Introduction
In 2020, the global number of deaths from head and
neck cancer (HNC), which includes cancer of different regions, such
as the salivary glands, lips and oral cavity, larynx, hypopharynx,
nasopharynx and oropharynx, reached 440,000 (1,2). Head
and neck squamous cell carcinoma (HNSCC), the predominant
histological type of HNC, originates from the mucosal epithelia of
the buccal cavity, pharynx and larynx, and accounts for >90% of
all cases of HNC (3,4). HNSCC can be categorized as human
papillomavirus (HPV)-positive or HPV-negative. The biological
characteristics of HNSCC notably differ between patients owing to
the impact of HPV (5). In a
previous study, patients with oropharyngeal squamous cell carcinoma
who tested positive for HPV had a 5-year overall survival (OS) rate
of 81.1%, while those with HPV-negative malignancies had a survival
rate of 39.7% (6).
The treatment of HNSCC is complicated by the
resistance of cancerous cells to chemotherapy. Resistance to
chemotherapy is primarily attributed to the inefficacy of most
chemotherapy medications for inducing cell death in resistant tumor
cells. This inefficacy is influenced by several factors, including
DNA damage repair, cell cycle regulation, immune invasion, tumor
progression, tumor recurrence induced by the impact of severe acute
respiratory syndrome coronavirus 2-mediated triggering of various
intracellular signaling axes, the presence of tumor-associated stem
cells, autophagy, epithelial-to-mesenchymal transition and the
effects of efflux pumps and metabolic rewiring (7–13).
Therefore, it is essential to develop new strategies to predict
clinical outcomes, and to design personalized treatment strategies
for patients with HNSCC, especially HPV-negative HNSCC.
In recent decades, several new non-apoptotic methods
of controlled cell death have been identified, such as pyroptosis,
ferroptosis, necroptosis, alkaliptosis and cuproptosis. These are
potentially useful tools in the context of cancer therapy (14–18).
Each of these processes has distinct biological mechanisms and
pathological characteristics. Ferroptosis, which was first proposed
by Dixon et al (19), is an
iron-dependent form of cell death that is characterized by the
intracellular generation of lipid reactive oxygen species (ROS).
Previous studies have shown that there are marked differences
between ferroptosis and other forms of cell death, such as
apoptosis, necrosis and autophagy, in terms of morphology (for
example, shrunken mitochondria with increased membrane density),
biochemistry (iron-mediated lethal lipid ROS accumulation and no
ATP depletion), and genetics (governed by unique genes such as
ribosomal protein L8 and iron responsive element binding protein 2,
independent of Bax/Bak) (19,20).
Ferroptotic agents could provide an effective
approach to address the inefficacy of apoptosis-inducing
chemotherapeutics, since ferroptosis differs completely from
apoptosis in terms of its mechanism for inducing cell death
(19). The importance of
ferroptosis in the development and advancement of diverse diseases
has been recognized such as in malignancies, neurodegenerative
conditions and ischemia-reperfusion damage (21). There has been an increased focus on
the potential applications of ferroptosis in cancer therapy for
improving prognosis and overcoming drug resistance (18,22,23).
In HNSCC cells, ferroptosis can decrease or reverse tumor cell
resistance to chemotherapy (24–26).
The immune system controls the therapeutic response of cancer and
impedes progression, infiltration and metastasis. Immune
surveillance functions as a mechanism to detect, regulate and
eradicate malignant cells (27–29).
In vivo, treatment with programmed death-ligand 1 blockers
and glutathione (GSH) elimination can induce ferroptosis in tumor
cells and activate the antitumor activity of T lymphocytes
(30). A previous study
demonstrated that immunological B cells triggered an immune
response that suppressed tumor growth and was associated with
ferroptosis by increasing the dendritic cell count in patients with
oral cancer (31). Thus,
stimulating ferroptosis in tumor cells could be an effective
approach to tumor treatment.
To date, most studies have primarily focused on the
differences in ferroptosis between healthy individuals and patients
with HNSCC (31,32). However, the potential differences
between HPV-positive and HPV-negative HNSCC in terms of
ferroptosis-related genes (FRGs) have received little attention.
Furthermore, previous studies (31,32)
have merely focused on verifying the molecular characteristics of
genes from the incorporated prognostic models, without an in-depth
analysis to determine their regulatory role in the ferroptosis of
HNSCC cells. Therefore, the present study aimed to identify the
FRGs with significant involvement in HPV-negative HNSCC (which has
a particularly poor prognosis), to develop a predictive model using
the FRG signatures of HPV-negative HNSCC and to explore the
associations of these FRGs with the immune microenvironment (IME).
The present study also aimed to verify the specific functions of
the most promising FRGs based on the prognostic model. The
successful identification of such genes is expected to assist
prognosis predictions and enable targeted treatment, thereby
enhancing the clinical outcomes of patients with HPV-negative
HNSCC.
Materials and methods
Data sources
The source of the clinical data was a publication by
Hoadley et al (33). The
HNSCC datasets containing the transcript data were attained from
The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) on February 17, 2024.
There were 487 HNSCC samples, including 415 HPV-negative and 72
HPV-positive samples, included in the TCGA-HNSCC dataset. Gene Set
Enrichment Analysis (GSEA) and hallmark datasets from the Molecular
Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb/) were
utilized to identify relevant genes and pathways. The database used
for researching ferroptosis and FRGs is available at http://www.zhounan.org/ferrdb. Genes associated
with ferroptosis, including driver, suppressor and marker genes,
were downloaded to avoid omission. Additionally, GSE83519 (34) data were downloaded from https://www.ncbi.nlm.nih.gov/geo/.
Identification of FRGs
First, the FRG expression matrix was obtained using
R software (RStudio 2022.07.2). The ‘limma’ package of R (version
3.52.4) was used to identify differences in the expression of FRGs
between the HPV-negative and HPV-positive samples from the
TCGA-HNSCC group.
Consensus clustering
Consensus clustering was conducted using the
k-means method in the ‘ConsensusClusterPlus’ package of R
(version 1.60.0) to identify distinct patterns related to the FRGs.
The clustering accuracy was confirmed using Uniform Manifold
Approximation and Projection (UMAP) in the ‘ggplot2’ package of R
(version 3.3.6).
Identification and validation of
prognostic signatures based on the FRGs
Least Absolute Shrinkage and Selection Operator
(LASSO) regression analysis was conducted using the ‘glmnet’
package of R (version 4.1.4) to identify genes associated with
survival. Next, 10-fold cross-validation was used to determine the
penalty regularization value (λ). The key genes and their
accompanying coefficients were then determined using multivariate
Cox regression analysis. In total, 10 FRGs were selected as risk
signatures based on the optimal λ values and their corresponding
coefficients. The following formula was used to determine the risk
score for each patient using the new FRG signature:
Riskscore=Σexpi × βi where β and exp
represent the coefficient and the expression level of the
corresponding gene in the multivariate Cox regression model,
respectively.
For external validation, it was found that most Gene
Expression Omnibus datasets either lacked explicit HPV status
annotations or did not include paired prognostic information,
making them unsuitable for validating the constructed
HPV-subtype-specific prognostic model. Thereafter, comprehensive
internal validation on the TCGA-HNSC cohort was performed by
randomly partitioning the data into three independent subsets:
Training, testing and combined cohorts. The predictive performance
of the model was assessed through time-dependent receiver operating
characteristic (ROC) curve analyses, and the area under the curve
(AUC) was defined as the area enclosed by the ROC curve and the
coordinate axes. Additionally, Kaplan-Meier survival analysis
curves were generated. The ROC curve and Kaplan-Meier survival
analyses were performed using R software (RStudio 2022.07.2)
through ‘timeROC’ (version 0.4) and ‘survival’ (version 3.4.0) R
packages, respectively.
Construction and examination of the
predictive nomogram
The nomogram was generated based on risk scores and
clinicopathological features. Internal validation of accuracy was
performed using a calibration curve. The predictive performance of
the nomogram was evaluated using the time C-index. To assess the
net clinical benefit, decision curve analysis (DCA) was performed
(35).
Associations between immune cell
infiltration and risk score
The quantification of infiltrated immune cell
percentages was conducted using both the CIBERSORT R script and the
single sample GSEA R script following previously reported methods
(36). The distributions of immune
cell types among the groups were compared using CIBERSORT R. Each
sample showed an aggregated inferred score of 1 for the immune cell
types. Score 1 indicates that the sum of the relative proportions
of all immune cell types inferred in the sample equals 1, which is
equivalent to 100%. Furthermore, Spearman's rank correlation
analysis was used to ascertain the associations between the
infiltration of immune cells and risk scores.
Drug sensitivity analysis
For determining how effectively a patient responded
to treatment, drug sensitivity, also known as the half maximum
inhibitory concentration (IC50), was an essential
statistic. Through the Genomics of Drug Sensitivity in Cancer
database, the IC50 measurements were utilized to
demonstrate the sensitivity of each drug to each sample. The R
package known as ‘oncoPredict’ (37) was utilized to make predictions
regarding the response of anticancer treatments for each HNSCC
sample that was differentiated into the low-risk and high-risk
groups. All statistical analyses are represented using the
‘ggplot2’ R package (version 3.3.6).
Cell culture
The HNSCC CAL27 (HPV-negative) and UPCI-SCC-090
(HPV-positive) cell lines were obtained from Procell Life Science
& Technology Co. Ltd., and Zhejiang Meisen Cell Technology Co.,
Ltd., respectively. CAL27 cells were cultured in high-glucose
Dulbecco's modified Eagle's medium (DMEM; Procell Life Science
& Technology Co. Ltd.) with 10% fetal bovine serum (FBS;
Procell Life Science & Technology Co. Ltd.), while UPCI-SCC-090
cells were cultured in DMEM/F-12 (Zhejiang Meisen Cell Technology
Co., Ltd.) with 10% FBS. The cells were maintained at 37°C in a
humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from CAL27 and UPCI-SCC-090
cells using the Total RNA extraction kit (cat. no. R1200; Beijing
Solarbio Science & Technology Co., Ltd.) and reverse
transcribed into complementary DNA using the reverse transcription
kit (cat. no. 11141ES60; Shanghai Yeasen Biotechnology Co., Ltd.)
following the manufacturer's instructions. qPCR was performed using
Realtime PCR fluorescence quantitative kit (Hieff® qPCR
SYBR Green Master Mix; cat. no. 11201ES08; Shanghai Yeasen
Biotechnology Co., Ltd.) and a Real-time Fluorescence Quantitative
PCR Instrument (Suzhou Molarray Co., Ltd.) and the following
thermocycler conditions: 95°C for 5 min, followed by 40 cycles at
95°C for 10 sec, 60°C for 20 sec and 72°C for 20 sec. The primer
sequences are shown in Table SI.
The 2−ΔΔCq method was used to calculate relative gene
expression (38). GAPDH was used as
an internal control to normalize the expression levels of target
genes, correcting for variations in RNA input and RT
efficiency.
Cell transfection of Tribbles
pseudokinase 3 (TRIB3) small interfering RNA (siRNA)
CAL27 cells were seeded into 6-well plates (cat. no.
F603201-9001; Sangon Biotech Co., Ltd.) at a density of
1.5×105 cells/ml, with 2 ml in each well, 24 h before
transfection. The siRNA was purchased from OBiO Technology
(Shanghai) Corp., Ltd. and Lipofectamine™ 3000 (cat. no. L3000008;
Thermo Fisher Scientific, Inc.) was used for transfection. The
final concentration of the siRNA knockdown reagent (siTRIB3) and
siRNA negative control reagent was 20 pmol/µl, and the experimental
volume used for each was 5 µl. The cells were divided into the
blank control, negative control and knockdown groups. The blank
control group was cells + complete medium, the negative control
group was cells + complete medium + transfection reagent + negative
control siRNA and the knockdown group was cells + complete medium +
transfection reagent + target siRNA. Next, the cells were placed
into a 5% CO2 incubator at 37°C for further culture. The
three TRIB3 siRNAs were transfected separately and the siRNA
resulting in the most significant knockdown was selected for
subsequent experiments. After culturing for 48 h, RT-qPCR was
performed to examine the transfection efficiency and the most
efficient siRNA was chosen for the next experiment. The siRNA
sequences are displayed in Table
SII.
Cell transfection for TRIB3
overexpression
UPCI-SCC-090 cells were digested and seeded into a
6-well plate 24 h prior to transfection at a density of
2×105 cells/ml, with 2 ml in each well. The
overexpression plasmid [pcDNA3.1(+)-EGFP] was purchased from SYNBIO
Technology (Suzhou) Co., Ltd. (Suzhou Hongxun Biotechnology Co.,
Ltd.). Lipofectamine™ 3000 (cat. no. L3000008; Thermo Fisher
Scientific, Inc.) was used to transfect the cells. The final
concentration of the overexpression plasmid and empty plasmid was 1
µg/µl, and the experimental volume used for each was 2 µl. The
cells were divided into the blank control, negative control and
overexpression groups. The blank control group was cells + complete
medium, the negative control group was cells + complete medium +
transfection reagent + empty vector plasmid, and the overexpression
group was cells + complete medium + transfection reagent +
overexpression plasmid. Next, the cells were incubated at 37°C in a
5% CO2 incubator. After 5 h, the medium was replaced
with 10% FBS-DMEM, and the cells were cultured for another 48 h
before subsequent experiments. The transfection efficiency was
verified by RT-qPCR.
Western blotting
Total protein was extracted from the HNSCC cells.
For protein extraction, RIPA lysis buffer (cat. no. P0013B;
Beyotime Biotechnology) was combined with a protease/phosphatase
inhibitor (cat. no. P1045; Beyotime Biotechnology) cocktail at a
1:100 ratio, and the mixture was used immediately. Next, for the
quantification of protein concentration, a BCA protein assay kit
(cat. no. PC0020; Beijing Solarbio Science & Technology Co.,
Ltd.) was employed, following the manufacturer's recommended
protocol. For electrophoresis, the processed samples were loaded
onto the gel with a total mass of 20 µg protein per well. Following
separation using 8% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, the extracted proteins were transferred onto
polyvinylidene fluoride membranes (cat. no. IPVH00010;
MilliporeSigma). Subsequently, the membranes were blocked using 5%
non-fat milk for 3 h on a shaker at room temperature and incubated
overnight at 4°C with primary antibodies targeting GAPDH (cat. no.
10494-1-AP; 1:5,000; Proteintech Group Inc.), TRIB3 (cat. no.
DF7844; 1:1,000; Affinity Biosciences, Ltd.) and acyl-CoA
synthetase long chain family member 1 (ACSL1; cat. no. GTX112430;
1:1,000; GeneTex, Inc.). The secondary antibody was a
HRP-conjugated goat anti-rabbit IgG antibody (cat. no.
bs-0295G-HRP; 1:5,000; BIOSS), which was incubated with the
membrane on a horizontal shaker at room temperature for 1.5 h.
Images were acquired with the Automated Chemiluminescent Image
Analysis System (Tanon 5200; Tanon Science & Technology Co.,
Ltd.) following ECL (cat. no. P0018S-2; Beyotime Biotechnology)
substrate incubation. Lastly, the intensity of the protein bands
was semi-quantified using ImageJ software (version 1.53; National
Institutes of Health).
Quantification of ROS
ROS were quantified using the ROS fluorometric assay
kit (cat. no. E-BC-K138-F, Wuhan Elabscience Biotechnology Co.,
Ltd.) according to the manufacturer's guidelines. CAL27 cells
(1×106 cells/well) were cultured in 6-well plates.
Cultured or transfected cells were treated in the dark with the ROS
solution for 20 min at 37°C, then the fluorescence intensity was
evaluated by an inverted fluorescence microscope [XD-RFL; Sunny
Optical Technology (Group) Co., Ltd.].
Measurement of intracellular Fe2+
content. The intracellular Fe2+ concentration was
quantified using Cell Ferrous Iron Colorimetric assay reagent (cat.
no. E-BC-K881-M; Wuhan Elabscience Biotechnology Co., Ltd.).
Briefly, the cultured or transfected cells (1×106) were
mixed with 0.2 ml reagent 1 and incubated in an ice bath for 10 min
to facilitate lysis, followed by centrifugation at 15,000 × g for
10 min at room temperature. For further analysis, the absorbance of
the supernatants at 593 nm was measured using a microplate reader
(Synergy H4; BioTek; Agilent Technologies, Inc.).
Measurement of cellular GSH
The GSH concentration in CAL27 cells was measured
using the Reduced GSH Colorimetric Assay Kit (cat. no. E-BC-K030-M;
Wuhan Elabscience Biotechnology Co., Ltd.) following the
manufacturer's protocol. The absorbance at 405 nm was measured
using a microplate reader (Synergy H4; BioTek; Agilent
Technologies, Inc.).
Measurement of cellular
malondialdehyde (MDA)
Cellular MDA was quantified using the
Malondialdehyde Colorimetric Assay Kit (cat. no. E-BC-K028-M; Wuhan
Elabscience Biotechnology Co., Ltd.) following the manufacturer's
instructions. Transfected cells were harvested using an extraction
solution. The test samples, blank tubes and standard tubes were
incubated at 100°C for 40 min and then cooled to room temperature
in a water bath. The resulting samples were centrifuged in
Eppendorf (EP) tubes at 1,078 × g for 10 min at room temperature
and 0.25 ml supernatant was transferred to the enzyme-labeled
plate. The absorbance of each well was measured at 532 nm using a
microplate reader (Synergy H4; BioTek; Agilent Technologies,
Inc.).
Flow cytometry
CAL27 cells (1×106) were cultured in
6-well plates. After transfection with siTRIB3, the cells were
incubated for 24 h before being treated with propidium iodide
(Beijing Solarbio Science & Technology Co., Ltd.) for 30 min at
37°C. Subsequently, the cell cycle distribution was analyzed by
flow cytometry (NL-CLC3000; Cytek Biosciences, Inc.) and FlowJo
software (10.8.1), with an excitation wavelength of 488 nm.
Transmission electron microscopy
CAL27 cells (both the control group and those
transfected with siTRIB3 together with the culture medium) were
subjected to centrifugation at 42-107 × g for 5 min at room
temperature, followed by careful removal of the resulting
supernatant with 1 ml remaining. The CAL27 cell pellet was gently
resuspended in the remaining media and transferred to a 1.5-ml EP
tube. After allowing the mixture to settle for 1 h, the supernatant
was meticulously extracted and 1 ml pre-chilled 2.5% glutaraldehyde
was gradually added to the tube along the periphery for fixation.
The tube was then placed in a refrigerator at 4°C for at least 2 h.
The cells were subsequently fixed in 1% osmic acid at 4°C for 2 h.
Following fixation, the cells were detached from the plastic
surface and dehydrated using ethanol and acetone, respectively. The
specimens were embedded in EPON812 embedding solution (SPI-Pon 812
Kit; cat. no. 02663-AB; SPI Supplies; Structure Probe, Inc.) for
2–3 h at 37°C and sectioned to 70 nm. Thereafter, double staining
was carried out: First with 2% uranyl acetate (staining time: 15–20
min at room temperature) and then with lead citrate (staining time:
10–15 min in a CO2-free environment to avoid
precipitation), ensuring sufficient contrast for ultrastructural
observation. The sections were then analyzed using an HT7800 (80
kV) transmission electron microscope (Hitachi, Ltd.).
Immunohistochemistry (IHC)
Samples from patients with primary HNSCC (with or
without comorbidities) who had undergone surgery at the Department
of Pathology, Heping Hospital Affiliated to Changzhi Medical
College (Changzhi, China) from January 1, 2018 to December 31, 2022
were included. The patient samples for IHC were used
retrospectively. All patients were aged >18 years and had not
undergone any other prior treatment (including radiation and
chemotherapy). Patients with a history of prior treatments
(radiotherapy or chemotherapy) or those with recurrent HNSCC were
excluded from the study. In accordance with the predefined
inclusion and exclusion criteria, a total of 179 eligible cases
were eventually enrolled. Among these patients, the age range was
38 to 87 years, with a mean age of 63.2 years. Regarding sex
distribution, 121 patients were men (accounting for 67.6% of the
total) and 58 were women (representing 32.4%).
For paraffin-embedded slides (0.4 µm), first the
sections were baked in a 60–65°C preheated oven for 15–20 min to
melt the surface paraffin. Then, the sections were sequentially
immersed in xylene to completely remove residual paraffin, followed
by gradual rehydration via gradient ethanol solutions (from high to
low concentration) and finally transitioned to an aqueous
environment. After antigen retrieval of paraffin sections (EDTA
buffer, 95–100°C, 20 min) and subsequent cooling to room
temperature followed by 2–3 washes with PBS (3–5 min each),
permeabilization was performed by immersing the sections in a
paraffin section-compatible permeabilization reagent (0.3% Triton
X-100 in PBS) for 10–15 min. Next, the sections were blocked with
10% bovine serum albumin (cat. no. C0221; Beijing Solarbio Science
& Technology Co., Ltd.) at room temperature for 30 min and then
incubated with primary antibodies against TRIB3 (cat. no. bs-7538R;
1:300; BIOSS) and p16 (cat. no. HA721415; 1:200; HUABIO) at 4°C
overnight. Next, the samples were incubated with goat anti-rabbit
IgG/biotin secondary antibody (cat. no. SHB134; 1:200; Beijing
Solarbio Science & Technology Co., Ltd.) for 2 h at 37°C.
Hematoxylin was used to counterstain the nucleus (1 min, room
temperature). Finally, the sections were incubated with 0.05–0.1%
3,3′-diaminobenzidine solution (containing 0.01%
H2O2, dissolved in Tris-HCl buffer) at room
temperature for 3–10 min to allow brown precipitates to form at the
target antigen-binding sites, followed by terminating the staining
reaction with distilled water. Then, the sections were scanned
using a panoramic slice scanner. The present study was approved by
the Human Ethical Committee of Heping Hospital Affiliated to
Changzhi Medical College [Changzhi, China; approval no. 2023(027)].
The requirement for informed consent was waived by the committee as
the risks involved in the study were very low and did not cause
significant harm to the participants.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism (version 10.0.3; Dotmatics) and R (version 4.2.1). The
results are presented as the mean ± standard deviation. Two-group
comparisons were performed using the unpaired t-test, while
multiple-group comparisons were performed using one-way ANOVA and
Tukey's HSD post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of FRGs associated with
prognosis
In total, 845 FRGs were identified in the
ferroptosis database. A comparison between HPV-negative and
HPV-positive samples identified 299 differentially expressed FRGs
from TCGA-HNSC. When HPV-negative was compared with HPV-positive
samples, 208 genes were downregulated, and 91 genes were
upregulated in the HPV-negative samples (Table SIII). Univariate Cox regression
analysis indicated that 25 of the 300 FRGs had a significant
association with survival, as depicted in the forest plot
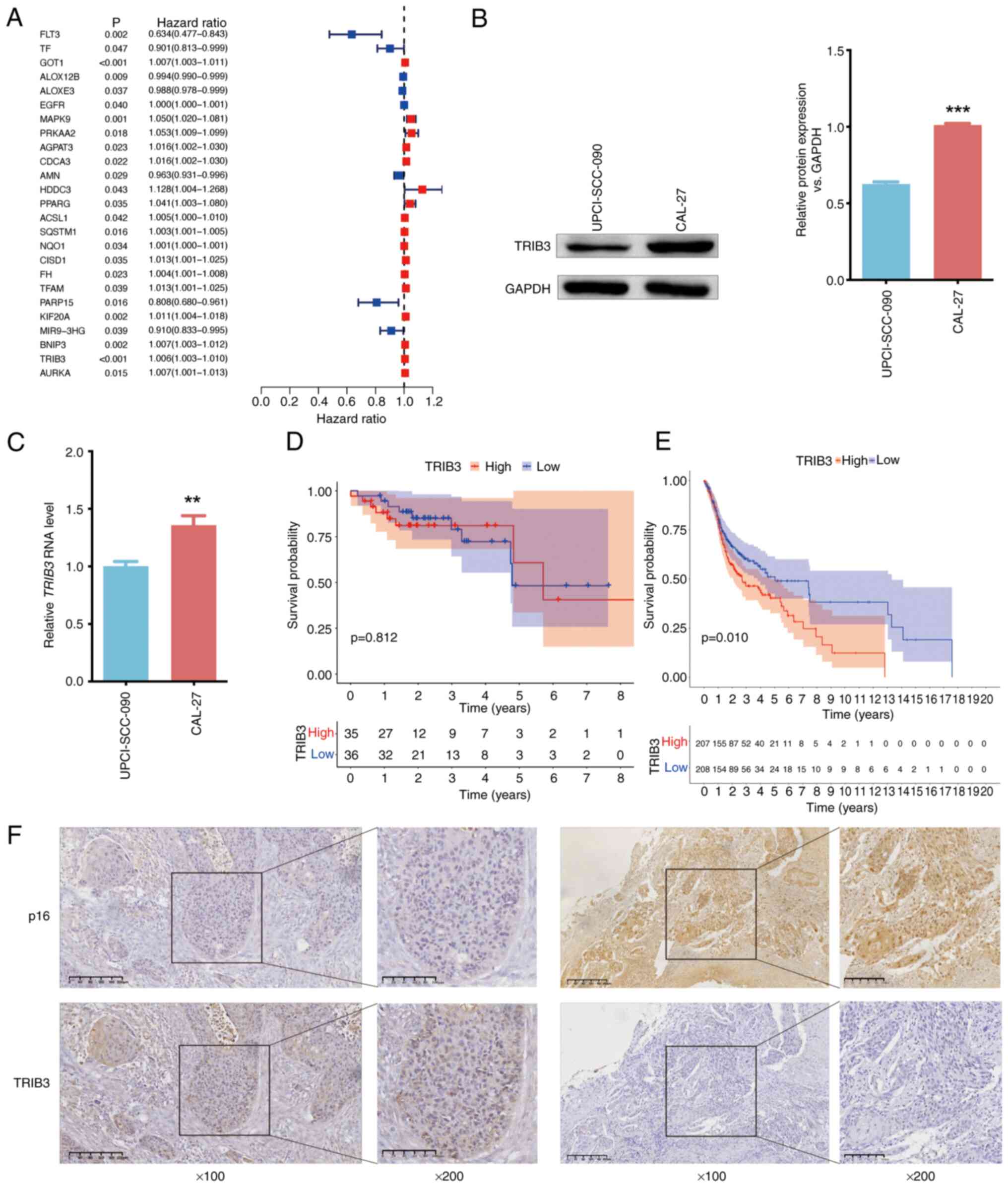
(P<0.05; Fig. 1A). TRIB3
was significantly elevated in HNSCC samples according to the
results of TCGA-HNSCC (Fig. S1)
and GSE83519 (Fig. S2) analyses.
TRIB3 was also more upregulated in HPV-negative samples than
in HPV-positive samples (Table
SIII and Fig. S3), suggesting
that it may be particularly important in HPV-negative HNSCC.
Western blotting (Fig. 1B) and
RT-qPCR (Fig. 1C) confirmed that
the difference in TRIB3 expression between HNSCC CAL27 (HPV
negative) and UPCI-SCC-090 (HPV positive) cells aligned with the
results of the bioinformatics analysis. Additionally, ACSL1
expression was also upregulated in HPV-negative cells (Fig. S4). TRIB3 was associated with
the prognostic outcome of patients with HPV-negative HNSCC, but not
with that of patients with HPV-positive HNSCC (Fig. 1D and E). The immunohistochemistry
results from paraffin-embedded slides of TRIB3 were consistent with
those of western blotting and RT-qPCR from cell culture experiments
(Fig. 1F).
Consensus clustering of the 25 FRGs in
HPV-negative HNSCC
To understand the significance of the FRGs in
HPV-negative HNSCC, consensus clustering on the 25
prognosis-associated FRGs was conducted using the
‘ConsensusClusterPlus’ package in R software. The HPV negative
TCGA-HNSC cohort was classified into three clusters at K=3
(Fig. 2A). Following this, UMAP was
used to verify the accuracy of the clustering, which indicated that
the three clusters were accurately distinguished at K=3 (Fig. 2B). The OS analysis revealed
significant variations in prognoses among the three subgroups and
cluster A had a favorable prognosis (P=0.029; Fig. 2C). The heatmap in Fig. 2D illustrates the expression of the
FRGs and the respective clinicopathological features of the three
clusters. For example, epidermal growth factor receptor
(EGFR) was significantly upregulated in cluster B, and T3
stage. Furthermore, the transcription patterns of the FRGs in the
three subgroups were depicted using boxplots. The expression levels
of EGFR and transcription factor A, mitochondrial were
markedly higher in cluster B than in clusters A and C, while the
expression levels of NAD(P)H quinone dehydrogenase 1, TRIB3,
glutamic-oxaloacetic transaminase 1, Sequestosome 1, CDGSH iron
sulfur domain 1 and fumarate hydratase were higher in cluster C
than in clusters A or B (Fig. 2E).
Therefore, these differentially expressed FRGs may serve as
important determinants for estimating the prognosis of patients
with HNSCC, and based on their association with OS, they may be
potentially useful therapeutic targets. The Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway enrichment analysis performed
using the GSEA package revealed different enrichment pathways
between clusters A and B. This analysis was performed to identify
significantly enriched pathways in clusters A and to clarify the
unique activation or inhibition of metabolic, signal transduction,
disease-related and other pathways in clusters A (Fig. 2F-I). Cluster A displayed significant
involvement in critical pathways, such as
‘KEGG_JAK_STAT_SIGNALING_PATHWAY’,
‘KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY’,
‘KEGG_GLUTATHIONE_METABOLISM’ and
‘KEGG_HEDGEHOG_SIGNALING_PATHWAY’, all of which play essential
roles in tumorigenesis.
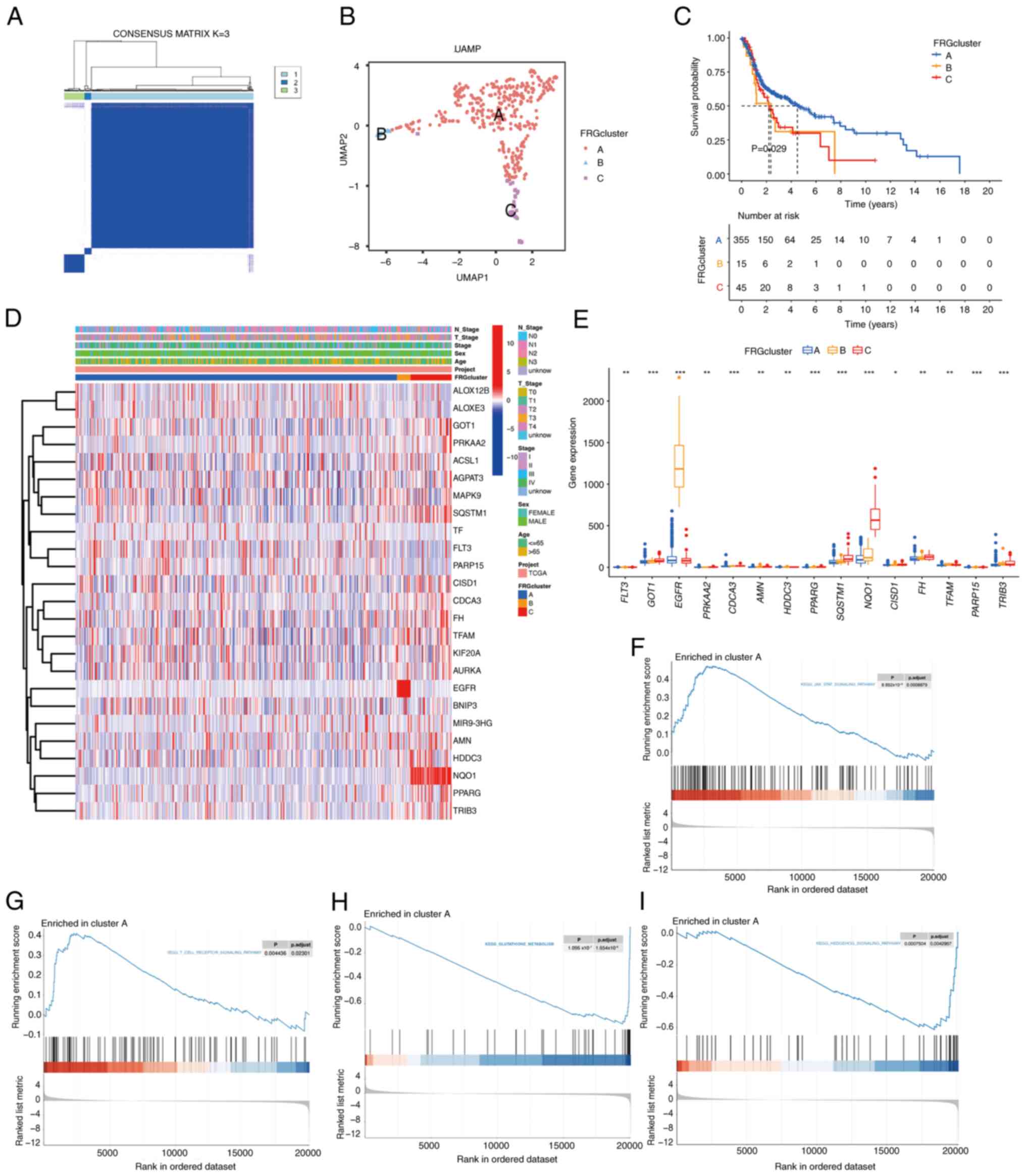 | Figure 2.FRG-related subgroups of HPV-negative
HNSCC samples from TCGA. (A) Consensus clustering. The consensus
matrix of K=3 was obtained. (B) UMAP identified three clusters
according to FRG expression. (C) Overall survival in the three
clusters (P=0.029). (D) Heatmap of the clinicopathological
characteristics and FRG expression associated with the three
clusters. (E) The expression of FRGs in the three clusters.
Comparative KEGG pathway enrichment between clusters A and B, as
evaluated by GSEA. Cluster A displayed significant involvement in
(F) KEGG_JAK_STAT_SIGNALING_PATHWAY, (G)
KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY, (H)
KEGG_GLUTATHIONE_METABOLISM and (I)
KEGG_HEDGEHOG_SIGNALING_PATHWAY. *P<0.05, **P<0.01,
***P<0.001. FRGs, ferroptosis related genes; HPV, human
papillomavirus; HNSCC, head and neck squamous cell carcinoma; TCGA,
The Cancer Genome Atlas; UMAP, Uniform Manifold Approximation and
Projection; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA,
Gene Set Enrichment Analysis. |
Identification of prognostic FRG
signatures using the FRG score model
The clinical utility of the 25 FRGs was evaluated
using the LASSO-penalized Cox analysis (Fig. 3A and B). Following the core steps of
LASSO Cox-based gene selection: First, non-zero coefficient FRGs
(that is FRGs contributing to survival prediction and free of
multicollinearity) were initially extracted from the 25 candidates;
second, these non-zero coefficient FRGs were further filtered by
prioritizing those with larger absolute regression coefficients. In
total, 10 prognostic FRGs, including TRIB3, EGFR,
1-acylglycerol-3-phosphate O-acyltransferase 3
(AGPAT3), peroxisome proliferator-activated receptor γ
(PPARG), lipoxygenase-3 (ALOXE3), BCL2-interacting
protein 3 (BNIP3), ACSL1, FMS-like tyrosine kinase 3
(FLT3), amnion associated transmembrane protein (AMN)
and MIR9-3 host gene (MIR9-3HG) were obtained from these 25
FRGs. The association coefficients are presented in Table SIV, and the final risk score
derived from the 10 FRG signatures is referred to as the
‘FRGscore.’ The prognostic index (FRGscore) was calculated
according to the following formula: (0.00078 × EGFR
expression) + (0.028 × AGPAT3 expression) + (0.043 ×
PPARG expression) + (0.006 × ACSL1 expression) +
(0.007 × BNIP3 expression) + (0.004 × TRIB3
expression)-(0.02 × ALOXE3 expression)-(0.798 × FLT3
expression)-(0.05 × AMN expression)-(0.098 × MIR9-3HG
expression). According to the FRGscore, the training, testing and
combined cohorts were divided into high- and low-risk groups.
Individuals or cases with scores above the median score were
categorized as high-risk, and those below or equal to the median
score as low-risk. The Kaplan-Meier survival curves indicated that
the prognoses of the training, testing and combined cohorts in the
high-risk group were poor (Fig.
3C-E). Using the FRGscore model, the time-dependent ROC curves
exhibited notable predictive efficacy by evaluating OS at 1-, 3-
and 5-year intervals. This was demonstrated by the AUC being
>0.7, except the AUC at 1 year in the testing (0.606) and
combined (0.698) cohorts (Fig.
3F-H). This supports the reliability of the predictive model
for assessing the predefined outcomes. The heatmap shown in
Fig. 3I illustrates the expression
of the FRGs across various risk scores: ALOXE3, FLT3, AMN
and MIR903GH showed greater expression levels in the
high-risk group, whereas EGFR, BNIP3, ACSL1, AGPAT3, PPARG
and TRIB3 exhibited higher expression in the low-risk group.
Additionally, the risk scores of the previously established three
clusters varied significantly from each other (Fig. 3J).
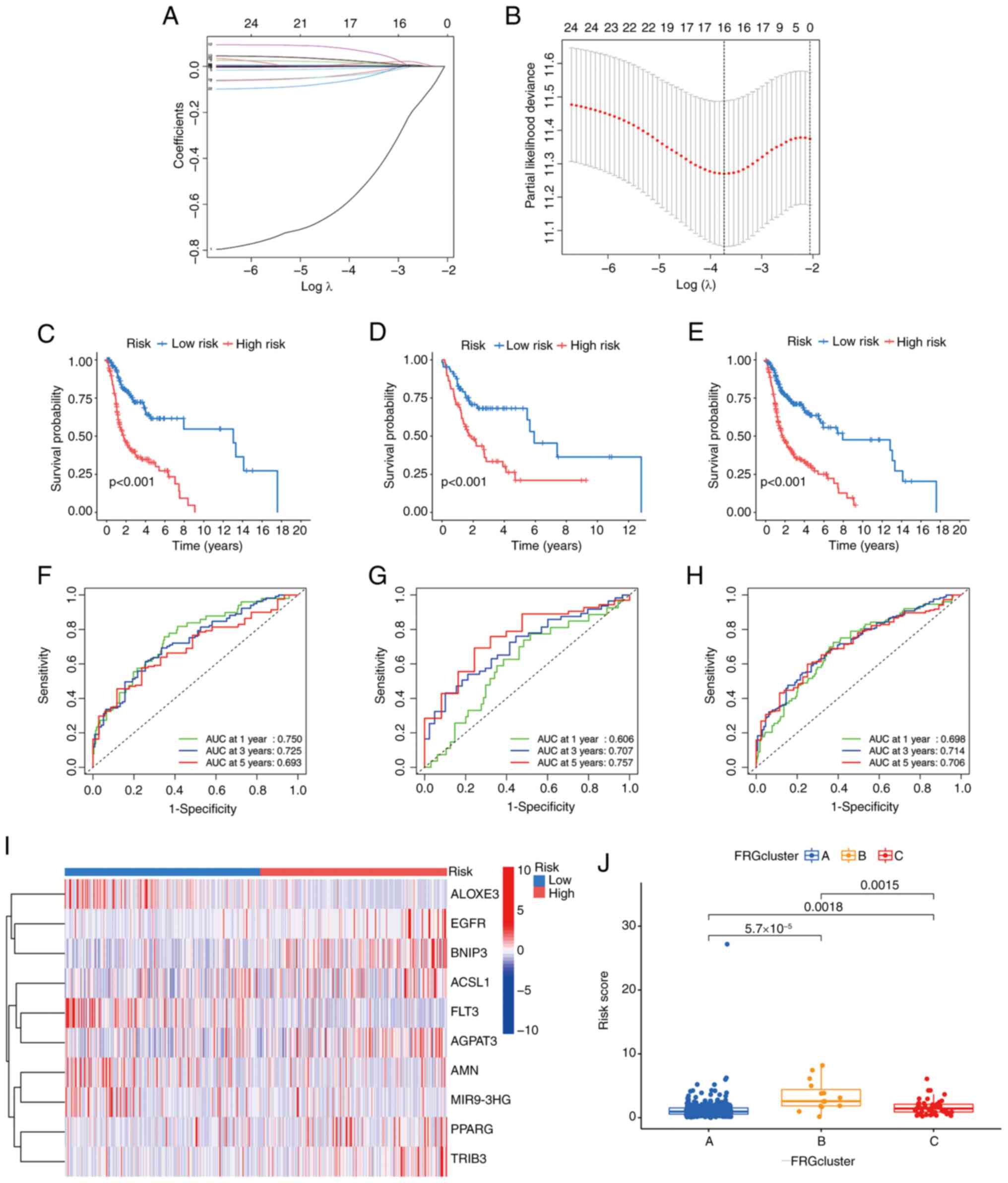 | Figure 3.Determination of the prognostic FRGs
in HPV-negative HNSCC samples from TCGA. (A) 10 prognostic FRGs
were determined by the LASSO regression analysis and validated by
10-fold cross-validation. (B) The graph of the coefficient profile
illustrated 10 prognostic FRGs. Kaplan-Meier survival curves
showing the prognoses of the different risk groups in the (C)
training, (D) testing and (E) combined cohorts. Time-dependent ROC
curves were generated for OS at 1, 3 and 5 years using the (F)
training, (G) testing and (H) combined cohorts. (I) The heatmap of
the 10 hub FRGs for both risk groups. (J) Evaluation of the risk
scores among the three previously established clusters. FRGs,
ferroptosis related genes; TCGA, The Cancer Genome Atlas; LASSO,
Least Absolute Shrinkage and Selection Operator; ROC, receiver
operating characteristic; HPV, human papillomavirus; ALOXE3,
lipoxygenase-3; EGFR, epidermal growth factor receptor; BNIP3,
BCL2-interacting protein 3; ACSL1, acyl-CoA synthetase long chain
family member 1; FLT3, FMS-like tyrosine kinase 3; AGPAT3,
1-acylglycerol-3-phosphate O-acyltransferase 3; AMN, amnion
associated transmembrane protein; MIR9-3HG, MIR9-3 host gene;
PPARG, peroxisome proliferator-activated receptor γ; TRIB3,
tribbles pseudokinase 3. |
Development of a prognostic nomogram
for patients with HPV-negative HNSCC
For the nomogram integrating FRGscore and clinical
data, the impact of clinicopathological variables was considered by
first screening prognosis-related variables (such as age and tumor
stage) via univariate Cox regression, then validating their
independent prognostic value by multivariate Cox regression and
including significant variables (P<0.05) together with FRGscore,
and finally assigning each variable a prognostic weight-based score
scale to quantify contributions for multi-dimensional outcome
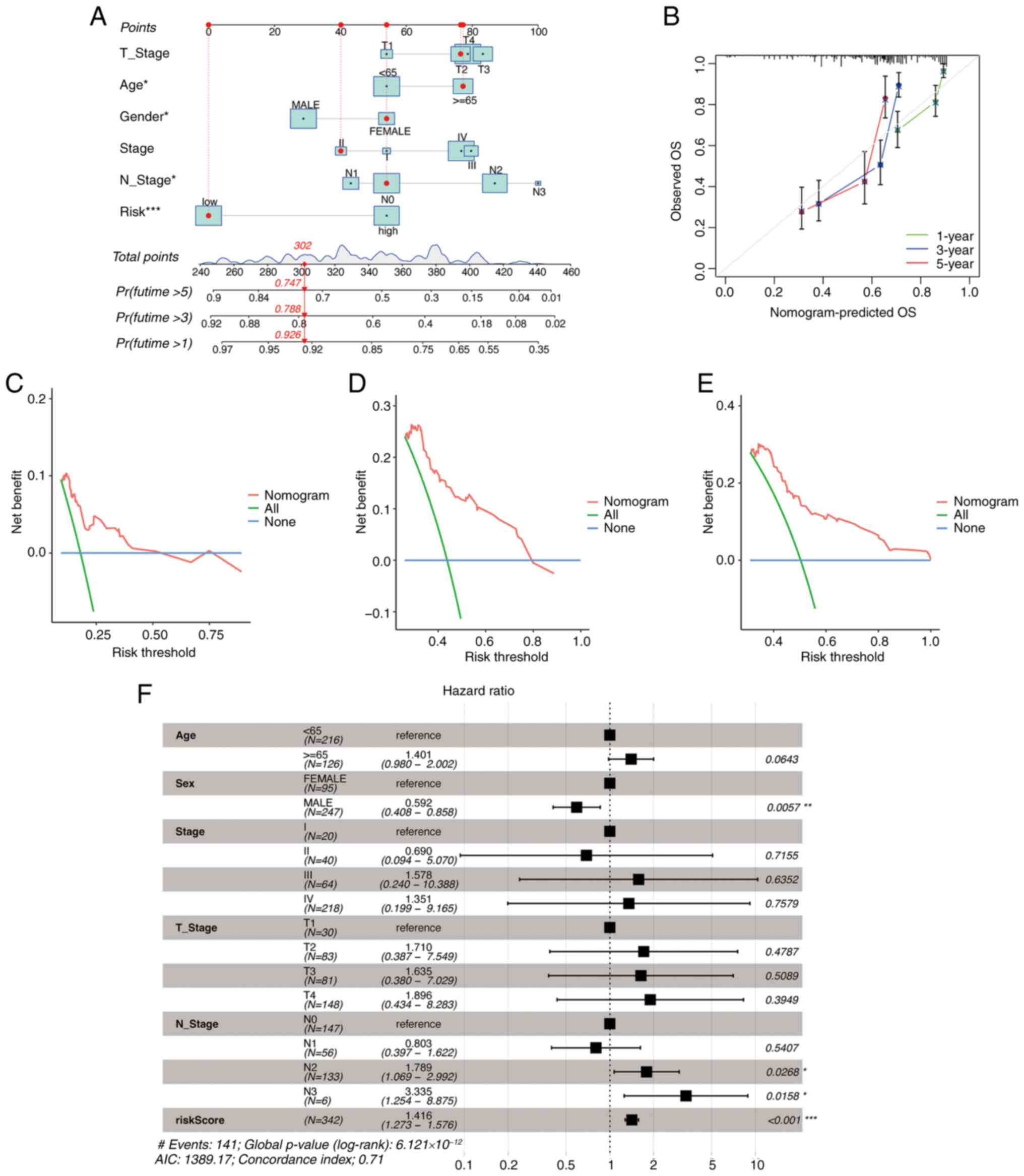
prediction (Fig. 4A). The accuracy
of the nomogram was verified through a calibration curve (Fig. 4B). DCA determines the possible
patient benefit of clinical prediction models, diagnostic
procedures and molecular markers (35). The reliability of the nomogram for
estimating the short- and long-term survival of patients with HNSCC
was confirmed by the DCA (Fig.
4C-E). The multivariable Cox regression analysis showed that
the primary influencing factors in the nomogram were sex, N stage
and risk score (Fig. 4F). The
constructed nomogram may thus be valuable for the prediction of
clinical prognosis.
Immune infiltration in patients with
HPV-negative HNSCC
Both tumor progression and the efficacy of
immunotherapy are influenced by the IME. To distinguish between the
risk groups of patients with HPV-negative HNSCC, the tumor IME was
investigated in more detail. The CIBERSORT R script was used to
assess the percentages of infiltrating immune cell types.
HPV-negative HNSCC tumors were ranked based on the risk score, and
associations between the immune cell types and risk scores were
observed (Fig. 5A). The low-risk
group represented a higher proportion of T cells, B cells and NK
cells, whereas the high-risk group represented a higher proportion
of macrophages, mast cells, eosinophils and neutrophils. The
correlations among immune cells in patients with HNSCC are
displayed in Fig. 5B. T cells CD4
memory activated was positively associated with T cells CD8
(r=0.58), while T cells CD8 was negatively associated with
macrophages M0 (r=−0.53). This may provide insights into the
specific IME associated with certain tumor types. In patients with
low-risk HNSCC without HPV infection, M0 macrophages accounted for
a lower proportion of immune cell components, as well as
CD8+ T cells, T cells CD4 memory activated, NK cells
resting and mast cells activated (Fig.
5C).
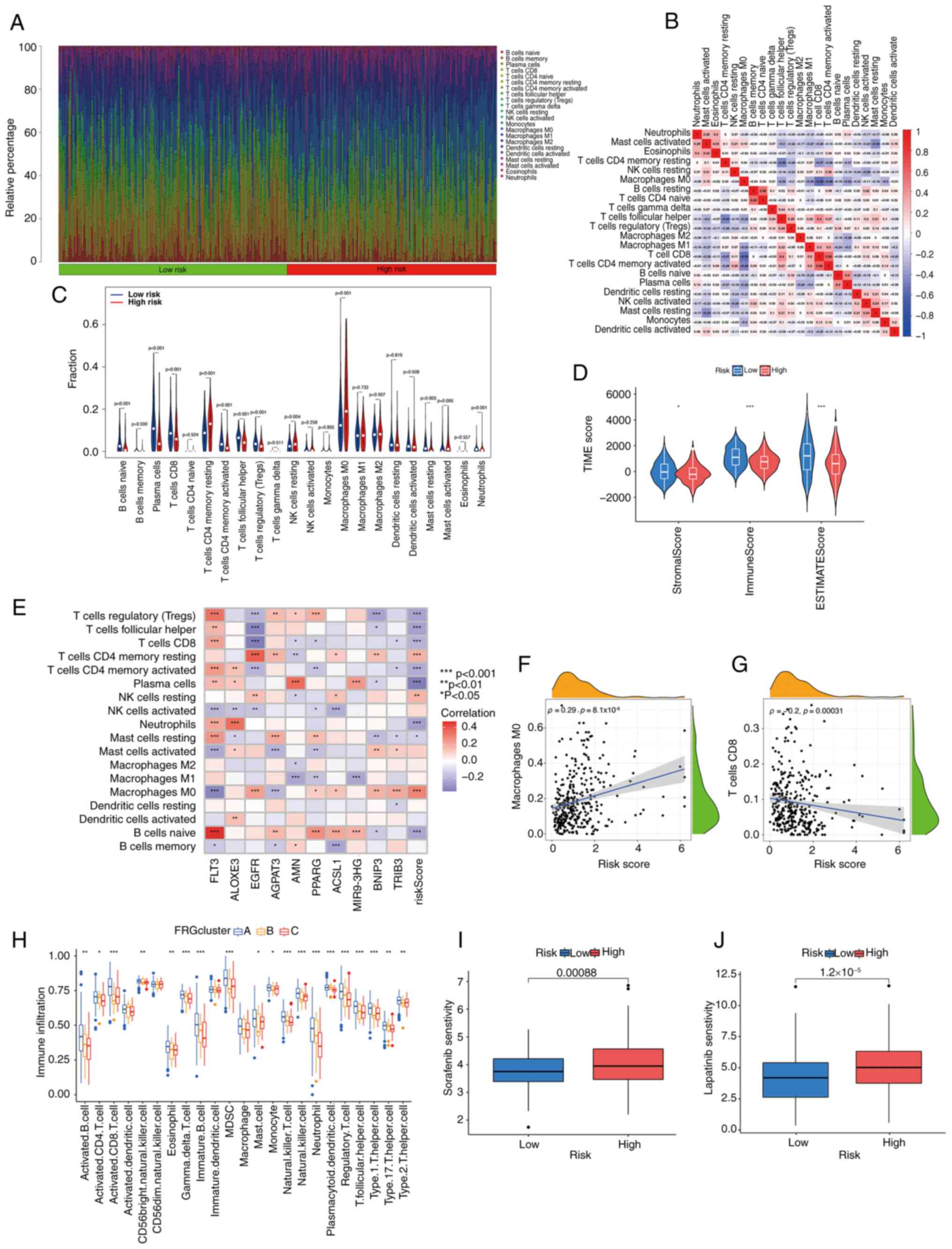 | Figure 5.Exploring the immune microenvironment
of HPV-negative HNSCC based on the various risk scores and clusters
in HPV-negative HNSCC samples from TCGA. (A) Different proportions
of infiltrating immune cells with distinct risk scores. (B) The
associations among immune cells. (C) Comparison of immune cell
components between the risk groups. (D) The predicted score of the
gene expression patterns in both risk groups. (E) The connection
between immune cells and the 10 main ferroptosis-related genes.
Associations of (F) the number of M0 macrophages and (G) the number
of CD8+ T cells in HNSCC-negative tissues with risk
scores. (H) The variation in the extent of immune cell infiltration
across the three FRG clusters. The susceptibility of both risk
groups to (I) sorafenib and (J) lapatinib. *P<0.05, **P<0.01,
***P<0.001. HNSCC, head and neck squamous cell carcinoma; HPV,
human papillomavirus; TCGA, The Cancer Genome Atlas; TME, tumor
microenvironment; FRG, ferroptosis related gene; FLT3, FMS-like
tyrosine kinase 3; ALOXE3, lipoxygenase-3; EGFR, epidermal growth
factor receptor; AGPAT3, 1-acylglycerol-3-phosphate
O-acyltransferase 3; AMN, amnion associated transmembrane protein;
PPARG, peroxisome proliferator-activated receptor γ; ACSL1,
acyl-CoA synthetase long chain family member 1; MIR9-3HG, MIR9-3
host gene; BNIP3, BCL2-interacting protein 3; TRIB3, tribbles
pseudokinase 3. |
Subsequently, the stromal and immune scores for both
risk groups were determined using the estimated score of the
expression profile. The core purpose of calculating stromal and
immune scores for both risk groups is to link the ‘FRGscore-based
molecular risk stratification’ with ‘tumor microenvironment
characteristics’, which not only provides TME-level support for the
mechanism by which FRGscore affects prognosis but also offers a
basis for formulating subsequent intervention strategies (such as
immunotherapy) for different risk groups. The StromalScore,
ImmuneScore and ESTIMATEScore represented a significant difference
between the high-risk and low-risk groups (Fig. 5D). Furthermore, the infiltration of
various immune cell types was relatively significantly correlated
with the 10 FRGs that were used to develop the FRGscore model
(Fig. 5E). Elevated risk scores
were linked to a greater number of M0 macrophages (Ρ=0.29; Fig. 5F) but fewer CD8+ T cells
(Ρ=−0.2; Fig. 5G). Furthermore,
there was notable variation in the extent of immune cell
infiltration across the three FRG clusters (Fig. 5H). Cluster B showed fewer activated
CD8 cells, MDSCs, mast cells, type 1 T helper cells and type 2 T
helper cells than the other two clusters, while cluster C showed
fewer activated B cells, activated CD4 T cells, gd T, immature B
cells, natural killer T cells, neutrophils, plasmacytoid dendritic
cells, regulatory T cells and type 17 T helper cells. The
‘pRRophetic’ package in R was used to assess therapeutic drug
susceptibility in the different risk groups. A significant
association was observed between the risk score and the
responsiveness to various drugs, for example, samples with high
risk were more sensitive to the two drugs sorafenib and lapatinib
(Fig. 5I and J).
TRIB3 knockdown induces ferroptosis in
HPV-negative HNSCC cells
To determine whether TRIB3 mediated
ferroptosis in HPV-negative HNSCC cells, TRIB3 was knocked
down in CAL27 cells using three different siRNA sequences. RT-qPCR
revealed that TRIB3 mRNA expression significantly decreased
after TRIB3 siRNA transfection (Fig. 6A), indicating successful
TRIB3 knockdown in CAL27 cells. Transfection with
TRIB3 siRNA-3 induced the largest effect. Therefore,
TRIB3 siRNA-3 was selected for further studies. The cellular
content of Fe2+, GSH and MDA, which can reflect the
ferroptosis state, were quantified, while the level of ROS (another
indicator of ferroptosis) was visualized via imaging; these
analyses were performed to investigate whether TRIB3 inhibits
ferroptosis in HNSCC cells. Upon TRIB3 knockdown in cancer
cells (CAL27), the concentration of GSH decreased (Fig. 6B), while the concentrations of
Fe2+ and MDA increased (Fig.
6C and D). Furthermore, intracellular ROS were upregulated
after TRIB3 knockdown, indicating ferroptosis induction
(Fig. 6E). Moreover, in CAL27 cells
with siRNA, transmission electron microscopy elucidated a
discernible decrease or complete absence of mitochondrial ridges,
concomitant with elevated mitochondrial membrane density (Fig. 6F). Finally, cell cycle analysis was
performed by flow cytometry, indicating that TRIB3 knockdown
decreased the population of cells in the S phase (Fig. 6G). These findings suggest that
TRIB3 knockdown may promote ferroptosis in HPV-negative
HNSCC cells.
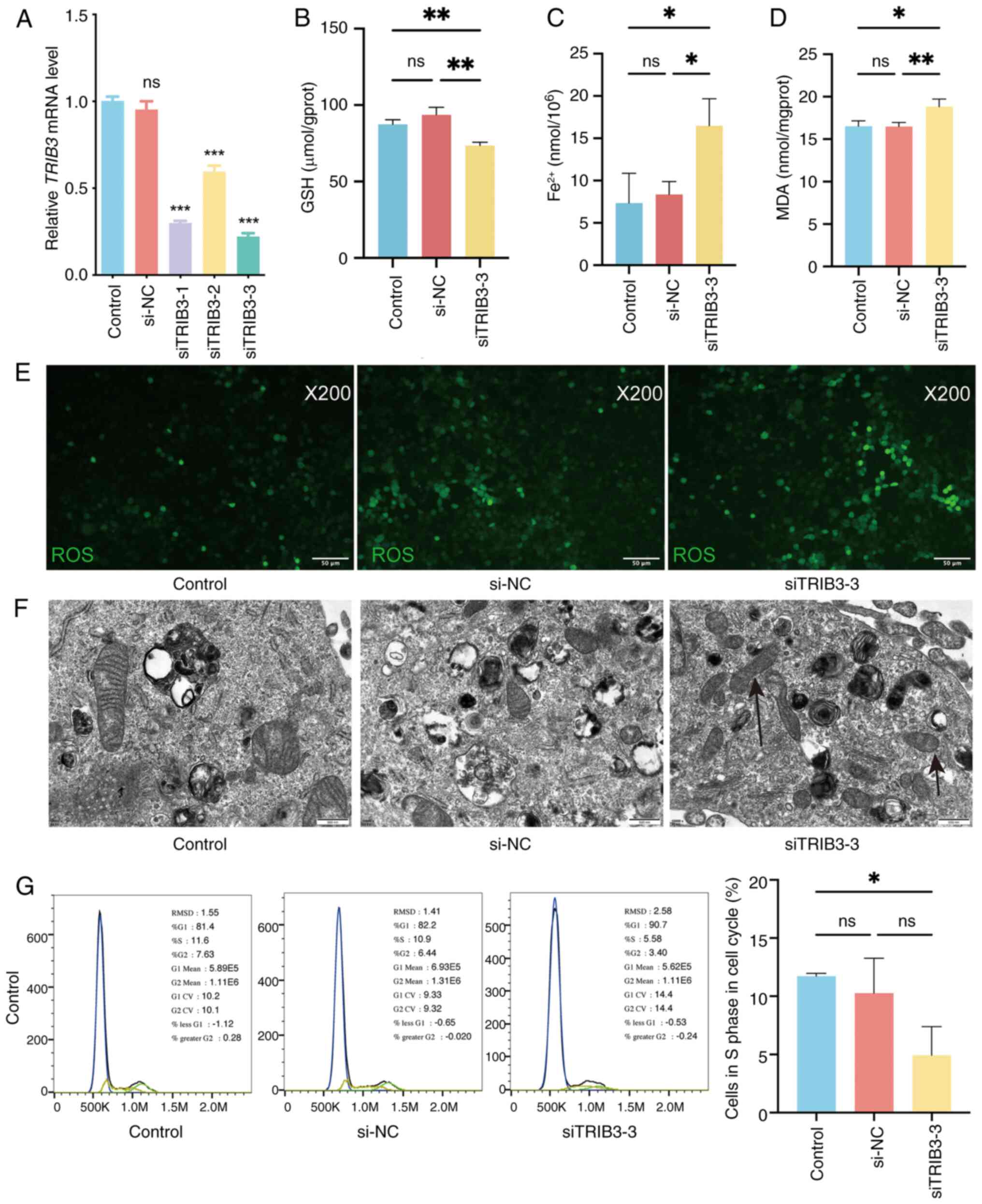 | Figure 6.Knockdown of TRIB3 promotes
ferroptosis in HPV-negative HNSCC cells. (A) Relative TRIB3
mRNA expression in CAL27 cells in the different groups measured by
reverse transcription-quantitative polymerase chain reaction after
siRNA transfection. (B) The intracellular GSH concentration was
measured using the reduced GSH Colorimetric Assay Kit. (C) The
intracellular Fe2+ concentration was measured using the
Cell Ferrous Iron Colorimetric Assay Kit. (D) The intracellular MDA
concentration was measured using the Cell Malondialdehyde
Colorimetric Assay Kit. (E) ROS (green fluorescence) were measured
in the cells. (F) Observation of the cells under transmission
electron microscopy. (G) Cell cycle distribution analysis using
flow cytometry. *P<0.05, **P<0.01, ***P<0.001. ns,
non-significant; HNSCC, head and neck squamous cell carcinoma; GSH,
glutathione; MDA, malondialdehyde; ROS, reactive oxygen species;
HPV, human papillomavirus; TRIB3, tribbles pseudokinase 3; si,
small interfering (RNA); NC, negative control. |
TRIB3 overexpression inhibits
ferroptosis in HPV-positive HNSCC cells
To determine whether TRIB3 mediates
ferroptosis in HPV-positive HNSCC cells, TRIB3 was
overexpressed in UPCI-SCC-090 cells. RT-qPCR revealed that
TRIB3 mRNA expression significantly increased after
TRIB3 transfection (Fig.
7A), indicating successful TRIB3 overexpression in
UPCI-SCC-090 cells. Furthermore, the concentrations of
Fe2+, GSH and MDA were quantified while the level of ROS
was visualized via imaging; these analyses were conducted to assess
the potential induction of ferroptosis by TRIB3 in HPV-positive
HNSCC cells. TRIB3 overexpression in HPV positive HNSCC cancer
cells (UPCI-SCC-090) led to an increase in the GSH concentration
(Fig. 7B), accompanied by a
decrease in the Fe2+ and MDA concentrations (Fig. 7C and D). Moreover, there was a
noticeable decline in intracellular ROS following TRIB3
overexpression, indicating ferroptosis suppression (Fig. 7E). These results suggest that TRIB3
expression may inhibit ferroptosis in HPV-positive HNSCC cells.
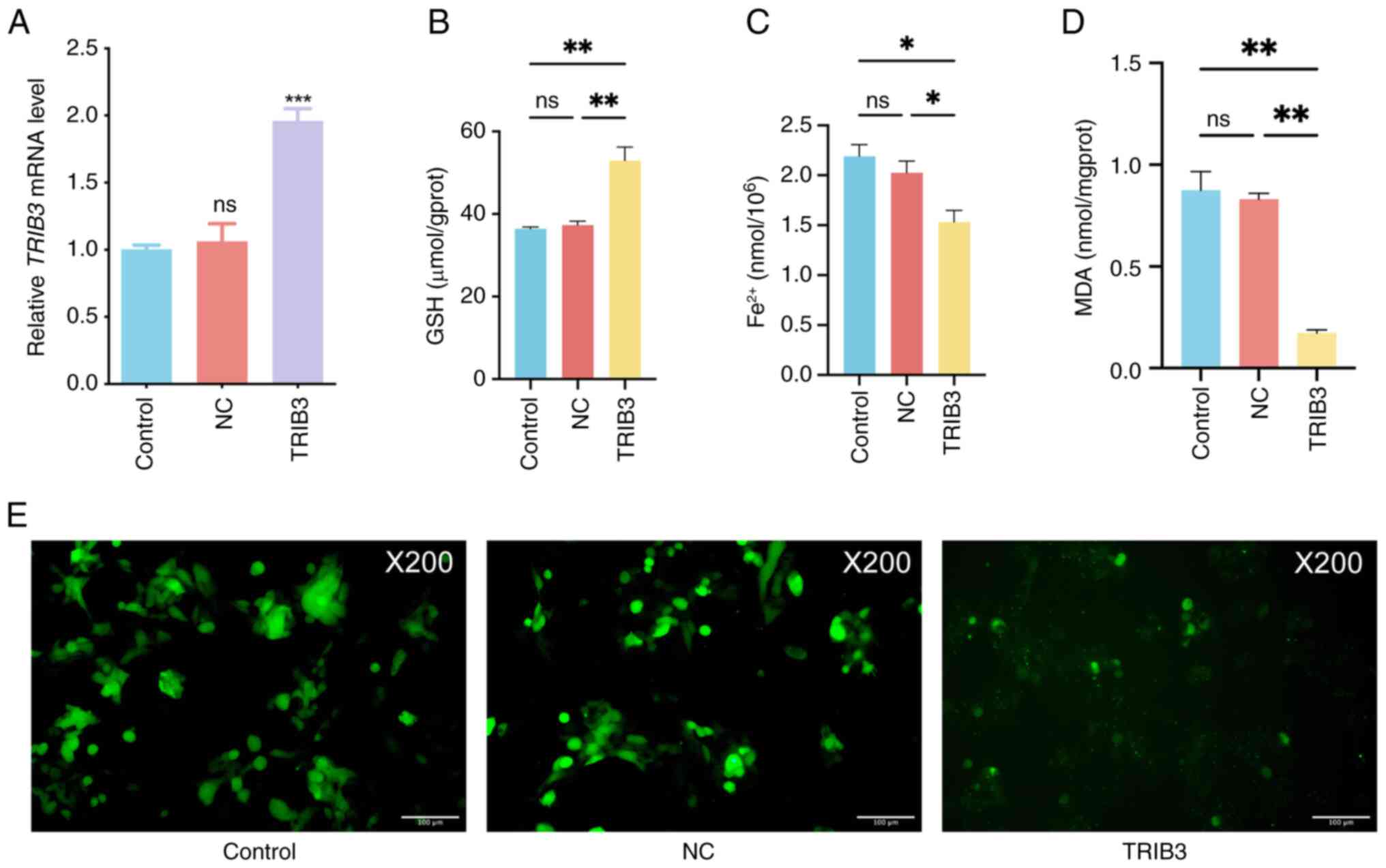 | Figure 7.TRIB3 overexpression inhibits
ferroptosis in HPV-positive HNSCC cells. (A) Relative TRIB3
mRNA expression in the different groups measured by reverse
transcription-quantitative polymerase chain reaction. (B) The
intracellular GSH concentration was measured using the reduced GSH
Colorimetric Assay Kit. (C) The intracellular Fe2+
concentration was measured using the Cell Ferrous Iron Colorimetric
Assay Kit. (D) The intracellular MDA concentration was measured
using the Cell Malondialdehyde Colorimetric Assay Kit. (E) ROS
(green fluorescence) were measured in the cells. *P<0.05,
**P<0.01, ***P<0.001. ns, non-significant; HPV, human
papillomavirus; HNSCC, head and neck squamous cell carcinoma; GSH,
glutathione; MDA, malondialdehyde; ROS, reactive oxygen species;
TRIB3, tribbles pseudokinase 3; NC, negative control. |
Discussion
Ferroptosis is a unique form of cell death (23) and a previous study demonstrated an
association between ferroptosis and HNC treatment: In murine
models, ferroptosis inducers and specific antitumor drugs, such as
cisplatin, have shown synergistic effects in suppressing the
progression of HNC (25). Although
a number of studies have compared FRGs between healthy tissues and
HNSCC tissues (31,32), the differences in FRG expression
between HPV-positive and HPV-negative HNSCC have not yet been well
reported. However, it has been demonstrated that HPV16 integration
confers ferroptosis resistance in cervical squamous cell carcinoma
via the c-Myc/microRNA-142-5p/Homeobox A5/solute carrier family 7
member 11 axis (39). Additionally,
HPV status has been identified as a key determinant of ferroptosis
sensitivity in HNSCC, with HPV E6/E7 interfering with NFE2-like
BZIP transcription factor 2 pathways in HPV-positive cases and
HPV-negative HNSCC relying more on glutathione peroxidase
4-dependent defense (40).
Additionally, a cross-tumor analysis has confirmed distinct
ferroptosis-related metabolic signatures between HPV subtypes,
including enhanced ROS detoxification in HPV-positive HNSCC and
pro-oxidative pathways with immune suppression in HPV-negative
patients (41). Collectively, these
findings, from the mechanistic regulation of ferroptosis by HPV
oncogenes and the subtype-specific ferroptosis sensitivity mediated
by HPV status, to the distinct ferroptosis-related metabolic
profiles, provide compelling evidence that FRG expression differs
between HPV-positive and HPV-negative HNSCC, highlighting the
necessity of stratifying HNSCC by HPV status when investigating
ferroptosis-associated mechanisms and therapeutic strategies.
In the present study, the expression of FRGs in
HPV-positive and HPV-negative HNSCC were investigated, given the
known biological differences between these two subtypes of HNSCC,
specifically in terms of etiopathogenesis, molecular genetic
characteristics, clinicopathologic presentation and prognosis
(5). To develop a prognostic model
for the HPV-negative group, 10 FRGs were specifically identified
(ALOXE3, ACSL1, TRIB3, FLT3, EGFR, AGPAT3, AMN, PPARG,
MIR9-3HG and BNIP3) as per their efficacy in the LASSO
Cox regression analysis. According to the findings, this model
could function as a reliable prognostic indicator for patients with
HPV-negative HNSCC. Furthermore, the RT-qPCR, western blotting and
immunohistochemistry analyses performed in the present study
demonstrated that among the 10 FRGs, TRIB3 exhibited higher
expression in HPV-negative samples than in HPV-positive
samples.
TRIB3, which belongs to the Tribbles-related
family, has been found to have biological associations with a range
of cancer types including oral squamous cell carcinoma, breast
cancer, lung adenocarcinoma, renal cell carcinoma, colorectal
cancer and glioblastoma, and it can act as either an oncogene or a
tumor suppressor (42–47). TRIB3 is involved in various
biological processes, including lipid and glucose metabolism,
epithelial-to-mesenchymal transition, cell proliferation,
differentiation and the cellular stress response (46,48,49).
As lipid peroxidation and iron overload are the main metabolic
characteristics of ferroptosis (50), whether TRIB3 regulated
ferroptosis in HPV-positive and HPV-negative cells was evaluated in
the present study. It was found that TRIB3 knockdown impeded
the survival of HPV-negative HNSCC cells. Moreover, TRIB3
knockdown led to elevated intracellular Fe2+, MDA and
ROS levels, along with a decrease in GSH. Conversely, TRIB3
overexpression in HPV-positive HNSCC cells yielded contrasting
outcomes. Additionally, after TRIB3 knockdown, transmission
electron microscopy revealed a notable reduction or complete loss
of mitochondrial ridges, along with a notable increase in
mitochondrial membrane density. The downregulation of TRIB3
resulting in the occurrence of ferroptosis in HPV-negative HNSCC
cells was thus highlighted by the results of the present study.
TRIB3 may become a new target that can trigger ferroptosis
in both HPV-positive and HPV-negative cancer cells. Nevertheless,
further investigation is necessary to understand the mechanism by
which TRIB3 influences ferroptosis. Additionally, in the
present study, the Kaplan-Meier analysis and prognostic model
demonstrated that TRIB3 had a more significant clinical
impact in patients with HPV-negative HNSCC compared with
HPV-positive. Furthermore, compared with HPV-positive HNSCC cells,
TRIB3 expression is upregulated in HPV-negative HNSCC. Given the
biological disparities between these two subtypes, the upregulation
of TRIB3 is not merely a quantitative elevation, but rather a
qualitative alteration intimately linked to their distinct
oncogenic drivers and regulatory mechanisms, which implies that
multiple distinct pathways may underlie this upregulation and, in
turn, modulate ferroptosis. Finally, despite TRIB3 mediating
ferroptosis in both HPV-positive and HPV-negative HNSCC cells, the
underlying mechanism of this effect remains unknown. Whether
TRIB3 functions through the same pathway in these two
subtypes of HNSCC requires further investigation.
Aside from TRIB3, the prognostic model
constructed in the present study included 9 other genes: EGFR,
AGPAT3, PPARG, ACSL1, BNIP3, ALOXE3, FLT3, AMN and
MIR9-3HG. EGFR, a promoter of HNSCC growth and
development, is upregulated in >90% of cases of HNSCC (51). Moreover, EGFR reduces
N6-methyladenosine and protects against ferroptosis in glioblastoma
by promoting ALKBH5 nuclear retention (52). ACSL1, which encodes an
essential enzyme involved in the modulation of lipid metabolism,
increases ferroptosis resistance and antioxidant capacity in
ovarian cancer by regulating the myristoylation of AIF family
member 2 (53). ACSL1 was also
observed to be upregulated in HPV-negative HNSCC in the present
study; however, to the best of our knowledge, no studies have yet
investigated the relationship between ACSL1 and HPV infection. In
addition, ALOXE3, a YAP-TEAD target gene, is associated with
YAP-induced ferroptosis and significantly increases the risk of
death induced by ferroptosis in hepatocellular carcinoma cells when
overexpressed (54). PPARG,
which encodes a nuclear receptor associated with the modulation of
lipid metabolism, mediates the ferroptosis of dendritic cells,
limiting antitumor immunity in mice (55). Moreover, research on
FLT3-mediated ferroptosis is primarily focused on
FLT3-mutant leukemia (56,57).
Bioinformatics analyses have also reported that BNIP3, AMN
and MIR9-3HG are involved in ferroptosis during
carcinogenesis (58–60). However, there is currently no
research on AGPAT3 and its role in mediating ferroptosis,
making this a novel finding of the present study.
The immune system modulates the response to therapy
and inhibits tumor growth, invasion and metastasis. Immune
surveillance provides a method for identifying, controlling and
eliminating tumor cells (27–29).
Lipid mediators may be generated by ferroptotic cells as
recognition signals to recruit antigen-presenting cells and other
immune cells into the microenvironment of the ferroptosis tumor
(61). However, the relationship
between infiltrating immune cells and ferroptosis in HNSCC remains
unclear. In the present study, increased infiltrating immune cells,
such as quiescent dendritic cells, plasma cells, mast cells,
regulatory T cells and naïve B cells, were found in the low-risk
group, while the high-risk group demonstrated elevated infiltration
of neutrophils, activated CD4+ T cells and mast cells.
Moreover, a significant correlation was observed among the 10 hub
FRGs, the risk score and various immune cell types. Thus, these 10
FRGs may induce ferroptosis and thus regulate immunological
therapy, positioning them as potential new targets in patients with
HPV-negative HNSCC. However, even when the scale of the
confirmation has been estimated, CIBERSORT predictions remain
limited. This is because deconvolution of bulk RNA-sequencing data
can be noisy, an issue that is particularly pronounced in
heterogeneous tumors. Experimental verification is needed to
confirm these results, and such validation could first extend to
prospective multicenter cohorts of patients with HPV-negative HNSCC
to verify the robustness of the 10-FRG-based prognostic model
across diverse populations, addressing the current reliance on
retrospective TCGA data.
The present study has some limitations that should
be considered. First, the prognostic model was developed by
analyzing publicly available datasets. Therefore, the findings
still require verification in prospective multicenter cohorts.
Second, the in vitro validation of TRIB3, was limited to
single HPV-negative (CAL27) and HPV-positive (UPCI-SCC-090) HNSCC
cell lines, risking biological variability-related confounding
factors. These results will be followed by experiments expanding
the cell panel to additional HPV-negative (such as SCC-25 and HN6)
and HPV-positive (such as SCC-47) cell lines and incorporating
patient-derived primary cells to verify the function of TRIB3 and
reduce variability. Third, only TRIB3, one of the risk
signature genes, was evaluated in vitro. More studies are
required to evaluate the functions of EGFR, AGPAT3, PPARG,
ACSL1, BNIP3, ALOXE3, FLT3, AMN and MIR9-3HG in the
pathogenesis and development of HNSCC. Moreover, studying the
mechanisms by which the identified FRGs regulate ferroptosis is
essential for comprehending their roles and potential implications
in cellular functions and disease processes. Last, the present
study is limited by the absence of in vivo data. Given that
in vivo validation is critical to verifying the biological
function of a target gene and evaluating its clinical translation
potential, future work should prioritize investigating the function
of TRIB3 in in vivo models to confirm its utility for
clinical translation.
In conclusion, the present study identified 10 hub
genes associated with ferroptosis, including TRIB3, EGFR,
AGPAT3, PPARG, ACSL1, BNIP3, ALOXE3, FLT3, AMN and
MIR9-3HG. A prognostic model was developed for the
HPV-negative patient group using these 10 hub FRGs. Thus, these 10
FRGs may serve as prognostic markers for patients with HPV-negative
HNSCC. TRIB3 expression was higher in HPV-negative clinical
samples than in HPV-positive clinical samples. TRIB3
knockdown resulted in increased intracellular Fe2+, MDA
and ROS, reduced GSH and the diminishment or absence of
mitochondrial ridges in HPV-negative HNSCC cells. Conversely,
TRIB3 overexpression resulted in decreased intracellular
Fe2+, MDA and ROS, but increased GSH in HPV-positive
HNSCC cells. These results indicate a novel association between
TRIB3 and ferroptosis in both HPV-positive and -negative
HNSCC cells. TRIB3 had a more significant clinical impact in
the HPV-negative HNSCC group than in the HPV-positive HNSCC group,
thereby providing a direction to improve clinical outcomes for
patients with HPV-negative HNSCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JL, NAR, SM and GY were responsible for conception.
JL and GY performed the interpretation and analysis of data. JL
prepared the manuscript. JL, NAR, SM and GY revised the manuscript
for important intellectual content. The study was supervised by GY.
JL and GY confirm the authenticity of all the raw data. All authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Ethical approval for this retrospective study using
de-identified data was granted by the Human Ethical Committee of
Heping Hospital Affiliated to Changzhi Medical College [Changzhi,
China; approval no. 2023(027)]. The committee also waived the
requirement for informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reid PA, Wilson P, Li Y, Marcu LG and
Bezak E: Current understanding of cancer stem cells: Review of
their radiobiology and role in head and neck cancers. Head Neck.
39:1920–1932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
3
|
Johnson DE, Burtness B, Leemans CR, Lui
VWY, Bauman JE and Grandis JR: Head and neck squamous cell
carcinoma. Nat Rev Dis Primers. 6:922020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mody MD, Rocco JW, Yom SS, Haddad RI and
Saba NF: Head and neck cancer. Lancet. 398:2289–2299. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma-an update. CA Cancer J
Clin. 65:401–421. 2015.PubMed/NCBI
|
|
6
|
Wuerdemann N, Wittekindt C, Sharma SJ,
Prigge ES, Reuschenbach M, Gattenlöhner S, Klussmann JP and Wagner
S: Risk factors for overall survival outcome in surgically treated
human Papillomavirus-Negative and positive patients with
oropharyngeal cancer. Oncol Res Treat. 40:320–327. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu W, Chen Y, Putluri N, Osman A, Coarfa
C, Putluri V, Kamal AHM, Asmussen JK, Katsonis P, Myers JN, et al:
Evolution of cisplatin resistance through coordinated metabolic
reprogramming of the cellular reductive state. Br J Cancer.
128:2013–2024. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Azharuddin M, Roberg K, Dhara AK, Jain MV,
Darcy P, Hinkula J, Slater NKH and Patra HK: Dissecting multi drug
resistance in head and neck cancer cells using multicellular tumor
spheroids. Sci Rep. 9:200662019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu H, Li B, Wang J, Tan Y, Xu M, Xu W and
Lu H: New advances into cisplatin resistance in head and neck
squamous carcinoma: Mechanisms and therapeutic aspects. Biomed
Pharmacother. 163:1147782023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mackenzie IC: Stem cell properties and
epithelial malignancies. Eur J Cancer. 42:1204–1212. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Norouzi A, Liaghat M, Bakhtiyari M,
Noorbakhsh Varnosfaderani SM, Zalpoor H, Nabi-Afjadi M and Molania
T: The potential role of COVID-19 in progression, chemo-resistance,
and tumor recurrence of oral squamous cell carcinoma (OSCC). Oral
Oncol. 144:1064832023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang SH, Oh SY, Lee KY, Lee HJ, Kim MS,
Kwon TG, Kim JW, Lee ST, Choi SY and Hong SH: Differential effect
of cancer-associated fibroblast-derived extracellular vesicles on
cisplatin resistance in oral squamous cell carcinoma via
miR-876-3p. Theranostics. 14:460–479. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Usman S, Waseem NH, Nguyen TKN, Mohsin S,
Jamal A, The MT and Waseem A: Vimentin is at the heart of
epithelial mesenchymal transition (EMT) mediated metastasis.
Cancers (Basel). 13:49852021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gong Y, Fan Z, Luo G, Yang C, Huang Q, Fan
K, Cheng H, Jin K, Ni Q, Yu X and Liu C: The role of necroptosis in
cancer biology and therapy. Mol Cancer. 18:1002019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hsu SK, Li CY, Lin IL, Syue WJ, Chen YF,
Cheng KC, Teng YN, Lin YH, Yen CH and Chiu CC: Inflammation-related
pyroptosis, a novel programmed cell death pathway, and its
crosstalk with immune therapy in cancer treatment. Theranostics.
11:8813–8835. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu J, Kuang F, Kang R and Tang D:
Alkaliptosis: A new weapon for cancer therapy. Cancer Gene Ther.
27:267–269. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tong X, Tang R, Xiao M, Xu J, Wang W,
Zhang B, Liu J, Yu X and Shi S: Targeting cell death pathways for
cancer therapy: Recent developments in necroptosis, pyroptosis,
ferroptosis, and cuproptosis research. J Hematol Oncol. 15:1742022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Liu X, Jin S, Chen Y and Guo R:
Ferroptosis in cancer therapy: A novel approach to reversing drug
resistance. Mol Cancer. 21:472022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie Y, Hou W, Song X, Yu Y, Huang J, Sun
X, Kang R and Tang D: Ferroptosis: Process and function. Cell Death
Differ. 23:369–379. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Conrad M and Pratt DA: Publisher
correction: The chemical basis of ferroptosis. Nat Chem Biol.
16:223–224. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stockwell BR: Ferroptosis turns 10:
Emerging mechanisms, physiological functions, and therapeutic
applications. Cell. 185:2401–2421. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xue Y, Jiang X, Wang J, Zong Y, Yuan Z,
Miao S and Mao X: Effect of regulatory cell death on the occurrence
and development of head and neck squamous cell carcinoma. Biomark
Res. 11:22023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim EH, Shin D, Lee J, Jung AR and Roh JL:
CISD2 inhibition overcomes resistance to sulfasalazine-induced
ferroptotic cell death in head and neck cancer. Cancer Lett.
432:180–190. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roh JL, Kim EH, Jang HJ, Park JY and Shin
D: Induction of ferroptotic cell death for overcoming cisplatin
resistance of head and neck cancer. Cancer Lett. 381:96–103. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ye J, Jiang X, Dong Z, Hu S and Xiao M:
Low-Concentration PTX And RSL3 inhibits tumor cell growth
synergistically by inducing ferroptosis in mutant p53
hypopharyngeal squamous carcinoma. Cancer Manag Res. 11:9783–9792.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dersh D, Hollý J and Yewdell JW: A few
good peptides: MHC class I-based cancer immunosurveillance and
immunoevasion. Nat Rev Immunol. 21:116–128. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mohme M, Riethdorf S and Pantel K:
Circulating and disseminated tumour Cells-mechanisms of immune
surveillance and escape. Nat Rev Clin Oncol. 14:155–167. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zou W: Immunosuppressive networks in the
tumour environment and their therapeutic relevance. Nat Rev Cancer.
5:263–274. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang W, Green M, Choi JE, Gijón M, Kennedy
PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al:
CD8+ T cells regulate tumour ferroptosis during cancer
immunotherapy. Nature. 569:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, Wang X, Qin R, Zhong Z and Sun C:
Identification of a ferroptosis gene set that mediates the
prognosis of squamous cell carcinoma of the head and neck. Front
Genet. 12:6980402021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu W, Wu Y, Huang S and Zhang D: A
Ferroptosis-related gene signature for predicting the prognosis and
drug sensitivity of head and neck squamous cell carcinoma. Front
Genet. 12:7554862021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hoadley KA, Yau C, Hinoue T, Wolf DM,
Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et
al: Cell-of-Origin patterns dominate the molecular classification
of 10,000 tumors from 33 types of cancer. Cell. 173:291–304.e6.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Martens-de Kemp SR BB, Smeets S, Voorham
Q, Rustenburg F, de Boer VD, Brink A, van Wieringen WN and
Brakenhoff RH: GEO accession number [GSE83519]. NCBI(GEO) (ed.), .
2017.
|
|
35
|
Vickers AJ, Cronin AM, Elkin EB and Gonen
M: Extensions to decision curve analysis, a novel method for
evaluating diagnostic tests, prediction models and molecular
markers. BMC Med Inform Decis Mak. 8:532008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen XJ, Guo CH, Yang Y, Wang ZC, Liang
YY, Cai YQ, Cui XF, Fan LS and Wang W: HPV16 integration regulates
ferroptosis resistance via the c-Myc/miR-142-5p/HOXA5/SLC7A11 axis
during cervical carcinogenesis. Cell Biosci. 14:1292024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang J and Gu Z: Ferroptosis in head and
neck squamous cell carcinoma: From pathogenesis to treatment. Front
Pharmacol. 15:12834652024. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hamid S, Khan MS, Khan MA, Muhammad N,
Singh M, Al-Shabeeb Akil AS, Bhat AA and Macha MA: Human papilloma
virus infection drives unique metabolic and immune profiles in head
and neck and cervical cancers: Implications for targeted therapies
and prognostic markers. Discov Oncol. 16:6762025. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen P, Zhang TY and Wang SY: TRIB3
promotes oral squamous cell carcinoma cell proliferation by
activating the AKT signaling pathway. Exp Ther Med. 21:3132021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wennemers M, Bussink J, Scheijen B,
Nagtegaal ID, van Laarhoven HW, Raleigh JA, Varia MA, Heuvel JJ,
Rouschop KM, Sweep FC and Span PN: Tribbles homolog 3 denotes a
poor prognosis in breast cancer and is involved in hypoxia
response. Breast Cancer Res. 13:R822011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhou H, Luo Y, Chen JH, Hu J, Luo YZ, Wang
W, Zeng Y and Xiao L: Knockdown of TRB3 induces apoptosis in human
lung adenocarcinoma cells through regulation of Notch 1 expression.
Mol Med Rep. 8:47–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hong B, Zhou J, Ma K, Zhang J, Xie H,
Zhang K, Li L, Cai L, Zhang N, Zhang Z and Gong K: TRIB3 promotes
the proliferation and invasion of renal cell carcinoma cells via
activating MAPK signaling pathway. Int J Biol Sci. 15:587–597.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miyoshi N, Ishii H, Mimori K, Takatsuno Y,
Kim H, Hirose H, Sekimoto M, Doki Y and Mori M: Abnormal expression
of TRIB3 in colorectal cancer: A novel marker for prognosis. Br J
Cancer. 101:1664–1670. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang Z, Chen H, Zhong D, Wei W, Liu L,
Duan Q, Han B and Li G: TRIB3 facilitates glioblastoma progression
via restraining autophagy. Aging (Albany NY). 12:25020–25034. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Izrailit J, Jaiswal A, Zheng W, Moran MF
and Reedijk M: Cellular stress induces TRB3/USP9×-dependent Notch
activation in cancer. Oncogene. 36:1048–1057. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hua F, Mu R, Liu J, Xue J, Wang Z, Lin H,
Yang H, Chen X and Hu Z: TRB3 interacts with SMAD3 promoting tumor
cell migration and invasion. J Cell Sci. 124:3235–3246. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang XD, Liu ZY, Wang MS, Guo YX, Wang
XK, Luo K, Huang S and Li RF: Mechanisms and regulations of
ferroptosis. Front Immunol. 14:12694512023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu X, Zhang F, Zhang W, He J, Zhao Y and
Chen X: Prognostic role of epidermal growth factor receptor in head
and neck cancer: A meta-analysis. J Surg Oncol. 108:387–397. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lv D, Zhong C, Dixit D, Yang K, Wu Q,
Godugu B, Prager BC, Zhao G, Wang X and Xie Q: EGFR promotes ALKBH5
nuclear retention to attenuate N6-methyladenosine and protect
against ferroptosis in glioblastoma. Mol Cell. 83:4334–4351.e7.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang Q, Li N, Deng L, Jiang X, Zhang Y,
Lee LTO and Zhang H: ACSL1-induced ferroptosis and platinum
resistance in ovarian cancer by increasing FSP1 N-myristylation and
stability. Cell Death Discov. 9:832023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Qin Y, Pei Z, Feng Z, Lin P, Wang S, Li Y,
Huo F, Wang Q, Wang Z, Chen ZN, et al: Oncogenic activation of YAP
signaling sensitizes ferroptosis of hepatocellular carcinoma via
ALOXE3-mediated lipid peroxidation accumulation. Front Cell Dev
Biol. 9:7515932021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Han L, Bai L, Qu C, Dai E, Liu J, Kang R,
Zhou D, Tang D and Zhao Y: PPARG-mediated ferroptosis in dendritic
cells limits antitumor immunity. Biochem Biophys Res Commun.
576:33–39. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sabatier M, Birsen R, Lauture L, Mouche S,
Angelino P, Dehairs J, Goupille L, Boussaid I, Heiblig M, Boet E,
et al: C/EBPα confers dependence to fatty acid anabolic pathways
and vulnerability to lipid oxidative Stress-induced ferroptosis in
FLT3-Mutant leukemia. Cancer Discov. 13:1720–1747. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cui Z, Fu Y, Yang Z, Gao Z, Feng H, Zhou
M, Zhang L and Chen C: Comprehensive analysis of a ferroptosis
pattern and associated prognostic signature in acute myeloid
leukemia. Front Pharmacol. 13:8663252022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Meng J, Du H, Lu J and Wang H:
Construction and validation of a predictive nomogram for
ferroptosis-related genes in osteosarcoma. J Cancer Res Clin Oncol.
149:14227–14239. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang J, Deng Y, Zhang H, Zhang Z, Jin X,
Xuan Y, Zhang Z and Ma X: Single-cell RNA-Seq analysis reveals
ferroptosis in the tumor microenvironment of clear cell renal cell
carcinoma. Int J Mol Sci. 24:90922023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang W, Song Y, Zhong Z, Gao J and Meng
X: Ferroptosis-related long Non-Coding RNA signature contributes to
the prediction of prognosis outcomes in head and neck squamous cell
carcinomas. Front Genet. 12:7858392021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Friedmann Angeli JP, Krysko DV and Conrad
M: Ferroptosis at the crossroads of Cancer-acquired drug resistance
and immune evasion. Nat Rev Cancer. 19:405–414. 2019. View Article : Google Scholar : PubMed/NCBI
|