13-TRBJ2-5
and TRBV13-TRBJ2-1 (Fig. 6G).
Additionally, in the pre-selection (Fig. S3G-I) repertoires, the top three V-J
combinations in the 10 mGy-exposed group were TRBV31-TRBJ2-4,
TRBV13-TRBJ2-7 and TRBV13-TRBJ2-1, while those in the 100
mGy-exposed group were TRBV13-TRBJ2-7, TRBV5-TRBJ1-4 and
TRBV13-TRBJ2-4. The top three V-J combinations in the HC group were
TRBV13-TRBJ2-7, TRBV4-TRBJ2-7 and TRBV13-TRBJ2-5 (Fig. S3G).
Discussion
At present, cancer remains a primary public health
problem. According to the International Agency for Research on
Cancer, 19.88 million new global cancer cases and 9.74 million
cancer-related deaths were reported in 2022 (3). RT is one of the most important
treatment strategies for cancer (40). The clinical application of various
immunomodulatory drugs has contributed to the field of cancer
immunotherapy, achieving notable progress in the design of
effective immunotherapy regimens. According to relevant research
reports, there is a correlation between neoantigen burden and
clinical benefits when using CTLA-4 blocking monoclonal antibodies
to treat patients with melanoma (41,42),
and similar associations have also been reported in patients with
lung cancer receiving anti-PD-1 treatment (43). However, to the best of our
knowledge, there is currently no systematic understanding of the
heterogeneity in immune cells following the use of these treatment
methods for cancer. The present study analyzed the TCRβ repertoire
from a public dataset of tumor tissues from mice treated with RT
and/or anti-PD-1/anti-CTLA-4 therapies, as well as in peripheral
blood mononuclear cells of NSG mice 6 months after transplantation
with HSCs isolated from CBA/HmCherry mice exposed to
radiation doses of 0, 10 and 100 mGy for 7 days.
Analyses of the TCRβ repertoire of the RT and
anti-CTLA-4 treated cancer samples revealed that the diversity of
the TCRβ repertoire in the RT and RT + anti-CTLA-4 groups was
significantly lower than that in the control group, especially in
the RT + anti-CTLA-4 group. Although not all differences were
significant, some changes were observed. This may be due to the
synergistic inhibitory effects of RT and anti-CTLA-4 therapy on T
cells. The arrest of T cells in contact with tumor cells is
dependent on major histocompatibility complex class I (44). A previous study indicated that the
TCRβ repertoire diversity is positively associated with the
polymorphism of human leukocyte antigen class I loci (45) and the present study findings further
confirmed these conclusions. Furthermore, a short biased TCRβ CDR3
length may lead to an increased degree of sequence sharing as short
sequences (with limited variability in sequence combinations)
increase the probability of two TCRβ CDR3 sequences being
coincidentally identical. The present study results are contrary to
those presented by Rudqvist et al (26), who reported that the overlap between
mice receiving RT combined with anti-CTLA-4 therapy was
significantly reduced compared with mice receiving RT alone.
TCR diversity is key for neoantigen recognition
(46,47). In the present study, compared with
the control group, the TCRβ repertoire diversity of the in-field
tumors was lower in the RT and RT + anti-PD-1 groups, although not
all differences were statistically significant. However, the TCRβ
repertoire diversity in the anti-PD-1 group was comparable with
that in the control group. Additionally, analysis of the diversity
of the TCRβ repertoire in the out-of-field tumors revealed that the
TCRβ repertoire diversity in the RT group was higher than that in
the control and RT + anti-PD-1 groups. RT can enhance the mobility
of T cells toward the localized treatment site of tumors and
augment existing anticancer T cell responses. The combination of RT
and anti-PD-1 therapy facilitates the regression of both local and
distant tumor lesions (27). A
recent study reported that the combination of RT and anti-PD-1
therapy can promote the circulation of T cell clones between the
peripheral blood and tumor tissues (48). Peripheral blood serves as a
reservoir for T cell clones, which contributes to the diversity of
the TCR repertoire of tumor tissue. We hypothesize that the
diversity of the TCR in the in-field tumors of the RT and RT +
anti-PD-1 groups was lower than that in the control group due to
the RT-induced potent local inflammatory response against tumor
tissue. This leads to an enhanced antitumor immune response and
increased utilization of the TCR. Additionally, the antitumor
immune response of off-site tumors may be slightly delayed compared
with on-site tumors. A key feature of the TCRβ repertoire is the
distribution of the CDR3 length. Epitope-specific T cell
repertoires often have a bias in CDR3 length (37,49,50).
Compared with the control group, the clonotypes in the in-field
tumors of the anti-PD-1 and the RT + anti-PD-1 groups shifted
toward longer clonotypes. However, the CDR3 length in the RT group
was comparable with that in the control group. Consistent with the
CDR3 length in some in-field tumors, the CDR3 length in the
anti-PD-1 group was longer than that in the control group in the
out-of-field tumors. These results may be due to the random
insertion of a large number of nucleotides into the CDR3 region in
the presence of terminal deoxynucleotidase after cancer treatment,
which increases CDR3 length (16).
The diverse TCRs provide protection to the human
body, shielding it from various external pathogens and internal
cancer cells (51). However, in the
present study, further investigation of the TCRβ repertoire
characteristics in mice exposed to different radiation doses
revealed that, as the radiation dose increased, the TCRβ diversity
in mice decreased while the degree of clonal expansion increased.
This may be associated with immune system damage caused by reduced
TCRβ diversity after radiation exposure, further confirming the
findings by Hollingsworth et al (52), in which it was reported that
radiation damage leads to impaired immune response and a decrease
in T cell repertoire diversity. Additionally, in the present study,
the CDR3 length and InDel length distribution were not
significantly different between the radiation-exposed and HC
groups. Furthermore, as the radiation dose increased, the
percentage of private sequences was significantly increased and the
degree of overlapping sequences was significantly decreased. The
shared TCRβ clonotypes among individuals is associated with
enhanced cross-reactivity toward potentially related antigens,
which is considered key for pathogen-specific responses and
infection control (53–56). Furthermore, in the present study,
the usage frequencies of TRBV/J segments significantly varied
between the radiation-exposed and HC groups in both the
pre-selection and post-selection repertoires. The biased usage of
TRBV/J segments may result from radiation-induced DNA rearrangement
(57). Upon radiation exposure,
activated T cells undergo clonal expansion to maintain
physiological functions. However, immune cell composition remains
stable. An increase in the number of dominant T cells can suppress
the proliferation of other T cells (58). Therefore, radiation exposure may
have increased the proportions of some TRBV/J genes but decreased
the proportion of other genes.
The results of the present study indicated that
different treatments decreased the TCRβ repertoire diversity of
mice. RT may be the main factor contributing to this decreased
diversity of the TCRβ repertoire. The decreased TCRβ diversity may
weaken the ability of the immune system to recognize and clear
tumor cells, affecting treatment efficacy. RT may affect the
diversity of TCRβ repertoire through different mechanisms. First,
radiation may directly damage and kill the lymphocytes in the TME,
reduce the number of T cells and consequently decrease TCRβ
diversity (59). Second, radiation
may affect thymic function. The thymus is the site for T cell
development and maturation. RT-induced thymic tissue damage can
adversely affect T cell production and TCRβ rearrangement, which
decreases TCRβ diversity (60).
Third, RT may induce alterations in the TME, such as eliciting
inflammatory responses and increasing the number of
immunosuppressive cells. These changes may adversely affect the
infiltration and function of T cells, indirectly decreasing the
diversity of TCRβ (61,62). Future studies should consider
reducing the negative impact of RT on TCRβ diversity through the
optimization of RT. Radiation dose may be decreased to a level that
minimizes direct damage to lymphocytes without compromising
treatment efficacy (63). A
previous study has demonstrated that fractionated low-dose RT is
efficacious for the activation of the immune system, mitigating the
negative impact on TCRβ diversity (63). Furthermore, adoptive T cell therapy
can be used after RT. Adoptive T cell therapy involves re-infusing
tumor-specific T cells expanded in vitro into the body of
the patient to enhance TCRβ diversity and antitumor activity
(64). The management of TCRβ
diversity is dependent on multiple factors, such as tumor type,
staging and patient immune status. Thus, different strategies can
be employed to overcome these issues. For example, thymus function
can be enhanced. The thymus is a key organ for T cell development
and diversification. The improvement of the thymic microenvironment
(such as supplementing IL-7, growth hormone and thymosin) or the
suppression of age-related thymic atrophy will promote the
generation of initial T cells and the maintenance of TCR repertoire
diversity (65–67). Furthermore, strategies can be
adopted to regulate gut microbiota. The gut microbiota affects T
cell differentiation by modulating the immune microenvironment
(68). For example, probiotics or
fecal transplantation may indirectly enrich the TCR repertoire
through antigen cross-reactivity (69). Treatment effectiveness may vary
depending on the type of tumor, individual patient and treatment
plan, highlighting the importance of developing personalized
treatment plans. Treatment plans for patients with cancer can be
personalized to achieve optimal therapeutic effects using two
strategies: i) TCR sequencing technology can be used to analyze the
TCRβ diversity of the patient and evaluate their immune status
(70,71); and ii) the identification of
biomarkers associated with TCRβ diversity can be used to predict
the efficacy of RT and immunotherapy (72,73).
The present study has several limitations. The
sample size was small; hence, the conclusions of the present study
need to be validated using a larger sample size. Additionally, the
types of cancer involved in the present study are limited and
future research needs to be conducted on more categories of cancer
samples to confirm the conclusions. Furthermore, this was a
cross-sectional study that evaluated the TCRβ repertoire at a
specific time point. The dynamic changes of the TCRβ repertoire
during treatment should be evaluated in the future, which will aid
in the identification of the optimal treatment window. Finally, TCR
sequencing, which was used to determine TCR rearrangement, does not
fully represent the functionality of T cells. Therefore, future
research needs to combine more functional experiments, such as i)
Cytokine release experiments: detecting the types and quantities of
cytokines released by T cells after stimulation to evaluate the
activation status of T cells; and ii) cytotoxicity assay: Evaluate
the ability of T cells to kill tumor cells.
In conclusion, the present study demonstrated that
the TCRβ repertoire may serve as an ideal predictive biomarker for
the efficacy of combination therapies. In the future, further
research should be conducted on: i) The molecular mechanisms by
which the CDR3 length is significantly shorter under RT and
significantly longer under immunotherapy, as well as the role of
changes in CDR3 length in tumor treatment and prognosis; and ii)
identifying methods that can be used to maintain and improve the
diversity of TCR profiles in RT and immunotherapy. Future studies
may explore and evaluate the potential mechanisms underlying
antitumor immune responses to address key challenges in clinical
treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Guangxi
Natural Science Foundation (grant no. 2024GXNSFAA 010096),
Innovation Training Program for College Students (grant nos.
202410601013 and S202410601182), Guilin Science Research and
Technology Development Project (grant nos. 20210218-2 and
20220139-13-2) and Guangxi Key Laboratory of Tumor Immunology and
Microenvironmental Regulation (grant nos. 2023KF006, 3030302213 and
2021KF001).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XY and WW curated the data, performed the formal
analysis, prepared and wrote the original draft and analyzed and
interpreted the data. XY and WW confirm the authenticity of all the
raw data. ZF, BZ, YL and FY participated in data analysis and
reviewed and edited the manuscript. CZ and MO reviewed and edited
the manuscript as well as supervised and conceptualized the present
study. SL conceptualized the present study, performed the formal
analysis, designed the present study methodology and supervised and
aided with the visualization of data. XH conceptualized the present
study, performed the formal analysis, obtained funding, designed
methodology and supervised and conceived the present study. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RT
|
radiotherapy
|
|
TCRβ
|
T-cell receptor β chain
|
|
TME
|
tumor microenvironment
|
|
CDR3
|
complementarity-determining region
3
|
|
CTLA-4
|
cytotoxic T lymphocyte-associated
antigen-4
|
|
PD-1
|
programmed cell death-1
|
|
HSC
|
hematopoietic stem cell
|
|
NSG
|
NOD scid γ
|
|
TRBV
|
TCR β chain variable gene
|
|
TRBJ
|
TCR β chain joining gene
|
References
|
1
|
Chen Z, Ma Y, Hua J, Wang Y and Guo H:
Impacts from economic development and environmental factors on life
expectancy: A comparative study based on data from both developed
and developing countries from 2004 to 2016. Int J Environ Res
Public Health. 18:85592021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tchkonia T, Palmer AK and Kirkland JL: New
horizons: Novel approaches to enhance healthspan through targeting
cellular senescence and related aging mechanisms. J Clin Endocrinol
Metab. 106:e1481–e1487. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao W, Qin K, Li F and Chen W:
Socioeconomic inequalities in cancer incidence and mortality: An
analysis of GLOBOCAN 2022. Chin Med J (Engl). 137:1407–1413. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng R, Zhang S, Zeng H, Wang S, Sun K,
Chen R, Li L, Wei W and He J: Cancer incidence and mortality in
China, 2016. J Natl Cancer Cent. 2:1–9. 2022.PubMed/NCBI
|
|
6
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhattacharya S and Asaithamby A:
Repurposing DNA repair factors to eradicate tumor cells upon
radiotherapy. Transl Cancer Res. 6 (Suppl 5):S822–S839. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mortezaee K and Najafi M: Immune system in
cancer radiotherapy: Resistance mechanisms and therapy
perspectives. Crit Rev Oncol Hematol. 157:1031802021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watanabe T, Sato GE, Yoshimura M, Suzuki M
and Mizowaki T: The mutual relationship between the host immune
system and radiotherapy: Stimulating the action of immune cells by
irradiation. Int J Clin Oncol. 28:201–208. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hoffman RM: Back to the future: Are
tumor-targeting bacteria the next-generation cancer therapy?
Methods Mol Biol. 1317:239–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dewan MZ, Galloway AE, Kawashima N,
Dewyngaert JK, Babb JS, Formenti SC and Demaria S: Fractionated but
not single-dose radiotherapy induces an immune-mediated abscopal
effect when combined with anti-CTLA-4 antibody. Clin Cancer Res.
15:5379–5388. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Verbrugge I, Hagekyriakou J, Sharp LL,
Galli M, West A, McLaughlin NM, Duret H, Yagita H, Johnstone RW,
Smyth MJ and Haynes NM: Radiotherapy increases the permissiveness
of established mammary tumors to rejection by immunomodulatory
antibodies. Cancer Res. 72:3163–3174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hou X, Yang Y, Chen J, Jia H, Zeng P, Lv
L, Lu Y, Liu X and Diao H: TCRβ repertoire of memory T cell reveals
potential role for Escherichia coli in the pathogenesis of primary
biliary cholangitis. Liver Int. 39:956–966. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou XL, Wang L, Ding YL, Xie Q and Diao
HY: Current status and recent advances of next generation
sequencing techniques in immunological repertoire. Genes Immun.
17:153–164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou X, Lu C, Chen S, Xie Q, Cui G, Chen J,
Chen Z, Wu Z, Ding Y, Ye P, et al: High throughput sequencing of T
cell antigen receptors reveals a conserved TCR repertoire. Medicine
(Baltimore). 95:e28392016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hou X, Wang M, Lu C, Xie Q, Cui G, Chen J,
Du Y, Dai Y and Diao H: Analysis of the repertoire features of TCR
beta chain CDR3 in human by high-throughput sequencing. Cell
Physiol Biochem. 39:651–667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zvyagin IV, Pogorelyy MV, Ivanova ME,
Komech EA, Shugay M, Bolotin DA, Shelenkov AA, Kurnosov AA,
Staroverov DB, Chudakov DM, et al: Distinctive properties of
identical twins' TCR repertoires revealed by high-throughput
sequencing. Proc Natl Acad Sci USA. 111:5980–5985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Larimore K, McCormick MW, Robins HS and
Greenberg PD: Shaping of human germline IgH repertoires revealed by
deep sequencing. J Immunol. 189:3221–3230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robins HS, Campregher PV, Srivastava SK,
Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH and Carlson
CS: Comprehensive assessment of T-cell receptor beta-chain
diversity in alphabeta T cells. Blood. 114:4099–4107. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robins HS, Srivastava SK, Campregher PV,
Turtle CJ, Andriesen J, Riddell SR, Carlson CS and Warren EH:
Overlap and effective size of the human CD8+ T cell receptor
repertoire. Sci Transl Med. 2:47ra642010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Ji Z and Smith KN: Analysis of
TCR β CDR3 sequencing data for tracking anti-tumor immunity.
Methods Enzymol. 629:443–464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li S, Sun J, Allesøe R, Datta K, Bao Y,
Oliveira G, Forman J, Jin R, Olsen LR, Keskin DB, et al: RNase
H-dependent PCR-enabled T-cell receptor sequencing for highly
specific and efficient targeted sequencing of T-cell receptor mRNA
for single-cell and repertoire analysis. Nat Protoc. 14:2571–2594.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baumeister SH, Freeman GJ, Dranoff G and
Sharpe AH: Coinhibitory pathways in immunotherapy for cancer. Annu
Rev Immunol. 34:539–573. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Candéias SM, Mika J, Finnon P, Verbiest T,
Finnon R, Brown N, Bouffler S, Polanska J and Badie C: Low-dose
radiation accelerates aging of the T-cell receptor repertoire in
CBA/Ca mice. Cell Mol Life Sci. 74:4339–4351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rudqvist NP, Pilones KA, Lhuillier C,
Wennerberg E, Sidhom JW, Emerson RO, Robins HS, Schneck J, Formenti
SC and Demaria S: Radiotherapy and CTLA-4 blockade shape the TCR
repertoire of tumor-infiltrating T cells. Cancer Immunol Res.
6:139–150. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dovedi SJ, Cheadle EJ, Popple AL, Poon E,
Morrow M, Stewart R, Yusko EC, Sanders CM, Vignali M, Emerson RO,
et al: Fractionated radiation therapy stimulates antitumor immunity
mediated by both resident and infiltrating polyclonal T-cell
populations when combined with PD-1 blockade. Clin Cancer Res.
23:5514–5526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Robins H, Desmarais C, Matthis J,
Livingston R, Andriesen J, Reijonen H, Carlson C, Nepom G, Yee C
and Cerosaletti K: Ultra-sensitive detection of rare T cell clones.
J Immunol Methods. 375:14–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carlson CS, Emerson RO, Sherwood AM,
Desmarais C, Chung MW, Parsons JM, Steen MS,
LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, et al:
Using synthetic templates to design an unbiased multiplex PCR
assay. Nat Commun. 4:26802013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bolotin DA, Shugay M, Mamedov IZ,
Putintseva EV, Turchaninova MA, Zvyagin IV, Britanova OV and
Chudakov DM: MiTCR: Software for T-cell receptor sequencing data
analysis. Nat Methods. 10:813–814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Elhanati Y, Sethna Z, Callan CG Jr, Mora T
and Walczak AM: Predicting the spectrum of TCR repertoire sharing
with a data-driven model of recombination. Immunol Rev.
284:167–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pogorelyy MV, Minervina AA, Chudakov DM,
Mamedov IZ, Lebedev YB, Mora T and Walczak AM: Method for
identification of condition-associated public antigen receptor
sequences. Elife. 7:e330502018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Murugan A, Mora T, Walczak AM and Callan
CG Jr: Statistical inference of the generation probability of
T-cell receptors from sequence repertoires. Proc Natl Acad Sci USA.
109:16161–16166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gomez-Tourino I, Kamra Y, Baptista R,
Lorenc A and Peakman M: T cell receptor β-chains display abnormal
shortening and repertoire sharing in type 1 diabetes. Nat Commun.
8:17922017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hou X, Zeng P, Chen J and Diao H: No
difference in TCRβ repertoire of CD4+ naive T cell between patients
with primary biliary cholangitis and healthy control subjects. Mol
Immunol. 116:167–173. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hou X, Zeng P, Zhang X, Chen J, Liang Y,
Yang J, Yang Y, Liu X and Diao H: Shorter TCR β-chains are highly
enriched during thymic selection and antigen-driven selection.
Front Immunol. 10:2992019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang C, Li X, Wu J, Zhang W, Sun S, Lin
L, Wang X, Li H, Wu X, Zhang P, et al: The landscape and diagnostic
potential of T and B cell repertoire in immunoglobulin A
nephropathy. J Autoimmun. 97:100–107. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Zhu H, Zhang Q and Zhao Y: Clonal
analysis of peripheral blood T cells in patients with hashimoto's
thyroiditis at different stages using TCR sequencing.
Immunobiology. 230:1528902025. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Venturi V, Kedzierska K, Turner SJ,
Doherty PC and Davenport MP: Methods for comparing the diversity of
samples of the T cell receptor repertoire. J Immunol Methods.
321:182–195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Z, Tang Y, Tan Y, Wei Q and Yu W:
Cancer-associated fibroblasts in radiotherapy: Challenges and new
opportunities. Cell Commun Signal. 17:472019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Van Allen EM, Miao D, Schilling B, Shukla
SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger
SM, et al: Genomic correlates of response to CTLA-4 blockade in
metastatic melanoma. Science. 9:207–211. 2015. View Article : Google Scholar
|
|
42
|
Chan TA, Wolchok JD and Snyder A: Genetic
basis for clinical response to CTLA-4 blockade in melanoma. N Engl
J Med. 12:19842015. View Article : Google Scholar
|
|
43
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 3:124–128. 2015.
View Article : Google Scholar
|
|
44
|
Ruocco MG, Pilones KA, Kawashima N, Cammer
M, Huang J, Babb JS, Liu M, Formenti SC, Dustin ML and Demaria S:
Suppressing T cell motility induced by anti-CTLA-4 monotherapy
improves antitumor effects. J Clin Invest. 122:3718–3730. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Krishna C, Chowell D, Gönen M, Elhanati Y
and Chan TA: Genetic and environmental determinants of human TCR
repertoire diversity. Immun Ageing. 17:262020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Riaz N, Havel JJ, Makarov V, Desrichard A,
Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH,
et al: Tumor and microenvironment evolution during immunotherapy
with nivolumab. Cell. 171:934–949.e916. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hebeisen M, Allard M, Gannon PO, Schmidt
J, Speiser DE and Rufer N: Identifying individual T cell receptors
of optimal avidity for tumor antigens. Front Immunol. 6:5822015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yan C, Ma X, Guo Z, Wei X, Han D, Zhang T,
Chen X, Cao F, Dong J, Zhao G, et al: Time-spatial analysis of T
cell receptor repertoire in esophageal squamous cell carcinoma
patients treated with combined radiotherapy and PD-1 blockade.
Oncoimmunology. 11:20256682022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pickman Y, Dunn-Walters D and Mehr R: BCR
CDR3 length distributions differ between blood and spleen and
between old and young patients, and TCR distributions can be used
to detect myelodysplastic syndrome. Phys Biol. 10:0560012013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu S, Hou XL, Sui WG, Lu QJ, Hu YL and
Dai Y: Direct measurement of B-cell receptor repertoire's
composition and variation in systemic lupus erythematosus. Genes
Immun. 18:22–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Weng NP: Numbers and odds: TCR repertoire
size and its age changes impacting on T cell functions. Semin
Immunol. 69:1018102023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hollingsworth BA, Aldrich JT, Case CM Jr,
DiCarlo AL, Hoffman CM, Jakubowski AA, Liu Q, Loelius SG, PrabhuDas
M, Winters TA and Cassatt DR: Immune dysfunction from radiation
exposure. Radiat Res. 200:396–416. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Price DA, Asher TE, Wilson NA, Nason MC,
Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK,
Roederer M, et al: Public clonotype usage identifies protective
Gag-specific CD8+ T cell responses in SIV infection. J Exp Med.
206:923–936. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Venturi V, Quigley MF, Greenaway HY, Ng
PC, Ende ZS, McIntosh T, Asher TE, Almeida JR, Levy S, Price DA, et
al: A mechanism for TCR sharing between T cell subsets and
individuals revealed by pyrosequencing. J Immunol. 186:4285–4294.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhao Y, Nguyen P, Ma J, Wu T, Jones LL,
Pei D, Cheng C and Geiger TL: Preferential use of public TCR during
autoimmune encephalomyelitis. J Immunol. 196:4905–4914. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hou X, Wang G, Fan W, Chen X, Mo C, Wang
Y, Gong W, Wen X, Chen H, He D, et al: T-cell receptor repertoires
as potential diagnostic markers for patients with COVID-19. Int J
Infect Dis. 113:308–317. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Forrester HB and Radford IR: Ionizing
radiation-induced chromosomal rearrangements occur in
transcriptionally active regions of the genome. Int J Radiat Biol.
80:757–767. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sui W, Hou X, Zou G, Che W, Yang M, Zheng
C, Liu F, Chen P, Wei X, Lai L and Dai Y: Composition and variation
analysis of the TCR β-chain CDR3 repertoire in systemic lupus
erythematosus using high-throughput sequencing. Mol Immunol.
67:455–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kuehm LM, Wolf K, Zahour J, DiPaolo RJ and
Teague RM: Checkpoint blockade immunotherapy enhances the frequency
and effector function of murine tumor-infiltrating T cells but does
not alter TCRβ diversity. Cancer Immunol Immunother. 68:1095–1106.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Valença-Pereira F, Sheridan RM, Riemondy
KA, Thornton T, Fang Q, Barret B, Paludo G, Thompson C, Collins P,
Santiago M, et al: Inactivation of GSK3β by Ser(389)
phosphorylation prevents thymocyte necroptosis and impacts Tcr
repertoire diversity. Cell Death Differ. 32:880–898. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kershaw MH, Devaud C, John LB, Westwood JA
and Darcy PK: Enhancing immunotherapy using chemotherapy and
radiation to modify the tumor microenvironment. Oncoimmunology.
2:e259622013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Huang J and Li JJ: Multiple dynamics in
tumor microenvironment under radiotherapy. Adv Exp Med Biol.
1263:175–202. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
D'Alonzo RA, Keam S, Gill S, Rowshanfarzad
P, Nowak AK, Ebert MA and Cook AM: Fractionated low-dose
radiotherapy primes the tumor microenvironment for immunotherapy in
a murine mesothelioma model. Cancer Immunol Immunother. 74:442025.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Pituch KC, Miska J, Krenciute G, Li G,
Panek WK, Gottschalk S and Balyasnikova IV: P06.01 Functional
analysis of IL13Rα2-specific chimeric antigen receptor T cells in
immunocompetent mouse of glioblastoma. Neuro Oncol. 19:iii48–iii49.
2017. View Article : Google Scholar
|
|
65
|
Hsieh J, Ng S, Bosinger S, Wu JH, Tharp
GK, Garcia A, Hossain MS, Yuan S, Waller EK and Galipeau J: A GMCSF
and IL7 fusion cytokine leads to functional thymic-dependent T-cell
regeneration in age-associated immune deficiency. Clin Transl
Immunology. 4:e372015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Savino W, Mendes-da-Cruz DA, Lepletier A
and Dardenne M: Hormonal control of T-cell development in health
and disease. Nat Rev Endocrinol. 12:77–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Besman M, Zambrowicz A and Matwiejczyk M:
Review of thymic peptides and hormones: From their properties to
clinical application. International Journal of Peptide Research and
Therapeutics. 31:102024. View Article : Google Scholar
|
|
68
|
Kim KS: Regulation of T cell repertoires
by commensal microbiota. Front Cell Infect Microbiol.
12:10043392022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bessell CA, Isser A, Havel JJ, Lee S, Bell
DR, Hickey JW, Chaisawangwong W, Glick Bieler J, Srivastava R, Kuo
F, et al: Commensal bacteria stimulate antitumor responses via T
cell cross-reactivity. JCI Insight. 5:e1355972020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fang M, Miao Y, Zhu L, Mei Y, Zeng H, Luo
L, Ding Y, Zhou L, Quan X, Zhao Q, et al: Age-related dynamics and
spectral characteristics of the tcrβ repertoire in healthy
children: Implications for immune aging. Aging Cell. 24:e144602025.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen M, Su Z and Xue J: Targeting T-cell
aging to remodel the aging immune system and revitalize geriatric
immunotherapy. Aging Dis. 15:10.14336/AD.2025.0061. 2025.
|
|
72
|
Han J, Duan J, Bai H, Wang Y, Wan R, Wang
X, Chen S, Tian Y, Wang D, Fei K, et al: TCR repertoire diversity
of peripheral PD-1(+)CD8(+) T cells predicts clinical outcomes
after immunotherapy in patients with non-small cell lung cancer.
Cancer Immunol Res. 8:146–154. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang J, Bie Z, Zhang Y, Li L, Zhu Y, Zhang
Y, Nie X, Zhang P, Cheng G, Di X, et al: Prognostic value of the
baseline circulating T cell receptor β chain diversity in advanced
lung cancer. Oncoimmunology. 10:18996092021. View Article : Google Scholar : PubMed/NCBI
|