Introduction
Lung cancer is the most frequently diagnosed
malignant tumor and a major cause of cancer-related death globally
(1). Non-small cell lung cancer
(NSCLC) constitutes >85% of all lung cancer cases, which can be
further classified as lung adenocarcinoma (LUAD), large cell
carcinoma and lung squamous cell carcinoma (LUSC) (2,3). In
recent years, surgery, chemotherapy and medication have been the
main treatment strategies for lung cancer, but the prognosis for
patients with this disease remains poor (3). To improve patient survival, the
development of new anticancer drugs with lower toxicity and fewer
side effects are required for the treatment of lung cancer.
Tanshinone IIA (TSIIA) is a natural extract derived
from Danshen (Salviae miltiorrhizae Bunge). Sodium TSIIA
sulfate (STS) is an aqueous solution of TSIIA after sulfonation and
is the sodium sulfate salt form of TSIIA. Compared with the
lipophilic properties of TSIIA, the introduction of the sulfate
group in STS enhances its solubility in aqueous media, which
improves its absorption and distribution within the body. Despite
the structural differences between the molecules, STS, as a
derivative of TSIIA, retains certain pharmacological activities of
TSIIA, such as anti-inflammatory, antioxidant and antitumor effects
(4). As previously reported, TSIIA
exerts antitumor effects on various malignant tumors, such as
esophageal, colorectal, prostate and gastric cancer (4,5).
Notably, the therapeutic effects of TSIIA on lung cancer have been
reported, and TSIIA markedly decreases NSCLC cell viability and
colony formation (6). In addition,
STS treatment has been shown to reduce LUAD cell malignant
behaviors (7). However, the precise
mechanism by which TSIIA functions in lung cancer remains
unclear.
Histone acetylation is a specific type of
modification in which the lysine residue at the tail of a histone
is acetylated and deacetylated by histone acetyltransferase (HAT)
and histone deacetylase (HDAC), respectively (8). Alterations in histone acetylation are
closely related to cancer progression (9). For instance, upregulation of histone
H3 lysine 18 acetylation (H3K18ac) inhibits lung cancer cell
viability, colony formation and migration (10). Additionally, celestrol inhibits lung
cancer growth by inducing histone acetylation and synergistically
acting with a HDAC inhibitor (11).
As previously reported, TSIIA protects against cerebral ischemia
reperfusion injury by upregulating H3K18ac and H4K8ac (12). Therefore, it could be suggested that
TSIIA may inhibit lung cancer development by upregulating H3K18ac
and H4K8ac.
RING finger protein 123 (RNF123), also known as
KPC1, is an E3 ubiquitin ligase component of the
ubiquitin-proteasome system that controls certain important cancer
pathways and processes (13). The
tumor suppressor functions of RNF123 have been previously reported
(13). RNF123 is expressed at low
levels in glioblastoma tumors and its low expression is a
predictive factor for poor overall survival (14). Nevertheless, at present, the role of
RNF123 in lung cancer is not well understood. To address this gap,
the present study aimed to identify a possible novel therapy for
lung cancer and elucidate its mechanism of action by investigating
RNF123 expression in LUAD and LUSC and assessing the potential role
of promoter histone acetylation (H3K18ac, H4K8ac and H3K27ac) in
regulating its transcription using bioinformatic methods.
Materials and methods
Cell culture and treatment
The A549, H1975, H1299, H460 and PC-9 lung cancer
cell lines and the BEAS-2B normal human bronchial epithelial cell
line were obtained from ATCC. All cells were cultured in RPMI 1640
(Gibco; Thermo Fisher Scientific, Inc.) comprising 10% FBS (Gibco
Fisher Scientific, Inc.) with 5% CO2 at 37°C. For STS
treatment, A549 and H1975 cells were incubated with 0, 5, 10, 20,
40 and 80 µM STS (Sigma-Aldrich; Merck KGaA) for 24 h.
Cell transfection
The short hairpin RNAs [shRNAs; sh-negative control
(NC) sense, 5′-GTTCTCCGAACGTGTCACGT-3′ and antisense,
5′-ACGTGACACGTTCGGAGAAC-3′; sh-RNF123 sense,
5′-CCCTCAAAGATGACCTTGCTT-3′ and antisense,
5′-AAGCAAGGTCATCTTTGAGGG-3′; and sh-lysine acetyltransferase 2B
(KAT2B) sense, 5′-GCTGGGACAATTTCATACAA-3′ and antisense,
5′-TTGTATGAAATTGTCCCAGC-3′], the overexpression (oe) plasmids
(oe-RNF123 and oe-KAT2B), pCDNA3.1-CMV-RNF123 (human)-GFP-Neo and
pCDNA3.1-CMV-KAT2B (human)-GFP-Neo, and their respective negative
controls (empty pCDNA3.1 vector) were purchased from Shanghai
GenePharma Co., Ltd. The overexpression vectors and shRNAs were
transfected into cells using Lipofectamine 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Briefly, 2.5 µg of nucleic acid was used
per well of a 6-well plate. Transfection complexes were formed at
room temperature for 15 min and then added to cells, which were
incubated at 37°C with 5% CO2. Cells were incubated for
48 h post-transfection before subsequent experimentation. In
addition to sh-NC and oe-NC controls, mock-transfected control
cells (cultured in parallel with transfection reagent only, without
nucleic acid) were included in all assays to serve as the baseline
control group.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from A549 and H1975 cells
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA
synthesis was carried out using the SuperScript™ IV
First-Strand Synthesis System (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was performed using
the PowerUp™ SYBR™ Green Master Mix (Thermo
Fisher Scientific, Inc.) on a QuantStudio™ 3 Real-Time
PCR System with the following thermocycling conditions: Initial
denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 1 min, with a final melting curve analysis from
65°C to 95°C.GAPDH was employed as the reference gene. The data
were analyzed using the 2−ΔΔCq method (15). The primers used in the present study
were as follows: RNF123 forward (F), 5′-TCTTTCTCCCGCAAGAGCTAT-3′
and reverse (R), 5′-AACTGGTCCAAATGTTCTGGC-3′; KAT2B F,
5′-CGAATCGCCGTGAAGAAAGC-3′ and R, 5′-CTTGCAGGCGGAGTACACT-3′; and
GAPDH F, 5′-AGGTCGGAGTCAACGGATTT-3′ and R,
5′-TGACGGTGCCATGGAATTTG-3′.
Western blotting
For nuclear extraction, cells were lysed in
hypotonic buffer (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM
MgCl2, 0.1% NP-40, protease inhibitors) on ice for 15
min, nuclei were pelleted at 3,000 × g for 5 min at 4°C, washed
once with hypotonic buffer without NP-40 and then extracted with
high-salt buffer (20 mM HEPES pH 7.9, 420 mM NaCl, 1.5 mM
MgCl2, 0.2 mM EDTA, 25% glycerol, protease inhibitors)
for 30 min at 4°C with rotation. A BCA kit (Thermo Fisher
Scientific, Inc.) was used to measure the proteins after they had
been isolated using RIPA (Beyotime Institute of Biotechnology). The
total protein (10 µg per lane) was separated by 10% SDS-PAGE and
transferred to a PVDF membrane (MilliporeSigma). The membranes were
blocked with 5% non-fat milk (Amresco, LLC; cat. no. GRM1254-500G)
in TBST (0.1% Tween-20) for 1 h at room temperature, then incubated
overnight at 4°C with primary antibodies purchased from Abcam
against RNF123 (cat. no. ab221877; 1:1,000), H3 (cat. no. ab1791;
1:10,000), H4 (cat. no. ab31830; 1:5,000), H3K18ac (cat. no.
ab40888; 1:500), H4K8ac (cat. no. ab45166; 1:500), H3K27ac (cat.
no. ab4729; 1:1,000), KAT2B (cat. no. ab96510; 1:1,000), E-cadherin
(cat. no. ab227639; 1:2,000), Vimentin (cat. no. ab92547; 1:1,000)
and GAPDH (cat. no. ab9485; 1:5,000). The membranes were then
incubated with the horseradish peroxidase-conjugated anti-rabbit
secondary antibody (cat. no. ab7090; Abcam; 1:5,000) and anti-mouse
secondary antibody (cat. no. ab97023; Abcam; 1:5,000) for 60 min at
room temperature. The protein bands were visualized using ECL (cat.
no. P10060; NCM Biotech; Suzhou Xinsaimei Biotechnology Co., Ltd.)
and semi-quantified using ImageJ software (version 1.80; National
Institutes of Health). For normalization, the ratio of the
grayscale value of the target protein to that of the loading
control was calculated for each sample. The relative expression
level of the target protein was determined using the following
formula: Relative expression (%)=(target protein grayscale
value/loading control grayscale value) ×100.
Cell Counting Kit-8 (CCK-8) assay
Cells were cultured in 24-well plates
(1×104 cells/well) for 24 h. Cells were then treated for
3 h with 10 µl CCK-8 solution (Yeason Biotechnology). Subsequently,
the absorbance at 450 nm was measured.
5-ethynyl-20-deoxyuridine (EdU)
assay
Cells were seeded into confocal plates
(1×106 cells/well) and incubated with 50 µM EdU buffer
for 2 h (Guangzhou RiboBio Co., Ltd.). Cells were then fixed with
4% paraformaldehyde in PBS for 15 min at room temperature and
permeabilized with 0.5% Triton X-100 in PBS for 20 min at room
temperature. After adding EdU solution to the culture, the nuclei
were stained with Hoechst for 15 min at room temperature in the
dark. The samples were imaged using an Olympus fluorescence
microscope. EdU-positive cells were quantified using ImageJ
software.
Transwell assay
DMEM (cat. no. 11965092; Thermo Fisher Scientific,
Inc.) containing 1×104 cells (500 µl) was added to the
upper chamber of a Transwell plate precoated with Matrigel (1:8
ratio; Corning, Inc.) at 37°C for 4 h for invasion assays.
Migration assays were performed using uncoated Transwell plates.
Complete DMEM (1,000 µl) was added to the lower chamber. Cells in
the upper chamber were removed after incubation at 37°C for 12 h,
while cells in the lower chamber were fixed with 4%
paraformaldehyde for 20 min at room temperature and stained with
0.5% crystal violet for 20 min at room temperature. An Olympus
light microscope was used to image the cells. DMEM was selected
instead of RPMI 1640 as A549 and H1975 cells exhibited optimal
growth and migration in DMEM based on preliminary optimization.
Chromatin immunoprecipitation (ChIP)
assay
Using a pre-prepared confluent 10 cm dish
(~107 cells), the cells were fixed with 1% formaldehyde
for 5 min at room temperature to induce DNA-protein cross-linking.
Cell lysate was then sonicated on ice for 15 min (30 sec on/30 sec
off cycles at 20% amplitude) to produce chromatin fragments and
incubated overnight with anti-H3K18ac (cat. no. ab40888; 1:500;
Abcam), anti-H4K8ac (cat. no. ab45166; 1:200; Abcam), anti-KAT2B
(cat. no. ab96510; 1:100; Abcam) or anti-IgG (cat. no. ab172730;
1:100; Abcam) antibodies. Pierce protein A/G magnetic beads (Thermo
Fisher Scientific, Inc.; 30 µl bead slurry per reaction) were
incubated with the chromatin-antibody mixture for 4 h at 4°C with
rotation. Beads were collected on a magnetic rack and washed
sequentially at 4°C with 1 ml each of low-salt buffer (20 mM
Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100),
high-salt buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 2 mM EDTA, 1%
Triton X-100), LiCl buffer (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 1
mM EDTA, 1% NP-40, 1% sodium deoxycholate) and TE buffer (10 mM
Tris-HCl pH 8.0, 1 mM EDTA). Each wash step included rotation for 5
min at 4°C followed by centrifugation at 1,000 × g for 1 min at
4°C. The chromatin-antibody complexes were eluted by incubating the
beads in 200 µl elution buffer (1% SDS, 100 mM NaHCO3)
at 65°C for 30 min with frequent vortexing. DNA was purified using
the QIAquick PCR Purification Kit (cat. no. 28106; Qiagen
Diagnostics) according to the manufacturer's instructions and
analyzed using RT-qPCR with primers specific for the RNF123
promoter region (forward: 5′-GCATCTGTGTGGTCCTGACA-3′; and reverse:
5′-TCTTGAGCACAGCTGGGAAG-3′).
Data retrieval and statistical
analysis
The StarBase database (http://starbase.sysu.edu.cn/) was used to retrieve and
analyze the expression levels of RNF123 and KAT2B in LUAD and LUSC.
The method used was as follows: i) Select the pan-cancer option,
then click on the Gene Differential Expression option and finally
search for the target gene in the search box; ii) select the chart
type, box plot; iii) select the data scale method, log2(FPKM +
0.01); and iv) select the cancer name on the pan-cancer table to
browse the corresponding differential profile. It should be noted
that the StarBase database does not provide patient demographic
data, which is a limitation of the present study. All data was
obtained from three separate trials. The enrichment of histone
acetylation marks on H3K18ac, H4K8ac and H3K27ac within gene
promoter regions was analyzed using the UCSC Genome Browser
(https://genome.ucsc.edu/) and the human genome
assembly GRCh38/hg38 (GSE16256) (16). The promoter region was defined as 2
kb upstream to 500 bp downstream of the transcription start site
for each gene of interest.
The statistical data, which were reported as the
mean ± SD, were examined using GraphPad Prism (version 7.0;
Dotmatics). The differences between two groups were investigated
using paired and unpaired Student's t-tests. One-way ANOVA was
performed to compare differences between >2 groups, followed by
Tukey's Honestly Significant Difference test to determine which
specific group means were significantly different from each other.
P<0.05 was considered to indicate a statistically significant
difference.
Results
RNF123 is a critical mediator of NSCLC
cell proliferation
The starBase database projected that RNF123 was
weakly expressed in LUAD and LUSC samples (Fig. 1A). Concurrently, it was observed
that RNF123 was significantly downregulated in lung cancer cells
compared with BEAS-2B cells (Fig. 1B
and C). Among the lung cancer cell lines, the RNF123 expression
levels exhibited the most pronounced changes in A549 and H1975
cells, hence these two cell lines were chosen for subsequent
investigation. To elucidate the role of RNF123 in controlling lung
cancer cell malignant behaviors, RNF123 overexpression and
knockdown were induced in A549 and H1975 cells. oe-RNF123 and
sh-RNF123 transfection significantly elevated and reduced RNF123
expression in both A549 and H1975 cells, respectively (Fig. 1D and E). Functional experiments
showed that RNF123 overexpression markedly inhibited A549 and H1975
cell viability (Fig. 1F),
proliferation (Figs. 1G and
S1A), migration (Figs. 1H and S1B) and invasion (Figs. 1I and S1C), while RNF123 knockdown had the
inverse effects. Epithelial-mesenchymal transition (EMT) is a
process linked to cancer metastasis (17). During EMT, the reciprocal loss of
E-cadherin (an adhesion molecule maintaining epithelial integrity
via cell-cell junctions) and gain of Vimentin (a mesenchymal
cytoskeletal protein promoting motility and invasive morphology)
creates an inverse profile that serves as a hallmark of EMT and a
key biomarker for tumor metastasis (18). RNF123 upregulation decreased
Vimentin expression levels and elevated E-cadherin expression
levels in lung cancer cells, while RNF123 downregulation had the
opposite effect (Fig. 1J).
Collectively, RNF123 upregulation suppressed lung cancer cell
malignant behaviors.
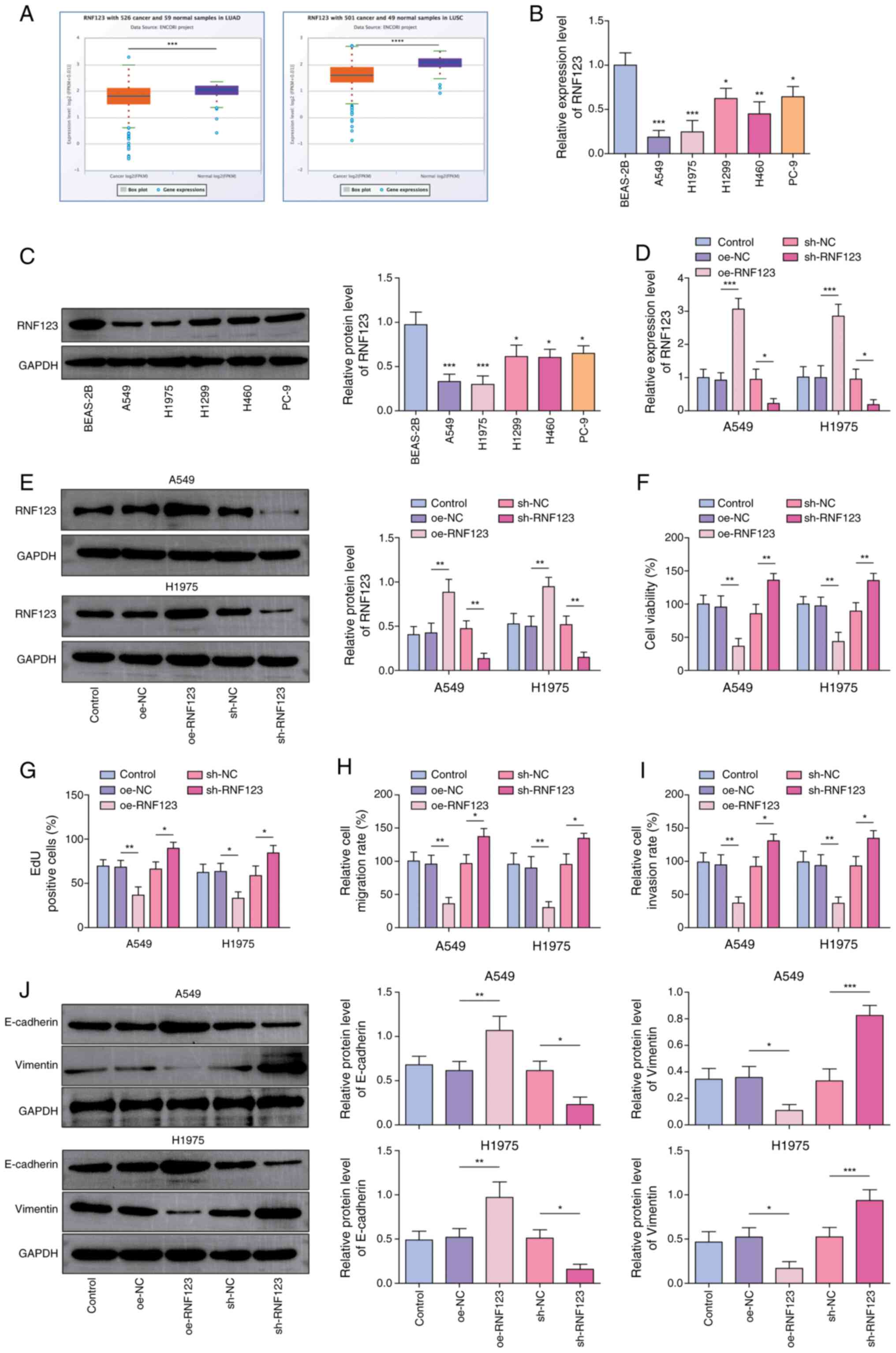 | Figure 1.RNF123 is a critical mediator of
non-small cell lung cancer cell proliferation, migration and
invasion. (A) RNF123 expression in LUAD and LUSC was predicted
using the starBase database. The mRNA and protein expression levels
of RNF123 in lung cancer cells (A549, H1975, H1299, H460 and PC-9
cells) and the normal human bronchial epithelial cell line (BEAS-2B
cells) were detected by (B) RT-qPCR and (C) western blotting,
respectively. A549 and H1975 cells were transfected with
oe-RNF123/sh-RNF123 or oe-NC/sh-NC. The mRNA and protein expression
levels of RNF123 in cells were detected by (D) RT-qPCR and (E)
western blotting, respectively. (F) Cell viability was examined
using the Cell Counting Kit-8 assay. (G) The EdU assay was
performed to determine cell proliferation. Cell (H) migration and
(I) invasion were assessed using the Transwell assay in cells with
RNF123 knockdown (sh-RNF123) or overexpression (oe-RNF123). Results
are expressed as percentages relative to the control group (set as
100%). (J) E-cadherin and Vimentin protein expression levels in
cells were measured by western blotting. Data are expressed as mean
± SD (n=3). *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001. RNF123, RING finger protein 123; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; RT-qPCR,
reverse transcription-quantitative PCR; oe, overexpression; sh,
short hairpin RNA; NC, negative control; EdU,
5-ethynyl-20-deoxyuridine. |
RNF123 knockdown reverses the
inhibitory effects of STS on NSCLC cells
STS, a water-soluble derivative of TSIIA with a
relative molecular mass of 396.39, dose-dependently reduced lung
cancer cell viability, as determined using the CCK-8 assay
(Fig. 2A). Additionally, STS
elevated RNF123 expression levels in lung cancer cells in a
dose-dependent manner (Fig. 2B). A
concentration 40 µM STS was chosen for use in subsequent
experiments due to its ability to reduce cell viability by ~50%.
STS significantly upregulated RNF123 expression levels in A549 and
H1975 cells; however, this effect was abrogated by sh-RNF123
transfection (Fig. 2C and D). In
addition, STS treatment significantly reduced A549 and H1975 cell
viability (Fig. 2E), proliferation
(Figs. 2F and S2A), migration (Figs. 2G and S2B) and invasion (Figs. 2H and S2C), while RNF123 knockdown reversed
these effects. Moreover, STS reduced Vimentin protein expression
levels and elevated E-cadherin protein expression levels in A549
and H1975 cells. These changes in protein expression levels were
reversed by RNF123 knockdown (Fig.
2I). Collectively, STS inhibited lung cancer cell malignant
phenotypes potentially by upregulating RNF123 expression.
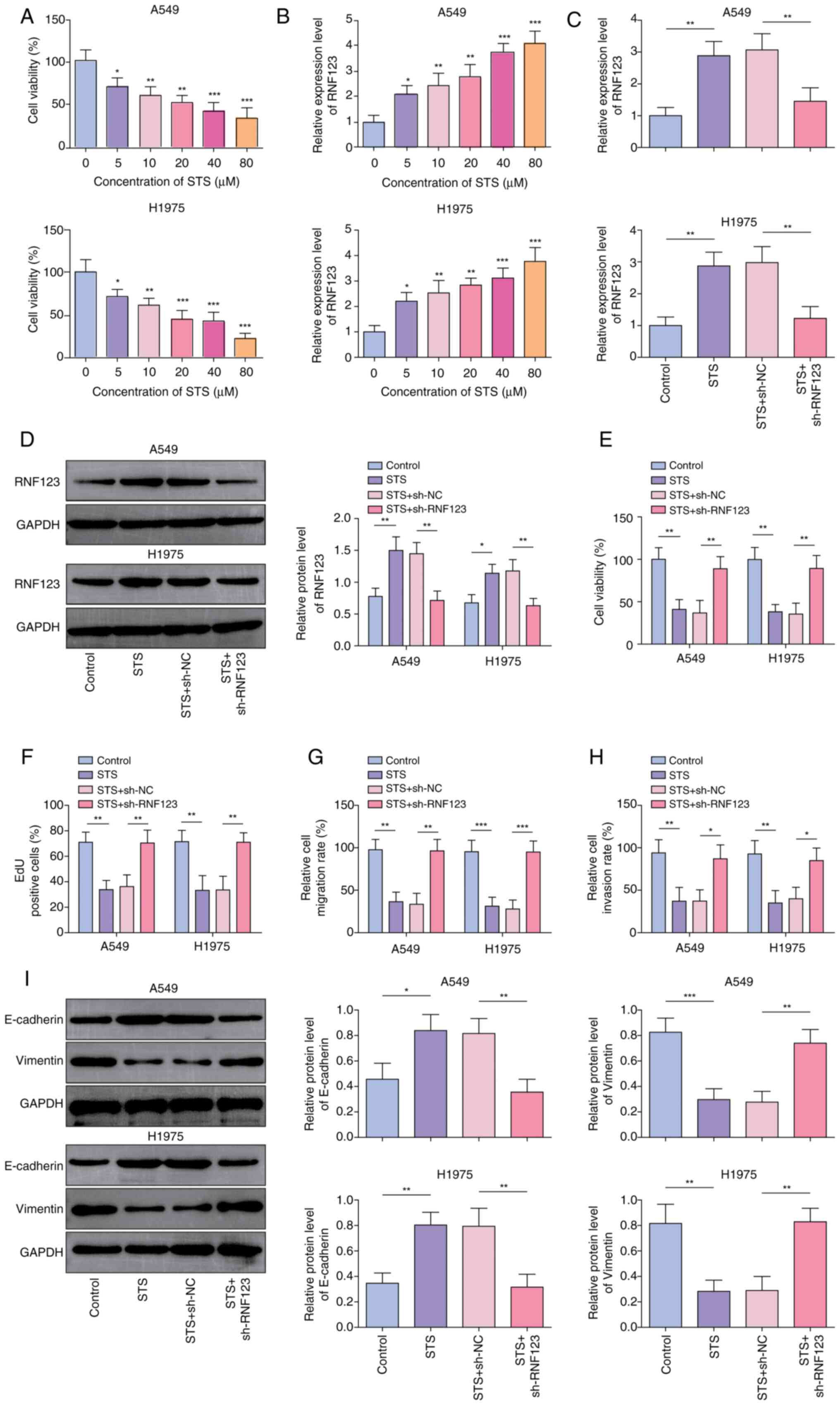 | Figure 2.RNF123 knockdown reverses the
inhibitory effects of STS on non-small cell lung cancer cells. A549
and H1975 cells were treated with 5, 10, 20, 40 and 80 µM STS for
24 h. (A) CCK-8 assay was employed to detect cell viability. (B)
RT-qPCR was conducted to examined RNF123 mRNA expression level in
cells. A549 and H1975 cells were treated with 40 µM STS for 24 h
combined with sh-NC or sh-RNF123 transfection. The mRNA and protein
expression levels of RNF123 in cells were detected by (C) RT-qPCR
and (D) western blotting, respectively. (E) Cell viability was
measured by CCK-8 assay. (F) EdU assay was performed to determine
cell proliferation. Cell (G) migration and (H) invasion were
detected by Transwell assay. Results are expressed as percentages
relative to the control group (set as 100%). (I) Western blotting
was performed to examine E-cadherin and Vimentin protein expression
levels in cells. Data are expressed as mean ± SD (n=3). *P<0.05,
**P<0.01, ***P<0.001. RNF123, RING finger protein 123; STS,
sodium tanshinone IIA sulfate; CCK-8, Cell Counting Kit-8; RT-qPCR,
reverse transcription-quantitative PCR; sh, short hairpin RNA; NC,
negative control; EdU, 5-ethynyl-20-deoxyuridine. |
STS promotes H3K18ac modification of
RNF123 in NSCLC cells
Histone acetylation alterations are linked to cancer
progression (9). H3K18ac and H4K8ac
expression levels were significantly reduced and H3K27ac expression
levels were elevated in A549 and H1975 cells compared with BEAS-2B
cells (Fig. 3A). As predicted by
the UCSC database, H3K18ac, H4K8ac and H3K27ac were enriched in the
RNF123 promoter region (Fig. S3).
Given the pivotal role of histone acetylation in promoting gene
transcription and the synchronous downregulation of RNF123, H3K18ac
and H4K8ac expression levels in lung cancer cells, it was
hypothesized that RNF123 may be modified by H3K18ac and H4K8ac.
ChIP assay results demonstrated that H3K18ac was enriched in the
RNF123 promoter, whereas enrichment of H4K8ac was not detected
(Fig. 3B). Moreover, STS treatment
significantly elevated H3K18ac and H4K8ac expression levels in A549
and H1975 cells (Fig. 3C). As
confirmed by the ChIP assay results, STS promoted H3K18ac
enrichment in the RNF123 promoter region but did not facilitate
H4K8ac enrichment in the RNF123 promoter region (Fig. 3D). These results suggested that STS
may increase the transcriptional activity of RNF123 through
promoting H4K8ac enrichment in the RNF123 promoter.
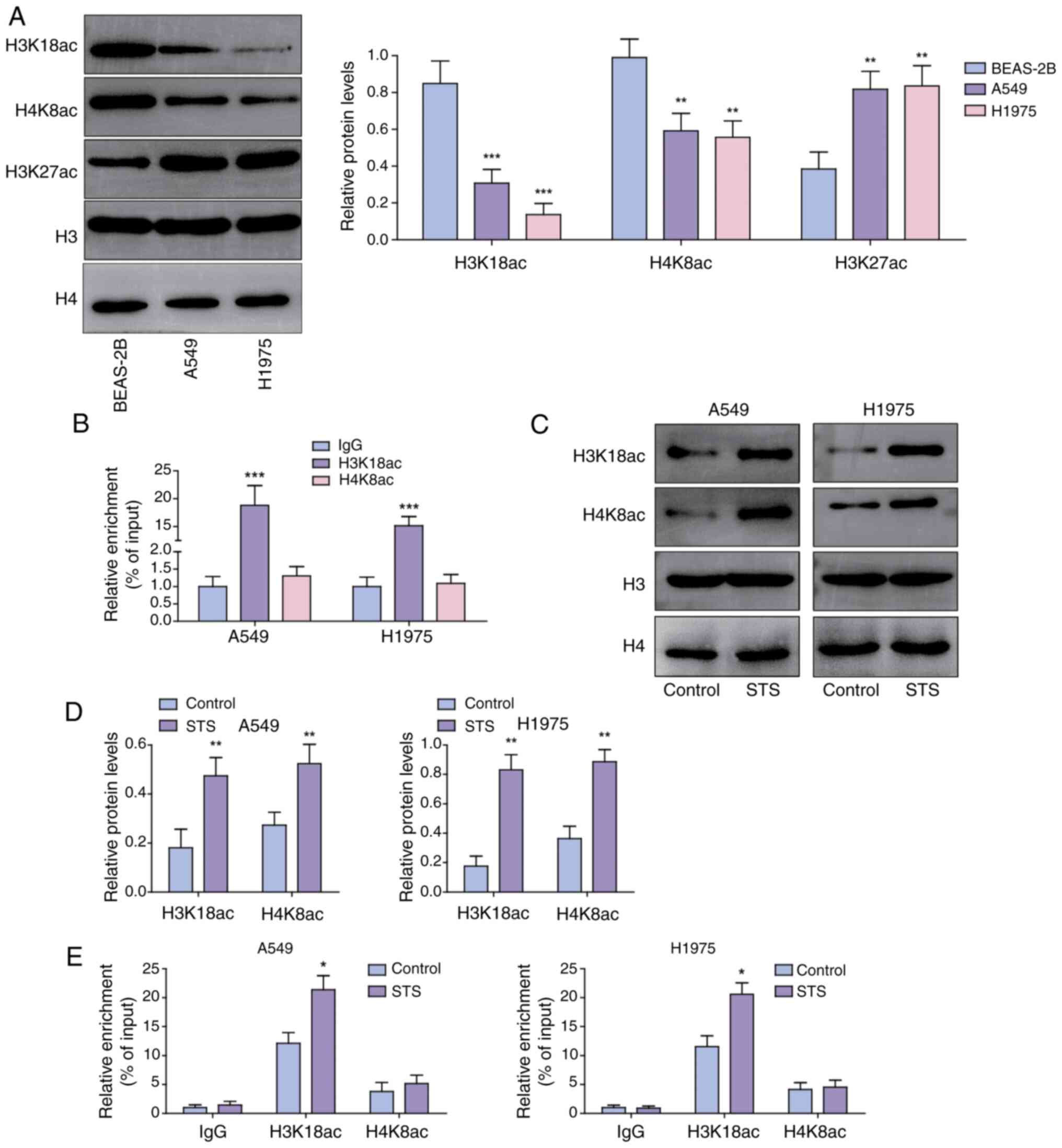 | Figure 3.STS promotes H3K18ac modification of
chromatin at the RNF123 locus in non-small cell lung cancer cells.
(A) H3K18ac, H4K8ac and H3K27ac levels in A549, H1975 and BEAS-2B
cells were assessed using western blotting. (B) The enrichment of
H3K18ac and H4K8ac in the RNF123 promoter was analyzed by ChIP
assay. A549 and H1975 cells were treated with 40 µM STS for 24 h.
(C) H3K18ac and H4K8ac protein expression levels in cells were
examined using western blotting and (D) semi-quantified. Histone
acetylation signals were normalized to total histone H3 or H4
levels, respectively. (E) H3K18ac and H4K8ac enrichment in the
RNF123 promoter was analyzed by ChIP assay. Data are expressed as
mean ± SD (n=3). *P<0.05, **P<0.01, ***P<0.001. STS,
sodium tanshinone IIA sulfate; RNF123, RING finger protein 123;
H3K18ac, histone H3 lysine 18 acetylation; H4K8ac, histone H4
lysine 8 acetylation; H3K127ac, histone H3 lysine 27 acetylation;
ChIP, chromatin immunoprecipitation. |
STS promotes the H3K18ac modification
and expression of RNF123 by upregulating the expression of
KAT2B
KAT2B, an essential HAT epigenetic factor, serves as
a biomarker for predicting prognosis in NSCLC (19,20).
Herein, it was demonstrated that KAT2B was expressed at low levels
in LUAD and LUSC using the starBase database (Fig. 4A). KAT2B expression levels were
lower in A549 and H1975 cells compared with BEAS-2B cells (Fig. 4B and C). Notably, KAT2B expression
in lung cancer cells was significantly increased by STS treatment
(Fig. 4D and E). Furthermore, it
was demonstrated that KAT2B was enriched in the RNF123 promoter
region (Fig. 4F). Next, A549 and
H1975 cells were transfected with oe-NC or oe-KAT2B, and the
subsequent RT-qPCR and western blotting results showed that
oe-KAT2B transfection significantly elevated the KAT2B expression
levels (Fig. 4G and H).
Additionally, KAT2B overexpression facilitated H3K18ac enrichment
in the RNF123 promoter region (Fig.
4I). Moreover, KAT2B overexpression significantly elevated
RNF123 expression levels in lung cancer cells (Fig. 4J and K). In conclusion, STS promoted
RNF123 expression in lung cancer cells potentially by increasing
the H3K18ac enrichment in the RNF123 promoter through upregulating
KAT2B.
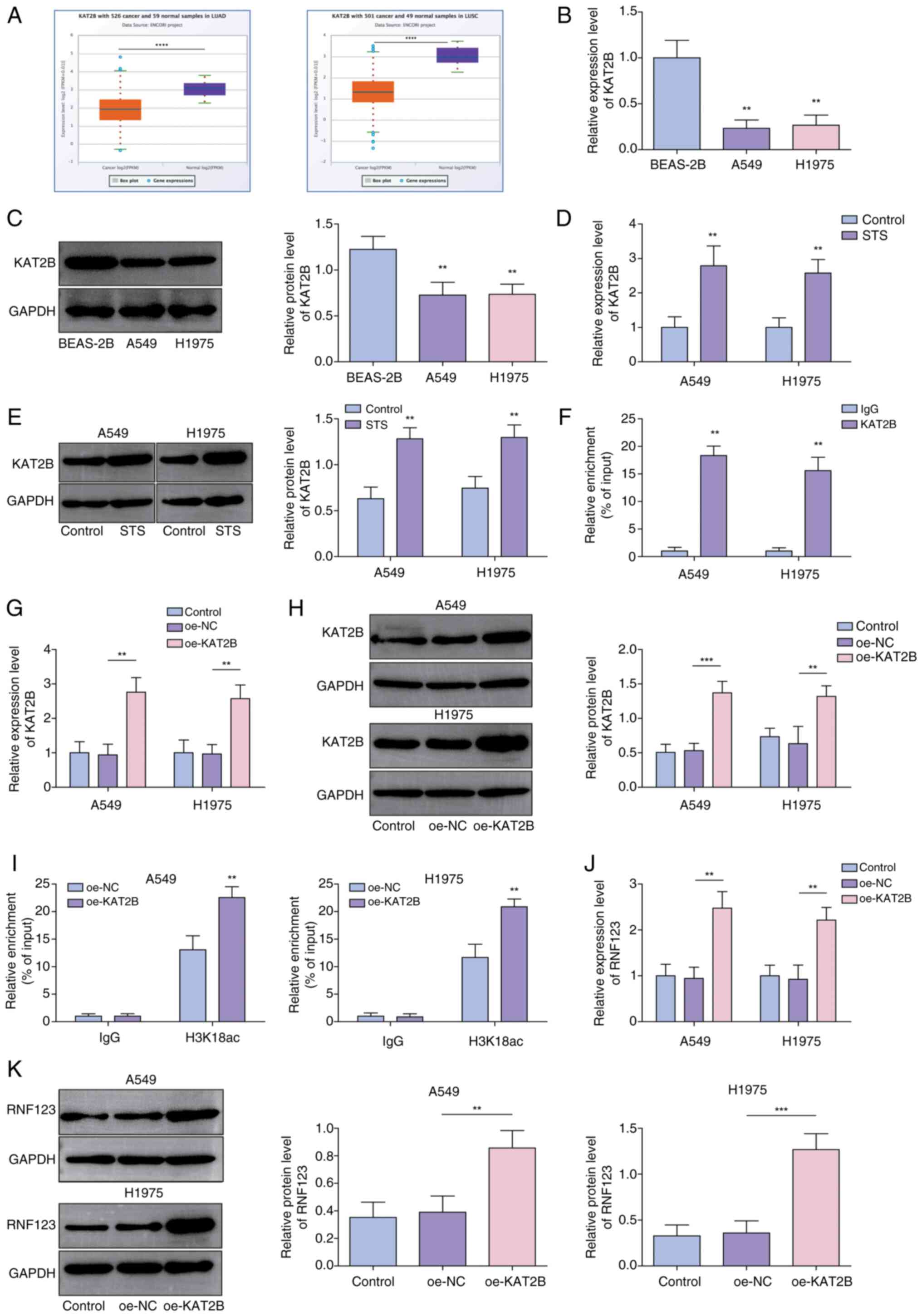 | Figure 4.STS promotes the H3K18ac modification
and expression of RNF123 by upregulating the expression of KAT2B.
(A) KAT2B expression in LUAD and LUSC was predicted using the
starBase database. The mRNA and protein expression levels of KAT2B
in A549, H1975 and BEAS-2B cells were detected by (B) RT-qPCR and
(C) western blotting, respectively. The mRNA and protein expression
levels of KAT2B in A549 and H1975 cells after STS treatment were
detected by (D) RT-qPCR and (E) western blotting, respectively. (F)
The interaction between KAT2B and RNF123 promoter was analyzed by
ChIP assay. A549 and H1975 cells were transfected with oe-NC or
oe-KAT2B. The mRNA and protein expression levels of KAT2B in cells
were detected by (G) RT-qPCR and (H) western blotting,
respectively. (I) H3K18ac enrichment in the RNF123 promoter was
analyzed by ChIP assay. The mRNA and protein levels of RNF123 in
cells were detected by (J) RT-qPCR and (K) western blotting,
respectively. Data are expressed as mean ± SD (n=3). **P<0.01,
***P<0.001, ****P<0.0001. STS, sodium tanshinone IIA sulfate;
RNF123, RING finger protein 123; H3K18ac, histone H3 lysine 18
acetylation; KAT2B, lysine acetyltransferase 2B; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; RT-qPCR,
reverse transcription-quantitative PCR; ChIP, chromatin
immunoprecipitation; oe, overexpression; NC, negative control. |
KAT2B knockdown reverses the
inhibitory effects of STS on the malignant phenotypes of NSCLC
cells
A549 and H1975 cells were subjected to STS treatment
combined with sh-KAT2B or sh-NC transfection to elucidate the role
of KAT2B in the STS-mediated anticancer effects. Initially, it was
demonstrated that sh-KAT2B transfection significantly downregulated
the KAT2B expression levels in A549 and H1975 cells (Fig. 5A and B). KAT2B expression levels in
lung cancer cells were also significantly increased by STS
administration and this upregulation was reversed by KAT2B
knockdown (Fig. 5C-E). Furthermore,
the knockdown of KAT2B counteracted the inhibitory effects of STS
on cell viability (Fig. 5F),
proliferation (Figs. 5G and
S4A), migration (Figs. 5H and S4B) and invasion (Figs. 5I and S4C). It was also demonstrated that the
promoting effect of STS on E-cadherin expression levels and the
inhibitory effect on Vimentin expression levels in lung cancer
cells were abolished following KAT2B knockdown (Fig. 5J). In summary, KAT2B knockdown
neutralized the inhibitory effects of STS on the malignant
phenotypes of lung cancer cells.
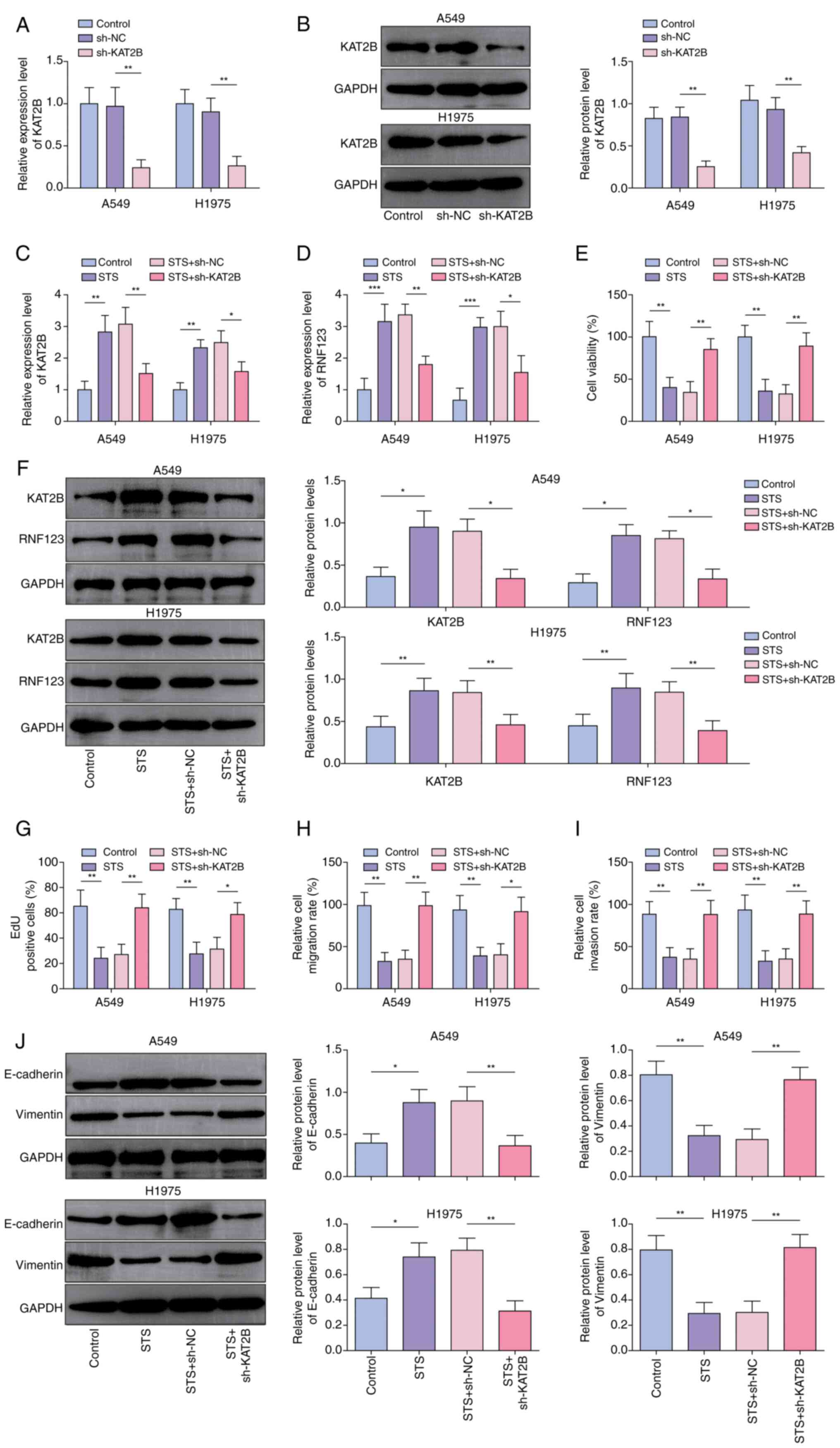 | Figure 5.KAT2B knockdown reverses the
inhibitory effects of STS on the malignant phenotypes of non-small
cell lung cancer cells. KAT2B knockdown was induced in STS-treated
A549 and H1975 cells. The mRNA and protein expression levels of
KAT2B in A549 and H1975 cells after sh-NC or sh-KAT2B transfection
were detected by (A) RT-qPCR and (B) western blotting,
respectively. A549 and H1975 cells were treated with 40 µM STS for
24 h combined with sh-NC or sh-KAT2B transfection. The mRNA
expression levels of (C) KAT2B and (D) RNF123 in cells were
detected by RT-qPCR. (E) Cell viability was measured by Cell
Counting Kit-8 assay. (F) The protein expression levels of RNF123
and KAT2B in cells were detected by western blotting. (G) EdU assay
was performed to determine cell proliferation. Cell (H) migration
and (I) invasion were detected by Transwell assay. Results are
expressed as percentages relative to the control group (set as
100%). (J) E-cadherin and Vimentin protein levels in cells were
examined by western blotting. Data are expressed as mean ± SD
(n=3). *P<0.05, **P<0.01, ***P<0.001. KAT2B, lysine
acetyltransferase 2B; STS, sodium tanshinone IIA sulfate; sh, short
hairpin RNA; RT-qPCR, reverse transcription-quantitative PCR; NC,
negative control; RNF123, RING finger protein 123; EdU,
5-ethynyl-20-deoxyuridine. |
RNF123 overexpression reverses the
effects of KAT2B knockdown on the STS-treated NSCLC cell malignant
phenotypes
To investigate the interaction between KAT2B and
RNF123 in the STS-mediated anticancer effects, A549 and H1975 cells
treated with STS were co-transfected with RNF123 overexpression and
KAT2B knockdown constructs. Co-transfection with oe-RNF123 rescued
the suppressive effect of KAT2B knockdown on RNF123 expression
levels in STS-treated A549 and H1975 cells (Fig. 6A and B). Additionally, RNF123
overexpression mitigated the sh-KAT2B-induced enhancement of the
viability (Fig. 6C), proliferation
(Figs. 6D and S5A), migration (Figs. 6E and S5B) and invasion (Figs. 6F and S5C) of STS-treated A549 and H1975 cells.
Moreover, KAT2B knockdown reduced E-cadherin protein expression
levels and elevated Vimentin expression levels in STS-treated A549
and H1975 cells, whereas RNF123 upregulation restored these effects
(Fig. 6G). In conclusion, STS
inhibited the lung cancer cell malignant phenotypes via modulating
the KAT2B/RNF123 axis.
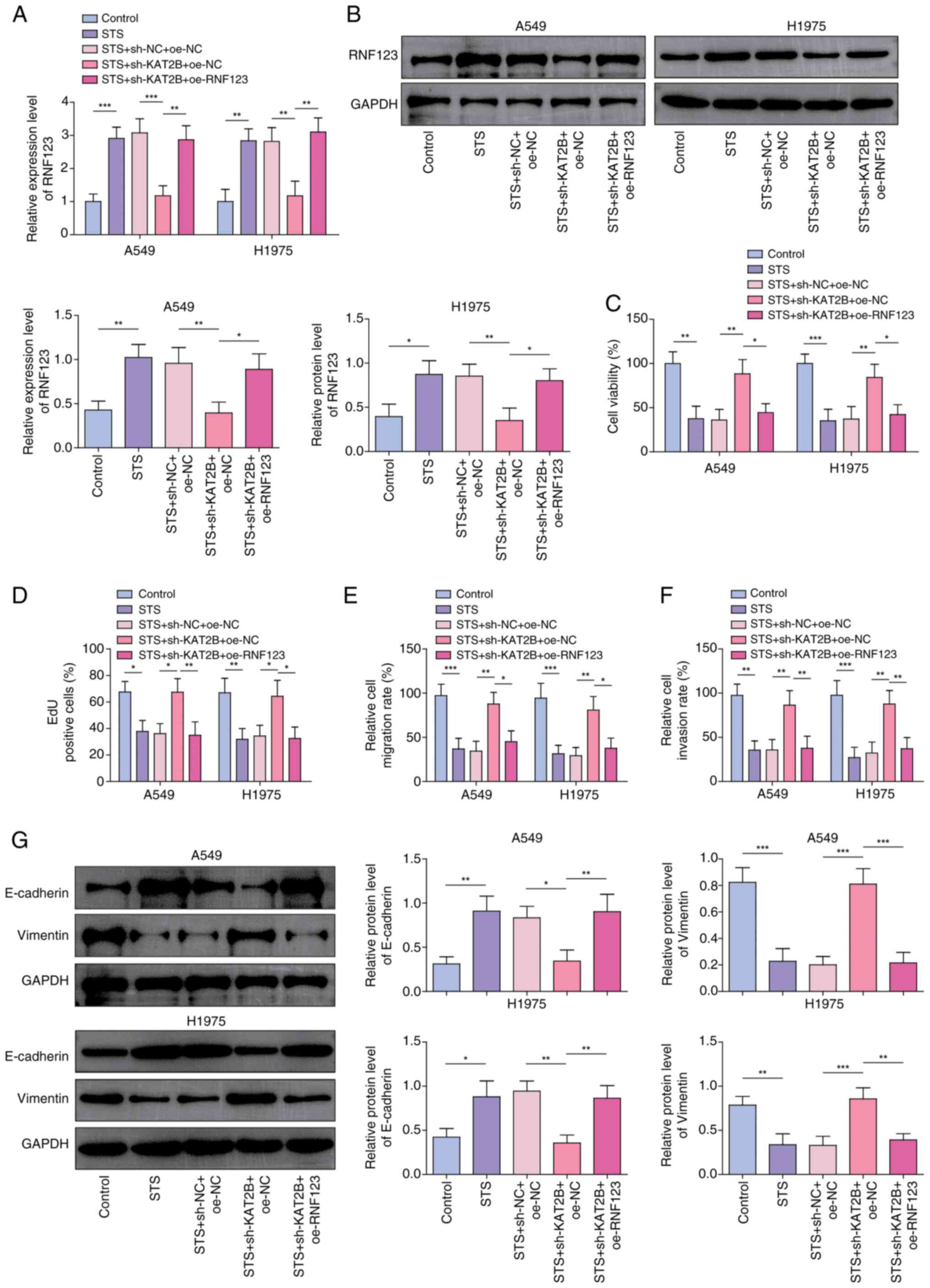 | Figure 6.RNF123 overexpression reverses the
effects of KAT2B knockdown on the malignant phenotypes of non-small
cell lung cancer cells treated with STS. Both RNF123 overexpression
and KAT2B knockdown were induced in STS-treated A549 and H1975
cells. The mRNA and protein expression levels of RNF123 in cells
were detected by (A) RT-qPCR and (B) western blotting,
respectively. (C) Cell viability was examined using Cell Counting
Kit-8 assay. (D) EdU assay was performed to determine cell
proliferation. Cell (E) migration and (F) invasion were assessed
using Transwell assay. Results are expressed as percentages
relative to the control group (set as 100%). (G) E-cadherin and
Vimentin protein levels in cells were measured by western blotting.
Data were expressed as mean ± SD (n=3). *P<0.05, **P<0.01,
***P< 0.001. STS, sodium tanshinone IIA sulfate; RNF123, RING
finger protein 123; KAT2B, lysine acetyltransferase 2B; RT-qPCR,
reverse transcription-quantitative PCR; NC, negative control; sh,
short hairpin RNA; oe, overexpression; EdU,
5-ethynyl-20-deoxyuridine. |
Discussion
Lung cancer is a major challenge to global health.
According to global cancer statistics, there were 2.481 million new
cases of lung cancer in 2022, accounting for 12.4% of all new
cancer cases globally (1).
Historically, the prognosis for patients with lung cancer has been
poor (9). Over the past decade,
significant advancements have been made in the field of lung cancer
treatment, particularly in the areas of molecular targeted therapy
and immunotherapy. For instance, targeted therapy against ROS
proto-oncogene 1, receptor tyrosine kinase (ROS1) gene fusions have
provided a new option for precision treatment for patients with
advanced NSCLC. ROS1-positive patients tend to be younger (median
onset ~50 years) and are frequently non-smoking women with
adenocarcinoma (3). However,
despite these advancements, lung cancer remains the leading cause
of cancer-related mortality (21).
As a result, the discovery of novel therapeutic options to improve
the survival of patients with lung cancer is required. The findings
of the present study indicate that TSIIA suppresses lung cancer
cell malignant phenotypes in vitro by increasing RNF123
expression levels through promoting KAT2B-mediated H3K18ac
modification.
TSIIA, the predominant diterpenoid quinone derived
from Salvia miltiorrhiza, has been utilized in China for
>2,000 years to treat cardiovascular disorders (22). In the past decade, research interest
in the protective effects of TSIIA on cancer has increased. TSIIA
suppresses EMT and migration in colorectal and gastric cancer cells
(23,24), inhibits NSCLC proliferation through
VEGF/VEGFR2 downregulation (25)
and triggers SCLC apoptosis by upregulating Bax/Bcl-2 and reducing
mitochondrial membrane potential (26). It has been previously shown that STS
has potential in anticancer research as STS can effectively inhibit
the cell viability and colony formation of NSCLC cells (6,7).
Furthermore, STS has been reported to exert antitumor effects in
the treatment of colorectal cancer (5). By targeting specific proteins and
signaling pathways, STS may inhibit tumor proliferation and
metastasis (25,27). However, the role of STS or TSIIA in
controlling lung cancer migration, invasion and EMT is not well
understood. The findings of the present study showed that STS
treatment significantly inhibited lung cancer cell viability,
proliferation, migration, invasion and EMT. TSIIA, a drug with
multiple pharmacological effects, has a high efficiency, low
toxicity and a natural source compared with chemotherapy drugs, and
has significant potential clinical value (27). These findings provide a further
foundation for TSIIA as a potential anti-lung cancer drug.
EMT is a critical process by which cancer cells
acquire invasive and metastatic capabilities (17,18).
In the present study, it was observed that treatment with STS
significantly reduced the expression of the EMT marker, Vimentin,
and increased the expression of E-cadherin in NSCLC cells. This
suggests that STS may inhibit the invasiveness and metastatic
potential of NSCLC cells by suppressing the EMT process. In recent
years, EMT has been increasingly recognized not only as a
biological process but also as a potential ‘ecological process’.
According to the nasopharyngeal carcinoma ecology theory proposed
by Luo (28), cancer can be viewed
as a multidimensional spatiotemporal ‘unity of ecology and
evolution’ pathological ecosystem. Within this ecosystem, cancer
cells interact dynamically with the tumor microenvironment and
these interactions not only influence the biological behavior of
cancer cells but may also shape tumor heterogeneity and
invasiveness through ecological selection and evolutionary
processes. During EMT, cancer cells exhibit mesenchymal
characteristics, such as increased migratory and invasive
abilities, which enable them to better adapt and survive in the
competitive and resource-limited tumor microenvironment. This
adaptive change can be seen as an ecological strategy that helps
cancer cells gain a survival advantage (28). Moreover, EMT-associated cancer cells
may interact dynamically with surrounding stromal cells (such as
cancer-associated fibroblasts and immune cells) through the
secretion of cytokines and exosomes, thereby further promoting
tumor progression (17,28).
Ubiquitin E3 ligases serve a key role in normal
human cells by regulating protein ubiquitination and degradation,
which is a necessary metabolic activity for life (29). The E3-ligase, RNF123, has been
reported to act as a tumor suppressor (13). Iida et al (30) demonstrated that RNF123 reduces
melanoma proliferation by processing NF-κB1 p105 into p50.
Furthermore, RNF123 is downregulated in aggressive glioblastoma and
its downregulation is associated with a poor prognosis (14). However, to the best of our
knowledge, the potential mechanism of action of RNF123 in lung
cancer has not been previously reported. The results of the present
study showed that RNF123 was expressed at low levels in lung cancer
cells and its overexpression inhibited the lung cancer cell
malignant phenotypes. Notably, it was also demonstrated that STS
increased the RNF123 expression levels in lung cancer cells and
RNF123 knockdown abolished the STS-induced inhibition of lung
cancer cell malignant phenotypes. Collectively, the results
suggested that STS inhibited lung cancer cell malignant phenotypes
by increasing RNF123 expression levels.
Histone acetylation is an important component of
transcriptional regulation, which depends on the balance of HAT and
HDAC activities (31). The
dysregulation of histone acetylation modifications promotes the
development of human malignant tumors (9). The findings of the present study
demonstrated that the H3K18ac and H4K8ac expression levels were
significantly reduced and the H3K27ac expression levels were
elevated in lung cancer cells. Moreover, H3K18ac and H4K8ac were
enriched in the RNF123 promoter. As previously reported, TSIIA
prevents cerebral ischemia reperfusion injury by regulating H3K18ac
and H4K8ac (12). In the present
study, STS increased the H3K18ac and H4K8ac expression levels in
lung cancer cells. Additionally, STS facilitated H3K18ac enrichment
in the RNF123 promoter. Collectively, the results of the present
study indicated that STS increased RNF123 expression levels in lung
cancer cells by promoting H3K18ac enrichment in the RNF123
promoter. KAT2B is a HAT that acetylates specific lysine residues
in histones and therefore primarily serves a role in regulating
chromatin remodeling, which is a key regulatory factor in signal
transduction during the occurrence of numerous diseases, including
cancer (32). KAT2B is an immune
infiltration-associated biomarker that predicts prognosis in
patients with NSCLC (20). The
findings of the present study showed that KAT2B was expressed at
low levels in lung cancer and STS increased KAT2B expression levels
in lung cancer cells. Notably, KAT2B increased RNF123 expression
levels in lung cancer cells by increasing the H3K18ac modification
in chromatin at the RNF123 locus. Furthermore, KAT2B knockdown
reversed the STS-induced inhibition of lung cancer cell malignant
phenotypes. Moreover, RNF123 upregulation abrogated the effects of
KAT2B knockdown on STS-treated lung cancer cell malignant
phenotypes. These findings may be significant for NSCLC treatment
as RNF123 was shown to suppress NSCLC proliferation and cellular
activity in A549 and H1975 NSCLC cell lines, potentially emerging
as a new target for treatment. STS inhibited NSCLC by upregulating
RNF123 expression through the KAT2B/H3K18ac pathway, presenting a
potential new treatment strategy. The low toxicity and lack of side
effects of STS make it a promising therapy, potentially improving
patient outcomes (27). The present
study may offer valuable insights and highlight the clinical
potential of this treatment against NSCLC.
However, there are a number of limitations of the
present study. At present, the hypothesis that Tanshinone IIA
targets RNF123 to inhibit NSCLC cell proliferation, migration and
invasion via KAT2B-mediated H3K18ac modification has only been
preliminarily verified at the cellular level and further
confirmation requires animal and clinical experiments.
Additionally, it was demonstrated that STS has effects on the
H3K18ac acetylation modification level; however, whether it also
affects other acetylation modification levels and whether it is
regulated by other acetylases requires further investigation.
Although the present study preliminarily revealed that STS inhibits
EMT by regulating RNF123 expression through KAT2B-mediated H3K18ac
modification, the detailed molecular mechanisms still need further
investigation. Specifically, whether TSIIA regulates other
epigenetic modifications or signaling pathways to influence the EMT
process should be explored. Moreover, the lack of patient
demographic data from StarBase is a limitation of the present
study, as it restricts the comprehensive understanding of the
clinical relevance of RNF123 and KAT2B in lung cancer. By
integrating the tumor ecology theory proposed by Luo (28), future studies could employ
multidimensional approaches (such as single-cell sequencing and
spatial transcriptomics) to comprehensively analyze the
spatiotemporal dynamics of the EMT process within the tumor
ecosystem. This will help us better understand the complexity of
tumors and provide new insights for developing precision medicine
strategies.
Taken together, the results of the present study
demonstrated that STS inhibited the lung cancer cell malignant
phenotypes by upregulating RNF123 through promoting KAT2B-mediated
H3K18ac modification of chromatin at the RNF123 locus. TSIIA may
have potential for the treatment of lung cancer in the future and
RNF123 may serve as an important target for the anticancer actions
of TSIIA.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Work was funded by the grant provided by the China Medicine
Education Association named ‘Construction of three-generation
sequencing database and data analysis platform’ (grant no.
2022KTZ017).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
The study and figures were designed by YH and KC. YH
and YZ conducted the experiments and interpreted the results with
JZ. YH, KC and JZ supervised the study. Bioinformatics analysis was
performed by YH. Funding was obtained by KC. YH wrote the
manuscript draft. All authors read and approved the final version
of the manuscript. YH, YZ, JZ and KC confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
LUAD
|
lung adenocarcinoma
|
|
LUSC
|
lung squamous cell carcinoma
|
|
TSIIA
|
tanshinone IIA
|
|
STS
|
sodium TSIIA sulfate
|
|
HAT
|
histone acetyltransferase
|
|
HDAC
|
histone deacetylase
|
|
RNF123
|
RING finger protein 123
|
|
KAT2B
|
lysine acetyltransferase 2B
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
CCK-8
|
Cell Counting Kit-8
|
|
EdU
|
5-ethynyl-20-deoxyuridine
|
|
ChIP
|
chromatin immunoprecipitation
|
|
H3K18ac
|
acetylation of histone H3 lysine 18
site
|
|
H4K8ac
|
acetylation of histone H4 lysine 8
site
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 71:7–33. 2022.
|
|
2
|
Feng J, Li J, Qie P, Li Z, Xu Y and Tian
Z: Long non-coding RNA (lncRNA) PGM5P4-AS1 inhibits lung cancer
progression by up-regulating leucine zipper tumor suppressor
(LZTS3) through sponging microRNA miR-1275. Bioengineered.
12:196–207. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bade BC and Dela Cruz CS: Lung cancer
2020: Epidemiology, etiology, and prevention. Clin Chest Med.
41:1–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang LJ, Jeng CJ, Kung HN, Chang CC, Wang
AG, Chau GY, Don MJ and Chau YP: Tanshinone IIA isolated from
Salvia miltiorrhiza elicits the cell death of human
endothelial cells. J Biomed Sci. 12:347–361. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu L, Gao H, Wen T, Gu T, Zhang S and
Yuan Z: Tanshinone IIA attenuates AOM/DSS-induced colorectal
tumorigenesis in mice via inhibition of intestinal inflammation.
Pharm Biol. 59:89–96. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao F, Li M, Liu W and Li W: Inhibition of
EGFR signaling and activation of mitochondrial apoptosis contribute
to tanshinone IIA-mediated tumor suppression in non-small cell lung
cancer cells. Onco Targets Ther. 13:2757–2769. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang B, Zou F, Xin G, Xiang BL, Zhao JQ,
Yuan SF, Zhang XL and Zhang ZH: Sodium tanshinone IIA sulphate
inhibits angiogenesis in lung adenocarcinoma via mediation of
miR-874/eEF-2K/TG2 axis. Pharm Biol. 61:868–877. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo P, Chen W, Li H, Li M and Li L: The
histone acetylation modifications of breast cancer and their
therapeutic implications. Pathol Oncol Res. 24:807–813. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng Z, Li X, Hou S, Wu Y, Sun Y and Liu
B: K-Ras-ERK1/2 accelerates lung cancer cell development via
mediating H3K18ac through the MDM2-GCN5-SIRT7 axis.
Pharm Biol. 57:701–709. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen G, Zhu X, Li J, Zhang Y, Wang X,
Zhang R, Qin X, Chen X, Wang J, Liao W, et al: Celastrol inhibits
lung cancer growth by triggering histone acetylation and acting
synergically with HDAC inhibitors. Pharmacol Res. 185:1064872022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma H, Hu ZC, Long Y, Cheng LC, Zhao CY and
Shao MK: Tanshinone IIA microemulsion protects against cerebral
ischemia reperfusion injury via regulating H3K18ac and H4K8ac in
vivo and in vitro. Am J Chin Med. 50:1845–1868. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gulei D, Drula R, Ghiaur G, Buzoianu AD,
Kravtsova-Ivantsiv Y, Tomuleasa C and Ciechanover A: The tumor
suppressor functions of ubiquitin ligase KPC1: From cell-cycle
control to NF-κB regulator. Cancer Res. 83:1762–1767. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Bustos MA, Zhang X, Ramos RI, Tan
C, Iida Y, Chang SC, Salomon MP, Tran K, Gentry R, et al:
Downregulation of the ubiquitin-E3 ligase RNF123 promotes
upregulation of the NF-κB1 target SerpinE1 in aggressive
glioblastoma tumors. Cancers (Basel). 12:10812020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heintzman ND, Hon GC, Hawkins RD,
Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW,
et al: Histone modifications at human enhancers reflect global
cell-type-specific gene expression. Nature. 459:108–112. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hashemi M, Arani HZ, Orouei S, Fallah S,
Ghorbani A, Khaledabadi M, Kakavand A, Tavakolpournegari A, Saebfar
H, Heidari H, et al: EMT mechanism in breast cancer metastasis and
drug resistance: Revisiting molecular interactions and biological
functions. Biomed Pharmacother. 155:1137742022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Serrano-Gomez SJ, Maziveyi M and Alahari
SK: Regulation of epithelial-mesenchymal transition through
epigenetic and post-translational modifications. Mol Cancer.
15:182016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou YS, Wang JZ, Shi S, Han Y, Zhang Y,
Zhi JX, Xu C, Li FF, Wang GY and Liu SL: Identification of
epigenetic factor KAT2B gene variants for possible roles in
congenital heart diseases. Biosci Rep. 40:BSR201917792020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou X, Wang N, Zhang Y, Yu H and Wu Q:
KAT2B is an immune infiltration-associated biomarker predicting
prognosis and response to immunotherapy in non-small cell lung
cancer. Invest New Drugs. 40:43–57. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Megyesfalvi Z, Gay CM, Popper H, Pirker R,
Ostoros G, Heeke S, Lang C, Hoetzenecker K, Schwendenwein A,
Boettiger K, et al: Clinical insights into small cell lung cancer:
Tumor heterogeneity, diagnosis, therapy, and future directions. CA
Cancer J Clin. 73:620–652. 2023.PubMed/NCBI
|
|
22
|
Xu S and Liu P: Tanshinone II-A: New
perspectives for old remedies. Expert Opin Ther Pat. 23:149–153.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song Q, Yang L, Han Z, Wu X, Li R, Zhou L,
Liu N, Sui H, Cai J, Wang Y, et al: Tanshinone IIA inhibits
epithelial-to-mesenchymal transition through hindering β-arrestin1
mediated β-catenin signaling pathway in colorectal cancer. Front
Pharmacol. 11:5866162020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan F, Zhao ZT, Jia B, Wang YP and Lei W:
TSN inhibits cell proliferation, migration, invasion, and EMT
through regulating miR-874/HMGB2/β-catenin pathway in gastric
cancer. Neoplasma. 67:1012–1021. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie J, Liu J, Liu H, Liang S, Lin M, Gu Y,
Liu T, Wang D, Ge H and Mo SL: The antitumor effect of tanshinone
IIA on anti-proliferation and decreasing VEGF/VEGFR2 expression on
the human non-small cell lung cancer A549 cell line. Acta Pharm Sin
B. 5:554–563. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng CY and Su CC: Tanshinone IIA may
inhibit the growth of small cell lung cancer H146 cells by
up-regulating the Bax/Bcl-2 ratio and decreasing mitochondrial
membrane potential. Mol Med Rep. 3:645–650. 2010.PubMed/NCBI
|
|
27
|
Fang ZY, Zhang M, Liu JN, Zhao X, Zhang YQ
and Fang L: Tanshinone IIA: A review of its anticancer effects.
Front Pharmacol. 11:6110872021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo W: Nasopharyngeal carcinoma ecology
theory: Cancer as multidimensional spatiotemporal ‘unity of ecology
and evolution’ pathological ecosystem. Theranostics. 13:1607–1631.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berndsen CE and Wolberger C: New insights
into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol.
21:301–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Iida Y, Ciechanover A, Marzese DM, Hata K,
Bustos M, Ono S, Wang J, Salomon MP, Tran K, Lam S, et al:
Epigenetic regulation of KPC1 ubiquitin ligase affects the NF-κB
pathway in melanoma. Clin Cancer Res. 23:4831–4842. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shvedunova M and Akhtar A: Modulation of
cellular processes by histone and non-histone protein acetylation.
Nat Rev Mol Cell Biol. 23:329–349. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li L, Zhang J and Cao S: Lysine
acetyltransferase 2B predicts favorable prognosis and functions as
anti-oncogene in cervical carcinoma. Bioengineered. 12:2563–2575.
2021. View Article : Google Scholar : PubMed/NCBI
|




















