Introduction
With increasing life expectancy, the morbidity of
prostate cancer (PCa) has risen rapidly. According to data from the
American Center for Cancer Research, PCa is now the most common
malignancy with 29% incidence rate and the second leading cause of
cancer-associated mortality with 11% death rate among men in the
United States in 2024 (1). Its
incidence is also increasing rapidly in China, where PCa has become
the fastest-growing malignancy with an incidence rate of 0.02% in
2022 among men (2). Effective
treatments such as ablative radiotherapy and radical prostatectomy
can markedly prolong survival (3).
Antiandrogen drugs can also effectively treat locally advanced and
metastatic PCa (4). By decreasing
androgen receptor (AR) expression (5), these drugs alleviate clinical symptoms
such as bone pain, difficulty with urinating and extend survival
(6). However, drug resistance
inevitably develops and the mechanisms underlying antiandrogen
resistance remain to be elucidated.
Antiandrogen therapy reduces androgen activity
through drugs (7), such as
bicalutamide (BIC), enzalutamide (ENZ) and apalutamide (APN)
(8). BIC is a non-steroidal
antiandrogen that inhibits the binding of testicular and adrenal
androgens to AR (9). ENZ prevents
nuclear translocation of activated AR, binding to androgen response
elements and coactivator recruitment, thereby suppressing PCa
proliferation (10). APN, another
non-steroidal antiandrogen, binds directly to the AR ligand-binding
domain and inhibits AR translocation, DNA binding and AR-mediated
transcription (11). These drugs
can inhibit PCa progression by decreasing AR expression. Although
these drugs provide clinical benefit, resistance eventually
develops and the underlying mechanisms require further study.
Genetic alterations are increasingly recognized as
key contributors to both PCa development and drug resistance. For
instance, microseminoprotein-β expression changes may influence PCa
risk in obese individuals (12),
while alterations in cyclin B1 and golgin A8 family member B have
been associated with chemotherapy resistance (13,14).
Bcl-2-binding component 3 mutations contribute to olaparib
resistance (15) and CDK6
dysregulation has been associated with ENZ resistance (16). Collectively, these studies
underscore the key role of genetic changes in PCa development and
therapeutic resistance, suggesting that additional genetic factors
may contribute to antiandrogen resistance.
The present study attempted to use bioinformatic
methods to find the hub genes which are important for PCa
resistance to antiandrogen drugs and verified their function using
different types of PCa cell lines. RNA-sequencing data of lymph
node carcinoma of the prostate (LNCaP) cells resistant to BIC, ENZ
and APN (GSE211781 dataset) were obtained to find the hub genes.
RNA-sequence data from public databases such as Gene Expression
omnibus (GEO), The Cancer Genome Atlast (TCGA) and Chinese Prostate
Cancer Genome and Epigenome Atlas (CPGEA) were used to analyze the
function of hub genes in affecting PCa progression and prognosis.
Finally, the role of hub genes in influencing PCa cells sensitive
to antiandrogen drugs were verified in two types of PCa cell lines:
LNCaP and C4-2.
Materials and methods
Data source
The GEO database (https://www.ncbi.nlm.nih.gov/) is a publicly available
repository containing microarray data for various diseases
(17). Meanwhile, TCGA (http://cancergenome.nih.gov/) is a large public
database that provides sequencing and clinical data for multiple
cancer types (18). Gene expression
data for prostate adenocarcinoma in TCGA database (TCGA-PRAD) was
searched. The Chinese Prostate Cancer Genome and Epigenome Atlas
(CPGEA; http://www.cpgea.com/) includes
microarray data from Chinese patients with PCa (19). Gene expression and clinical data
were obtained from these databases.
Data handling
The GSE211781 dataset (20) consists of RNA-sequencing data from
LNCaP PCa cells resistant to antiandrogen drugs, including BIC, ENZ
and APN, and was used to identify DEGs associated with drug
resistance. In addition, the GSE136129 and GSE81796 datasets
(21,22), which contain RNA-sequencing data
from C4-2 PCa cells resistant to ENZ, were also collected. TCGA
provides clinical information and microarray data from 497 patients
with PCa and 52 healthy controls. The CPGEA database contains
microarray data from 136 paired tumor and adjacent normal tissues
from Chinese patients with PCa. All primary data from these public
databases were normalized using the R package ‘limma’ (version,
3.40.2; R Development Core Team) (23).
Survival analysis
Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/) an
online tool, was used to evaluate the effects of genes on patient
survival based on TCGA data (24).
Disease-free survival (DFS) was assessed using GEPIA.
Identifying antiandrogen drug
resistance genes (ADRGs) and constructing the PPI network
ADRGs in the GSE211781 dataset were identified based
on the following criteria: i) Adjusted P<0.05 and
log2 fold-change (FC) >1; and ii) expression changes
observed in response to all three antiandrogen drugs.
A PPI network of these ADRGs was constructed using
the STRING database. Raw PPI data were downloaded from STRING
(http://string-db.org/) (25) and reconstructed with Cytoscape
(version, 3.9.1; http://cytoscape.org/) software (26). Hub genes were identified as those
exhibiting the highest connectivity within the network (16).
Cell culture and transduction
The C4-2 and LNCaP PCa cell lines were purchased
from the Chinese Academy of Sciences Cell Bank. Cell culture and
transduction were performed as previously described (27). Briefly, cells were maintained in
RPMI 1640 medium (cat. no. R8758; MilliporeSigma) supplemented with
10% FBS (cat. no. 10091; Gibco; Thermo Fisher Scientific, Inc.) in
a humidified atmosphere with 5% CO2 and 95% air at 37°C.
Antibiotics were not used and cells were passaged when density
reached 80%. All lines were tested using a mycoplasma detection kit
(cat. no. C0298; Beyotime Institute of Biotechnology) to confirm
that the cells are free of mycoplasma contamination (cat. no.
MP0030; MilliporeSigma; Merck KGaA).
Cell transduction
To generate PCa cells with MYH11 knockdown, the 2nd
generation system was utilized comprising the packaging plasmids
pMD2.G (plasmid cat. no. 12259; Addgene, Inc.) and psPAX2 (plasmid
cat. no. 12260; Addgene, Inc.). For each 6-well plate, 5.5 µg
pLVX-shMYH11 and 2.0 µg pMD2.G was used to transfect cells for 12 h
at 37°C by Lipofectamine™ 2000 (cat. no. 11668019; Thermo Fisher
Scientific, Inc.) and polyethyleneimine (PEI; cat. no. 408700;
MilliporeSigma; Merck KGaA), according to the manufacturers'
protocol. Short hairpin RNA (shRNA) targeting MYH11 and a negative
control (shControl) lentivirus were constructed by Hebei Youze
Biotechnology Co., Ltd. Lentiviral packaging plasmids were
transduced into 293T cells using PEI for 12 h at 37°C. The culture
medium was replaced at 24 h post-transduction and supernatants were
collected at 48 h. These were then used to infect PCa cells with
multiplicity of infection of 2 at 37°C for 48 h. Following
transduction, cells exhibiting stable integration were selected
using puromycin (cat. no. P8833; Sigma-Aldrich; Merck KGaA) at 1.0
µg/ml for 5 days. The shMYH11 sequences were sense (S),
5′-GCAAATTCATCCGCATCAACT-3′ and antisense (AS),
3′-AGTTGATGCGGATGAATTTGC-5′, and the shControl sequences were S,
5′-GCTCACACGGATGTAGG-3′ and AS, 3′-CCTACATCCGTGTGAGCA-5′, which do
not affect gene expression in these cells.
Drug treatment
BIC (cat. no. S1190) and ENZ (cat. no. S1250) were
purchased from Selleck Chemicals. Untreated C4-2 and LNCaP cells
were exposed to 40 µM BIC or ENZ for 10 days before subsequent
experiments. After transduction with lentivirus, cells were treated
with 80 µM BIC or ENZ for 4 days at 37°C for further experiments.
In addition, the control cells were treated with 0.5% DMSO for 4
days at 37°C.
Cell proliferation assay
Cell proliferation experiments were performed 48 h
after transduction, as previously described (26). Proliferation was assessed using the
Cell Counting Kit-8 kit (CCK-8; cat. no. CA1210; Beijing Solarbio
Science & Technology Co., Ltd.). Cells were seeded in 96-well
plates (3,000 cells/well) and cultured in 200 µl RPMI 1640 medium
supplemented with 10% FBS for 0, 24, 48 and 72 h. The cells were
treated with BIC or ENZ at 40 or 80 µM according to experimental
requirements. After incubation, cells were treated with CCK-8
reagent for 1 h following the manufacturer's instructions and
absorbance was measured at 450 nm using a multimode reader (cat.
no. LB942; Titertek-Berthold).
Western blotting
Western blotting experiments were performed as
previously described (28). Total
proteins were extracted from samples and cell lines using RIPA
lysis buffer with a protease inhibitor cocktail (MilliporeSigma;
Merck KGaA). Then, the protein concentration was detected by BCA
using a BCA kit (cat. no. P0012; Beyotime Institute of
Biotechnology). Then, protein samples were handled by Dual Color
Protein Loading Buffer (Thermo Fisher Scientific, Inc.). Equal
amounts of protein (24 µg per lane) were added and separated via
SDS-PAGE (7.5 and 10% gels) and then transferred to nitrocellulose
membranes (cat. no. 71078; Merck KGaA). Membranes were blocked with
Protein-Free Rapid Blocking Buffer at 37°C for 1 h (cat. no. 37584;
Thermo Fisher Scientific, Inc.) and incubated overnight at 4°C with
primary antibodies against MYH11 (1:1,000; cat. no. ab124679;
Abcam) and GAPDH (1:1,000; cat. no. ab9482; Abcam). The next day,
membranes were washed three times for 10 min each with 1X TBST with
0.1% Tween, then incubated at room temperature for 1 h with a
matched secondary antibody HRP-labeled Goat Anti-Human IgG (H+L)
(cat nos. A0216 and A0208; Beyotime Institute of Biotechnology).
Signals were visualized using X-ray exposure (Abcam). Protein band
intensities were quantified using ImageJ software (version no.
1.54h; National Institutes of Health) and bar charts were generated
to present the results.
Statistical analysis
Data were obtained from at least three independent
experiments and are presented as mean ± SD. Differences between two
groups were assessed using an unpaired, two-tailed Student's t-test
and differences among ≥3 groups were analyzed with one-way ANOVA
followed by the Scheffe post hoc test to account for unequal group
sizes. The box plots were made using an online tool: Sangerbox
(http://sangerbox.com/index.html). All
statistical analyses were performed with SPSS (version 22.0; IBM
Corp.) and data from public databases were analyzed using R
software (version 4.0.3; R Development Core Team) with appropriate
packages. P<0.05 was considered to indicate a statistically
significant difference.
Results
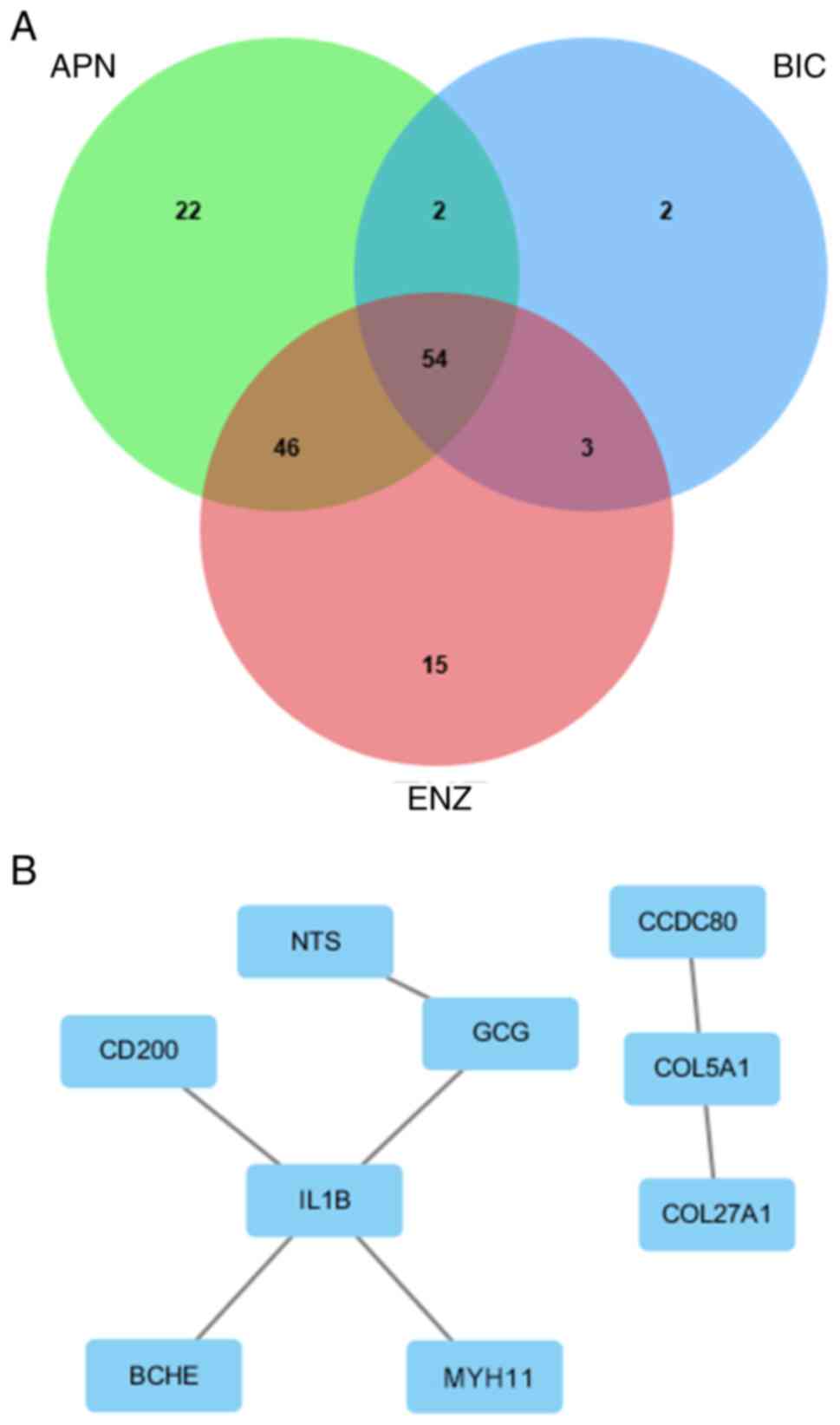
Identifying nine hub genes among 54
ADRGs from the GSE211781 dataset
The present study first analyzed the GSE211781
dataset to identify genes key for PCa resistance to antiandrogen
drugs. RNA-sequencing data from LNCaP cells resistant to BIC, ENZ
and APN were examined. In APN-resistant cells, 68 genes were
upregulated and 56 were downregulated. In BIC-resistant cells, 34
genes were upregulated and 27 were downregulated. In ENZ-resistant
cells, 70 genes were upregulated and 48 were downregulated.
Altogether, 54 genes were differentially expressed across all three
resistant cell lines, which were classified as ADRGs in the present
study (Fig. 1A). Table I summarizes the expression levels of
the nine hub genes identified among these 54 ADRGs and complete
expression data for all genes are presented in Table SI. A PPI network generated with
Cytoscape further revealed nine hub genes (NTS, GCG, CD200,
IL1B, BCHE, MYH11, CCDC80, COL5A1 and COL27A1) among
these ADRGs, each demonstrating strong connectivity with other
genes in the network (Fig. 1B).
 | Table I.Hub genes of 54 ADRGs among BIC, ENZ
and APN in GSE211781 dataset. |
Table I.
Hub genes of 54 ADRGs among BIC, ENZ
and APN in GSE211781 dataset.
|
| BIC | ENZ | APN |
|---|
|
|
|
|
|
|---|
| Gene name | Log2
FC | P-value | Log2
FC | P-value | Log2
FC | P-value |
|---|
| CD200 | 2.0458783 |
5.93×10−19 | 1.59821854 |
4.63×10−12 | 3.0572163 |
9.50×10−21 |
| GCG | 1.3191416 |
5.76×10−14 | 1.11484425 |
1.26×10−8 | 3.9088046 |
4.55×10−12 |
| NTS | 2.7808322 |
3.18×10−10 | 3.02708496 |
8.10×10−7 | 2.7487693 |
2.77×10−10 |
| IL1B | −2.409564 |
8.70×10−8 | −1.6599581 |
1.08×10−5 | −1.1746371 |
1.38×10−8 |
| CCDC80 | −1.814258 |
5.51×10−6 | −1.9233845 |
1.06×10−4 | −1.6778532 |
3.09×10−7 |
| BCHE | 1.3011477 |
2.06×10−4 | 2.91307066 |
8.68×10−4 | 2.4957071 |
7.11×10−6 |
| MYH11 | 1.4286021 |
6.93×10−4 | 1.08698911 |
3.41×10−3 | 2.3045734 |
4.55×10−5 |
| COL5A1 | −3.935565 |
1.43×10−3 | −3.6223903 |
5.24×10−3 | −1.5384003 |
1.06×10−4 |
| COL27A1 | −1.532002 |
2.39×10−3 | −1.1766088 |
1.06×10−2 | −2.6972333 |
2.81×10−4 |
Hub genes from ADRGs influence PCa
occurrence
The present study next investigated whether these
nine hub genes were associated with PCa occurrence. GEPIA analysis
of TCGA-PRAD data was used to compare hub gene expression between
PCa samples and healthy prostate tissues. Among the nine hub genes,
BCHE (P=2.3×10−12), CD200
(P=5.8×10−17) and MYH11 (P=2.3×10−17)
were significantly downregulated in PCa samples compared with
healthy tissues (Fig. 2).
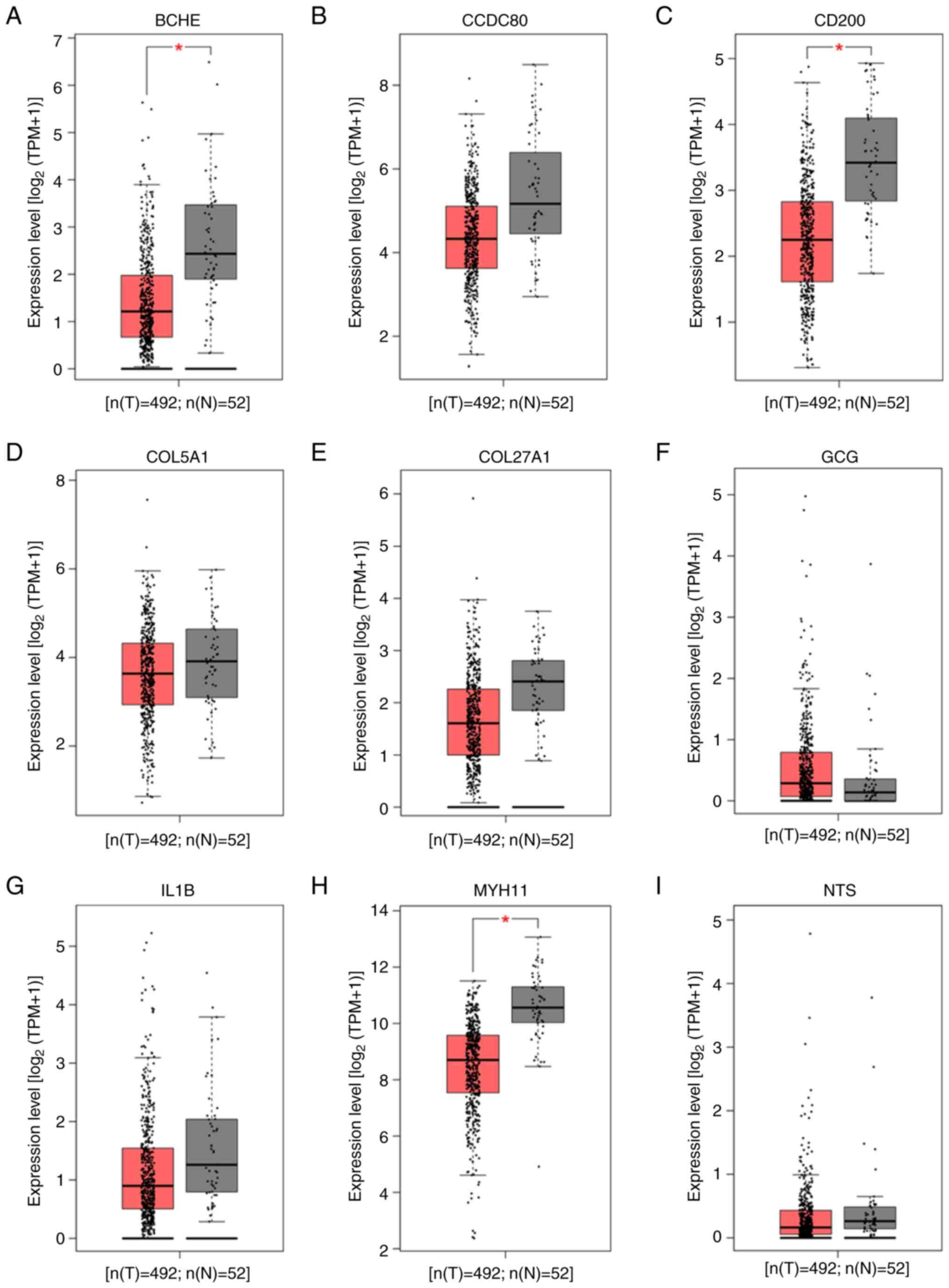 | Figure 2.Expression levels of nine hub genes
from ADRGs in PCa tissues and healthy prostate tissues analyzed
using the GEPIA online tool based on TCGA. Expression levels of (A)
BCHE, (B) CCDC80, (C) CD200, (D) COL5A1, (E) COL27A1, (F) GCG, (G)
IL1B, (H) MYH11 and (I) NTS. *P<0.05. ADRGs, antiandrogen drug
resistance genes; PCa, prostate cancer; GEPIA, Gene Expression
Profiling Interactive Analysis; TGCA, The Cancer Genome Atlas;
CD200, cluster of differentiation 200; BCHE, butyrylcholinesterase;
CCDC80, coiled-coil domain containing 80; COL5A1, collagen type V α
1 chain; COL27A1, collagen type XXVII α 1 chain; GCG, glucagon;
IL1B, interleukin 1β; MYH11, myosin heavy chain 11; NTS,
neurotensin; PRAD, prostate adenocarcinoma; T, tumor; N,
normal. |
Hub genes influence PCa occurrence in
Chinese patients
Since TCGA predominantly represents Western
populations, the present study also investigated gene expression in
a Chinese cohort using microarray data from the CPGEA database.
Furthermore, 8 out of the 9 hub genes (excluding NT; P=0.86)
were significantly differentially expressed in Chinese patients
with PCa (Fig. 3). These findings
suggested that the identified hub genes are key to PCa occurrence
in Chinese populations.
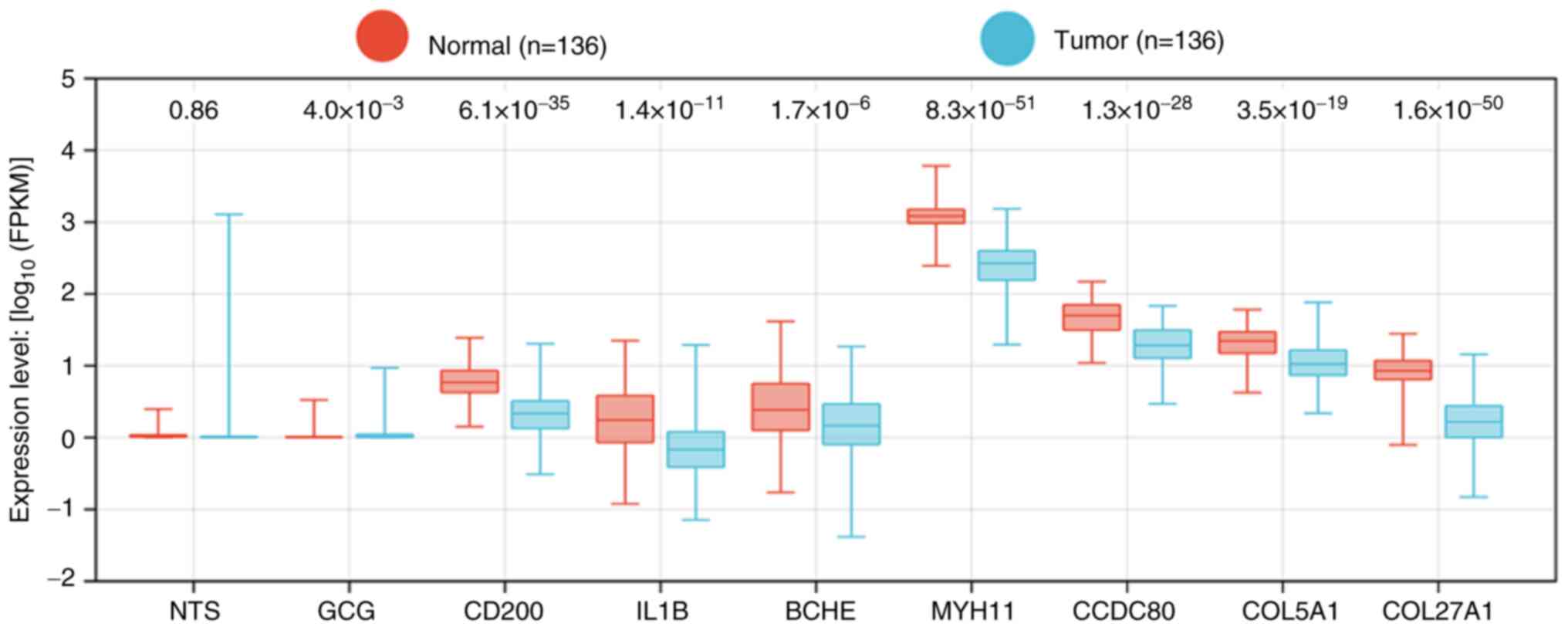 | Figure 3.Expression levels of nine hub genes
from ADRGs in the CPGEA database. CPGEA, Chinese Prostate Cancer
Genome and Epigenome Atlas; ADRGs, antiandrogen drug resistance
genes; NTS, neurotensin; GCG, glucagon; IL1B, interleukin 1β; BCHE,
butyrylcholinesterase; CCDC80, coiled-coil domain containing 80;
COL5A1, collagen type V α 1 chain; COL27A1, collagen type XXVII α 1
chain; MYH11, myosin heavy chain 11; CD200, cluster of
differentiation 200. |
Hub genes influence PCa
progression
Tumor, lymph node, metastasis (TNM) staging is
widely used to evaluate tumor severity (29). Therefore, the present study examined
whether hub genes associated with antiandrogen resistance also
influenced PCa progression. RNA-sequencing data and clinical
information were obtained from TCGA. Since metastasis data were
unavailable for most patients, only T and N stages were analyzed.
Hub gene expression was not associated with the T stage (Fig. 4A). However, MYH11 expression
was significantly reduced in patients with lymph node metastasis
(P=0.03; Fig. 4B). Thus, in
addition to its role in PCa occurrence, MYH11 also appears to
affect disease progression.
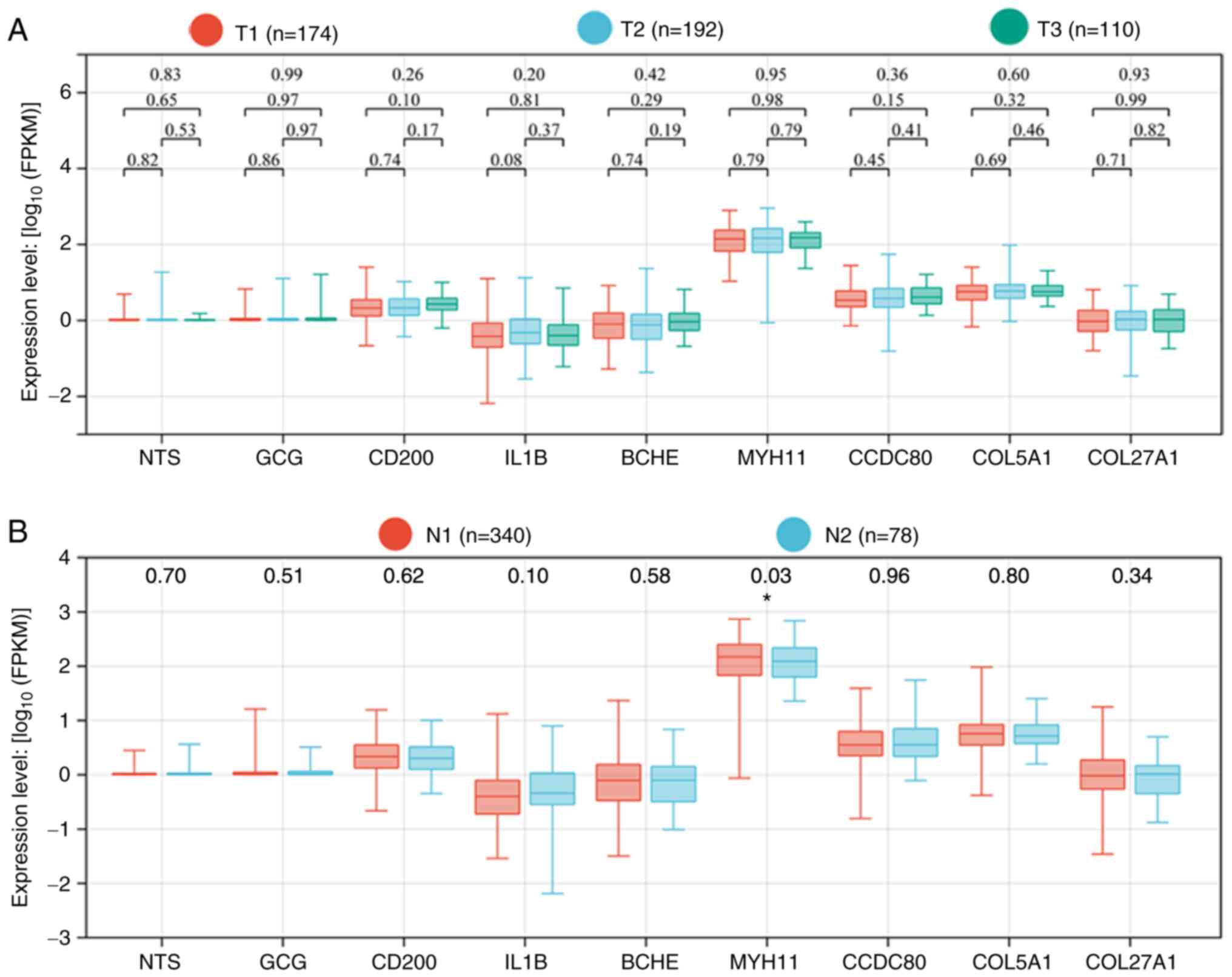 | Figure 4.Expression levels of nine hub genes
from ADRGs in patients with PCa and different tumor stages (data
from TGCA). (A) T stage (B) N stage. *P<0.05. ADRGs,
antiandrogen drug resistance genes; PCa, prostate cancer; TGCA, The
Cancer Genome Atlas; T, tumor; N, lymph node; NTS, neurotensin;
GCG, glucagon; IL1B, interleukin 1β; BCHE, butyrylcholinesterase;
CCDC80, coiled-coil domain containing 80; COL5A1, collagen type V α
1 chain; COL27A1, collagen type XXVII α 1 chain; MYH11, myosin
heavy chain 11; CD200, cluster of differentiation 200. |
MYH11 influences DFS in patients with
PCa
Next, the present study evaluated whether hub genes
were associated with patient survival. GEPIA analysis of TCGA data
was used to assess correlations between hub gene expression and
DFS. Only COL5A1 (P=0.011) and MYH11 (P=0.02)
expression significantly influenced DFS in patients with PCa
(Fig. 5). These results indicated
that MYH11 impacts both PCa progression and patient
survival.
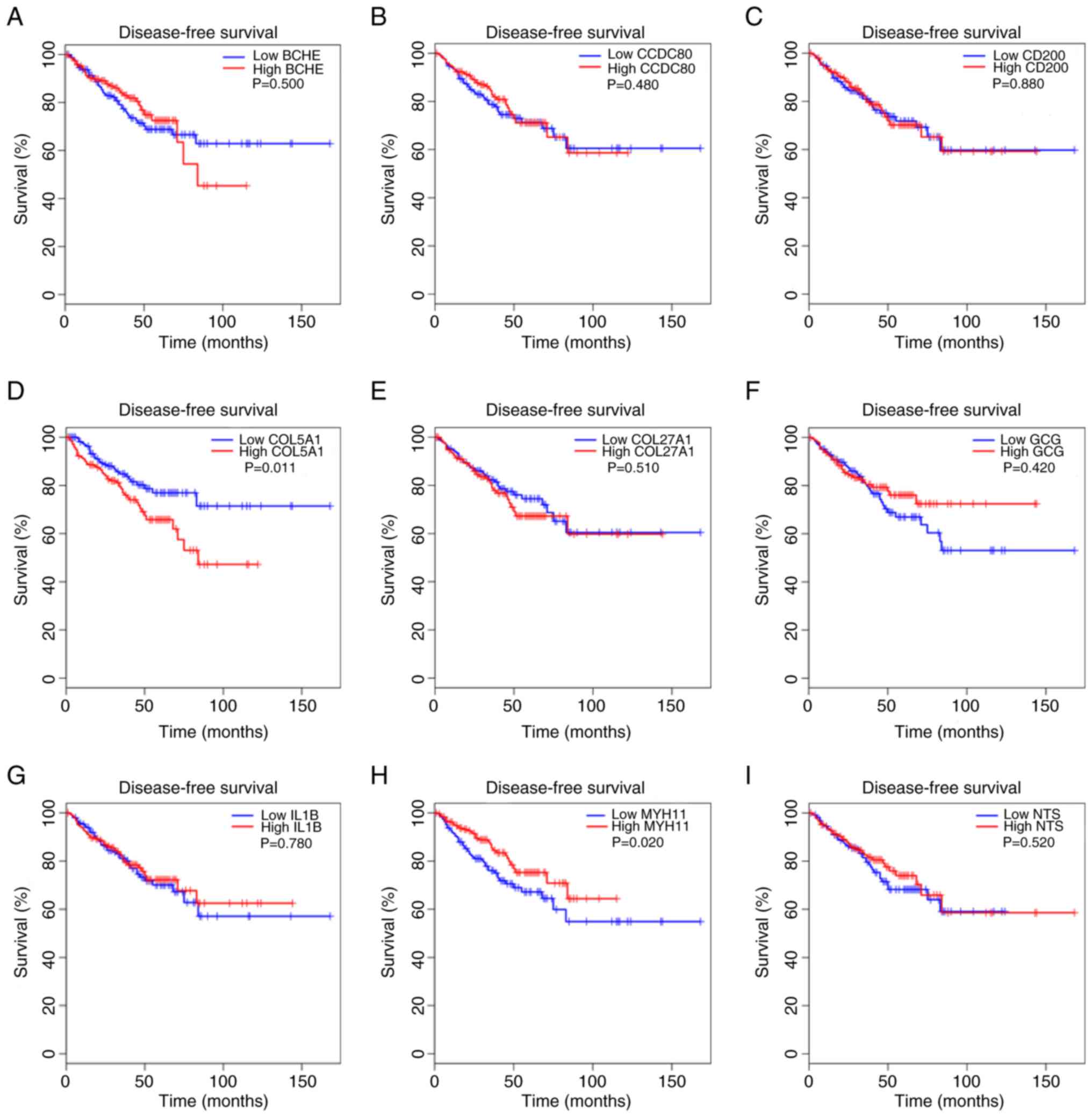 | Figure 5.Association between the expression
levels of nine hub genes from ADRGs and DFS status from the GEPIA
online tool based on TCGA. (A) BCHE (B) CCDC80 (C) CD200 (D) COL5A1
(E) COL27A1 (F) GCG (G) IL1B (H) MYH11 (I) NTS. DFS, disease-free
survival; ADRGs, antiandrogen drug resistance genes; GEPIA, Gene
Expression Profiling Interactive Analysis; TGCA, The Cancer Genome
Atlas; CD200, cluster of differentiation 200; BCHE,
butyrylcholinesterase; CCDC80, coiled-coil domain containing 80;
COL5A1, collagen type V α 1 chain; COL27A1, collagen type XXVII α 1
chain; GCG, glucagon, IL1B, interleukin 1 β; MYH11, myosin heavy
chain 11; NTS, neurotensin. |
MYH11 is central to PCa cells
resistance to antiandrogen drugs in vitro
The present study observed that MYH11 was
upregulated in LNCaP cells resistant to BIC, ENZ and APN, and that
it influenced PCa occurrence, progression, and prognosis based on
multiple databases. Therefore, we hypothesized that MYH11
serves a key role in PCa development and resistance to antiandrogen
drugs. To further investigate this, the present study collected two
additional datasets, GSE189129 and GSE81796, which include
RNA-sequencing data from C4-2 cells resistant to ENZ. Consistent
with results from LNCaP cells, MYH11 expression was also
significantly increased in ENZ-resistant C4-2 cells (GSE136129;
P=5.47×10−7 and GSE81796; P=0.0274; Fig. S1). These findings suggested that
MYH11 may be key in mediating resistance to antiandrogen
drugs in PCa cells.
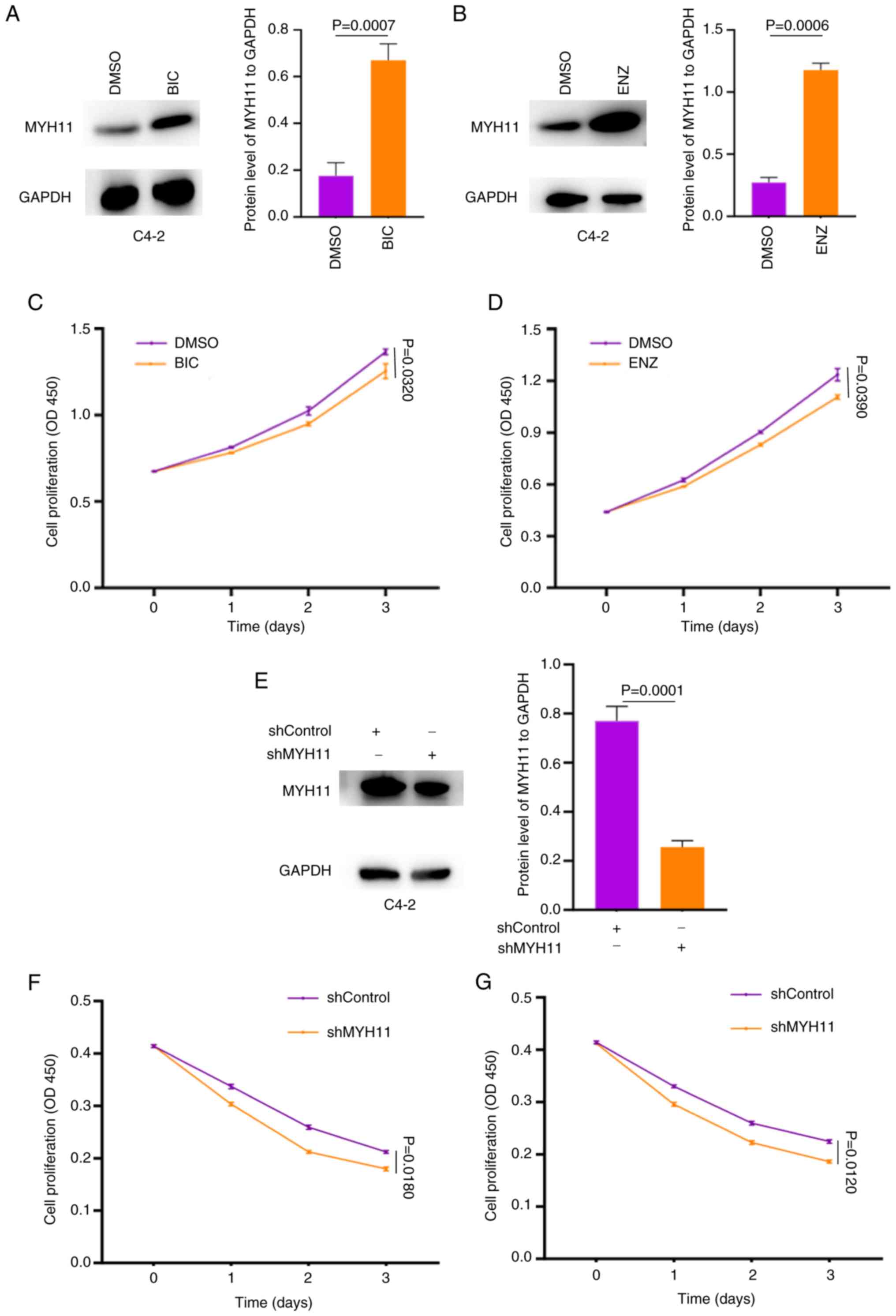
The present study next validated this function
experimentally. A shMYH11 lentivirus was constructed and transduced
into C4-2 cells, which were then treated with BIC or ENZ. First,
the present study identified MYH11 expression significantly
increased rapidly in C4-2 cells exposed to 40 µM BIC or ENZ for 10
days (BIC group, P=0.0007; ENZ group, P=0.006; Fig. 6A and B). Proliferation assays
demonstrated a significant reduction in cell proliferation after
treatment with different drugs (BIC group, P=0.0320; ENZ group,
P=0.0390; Fig. 6C and D). Knockdown
with shMYH11 lentivirus significantly reduced MYH11 protein
expression in C4-2 cells compared with shControl cells (P=0.0001;
Fig. 6E). When these cells were
subsequently treated with 80 µM BIC or ENZ, became more sensitive
to the drugs, indicating higher rates of cell death compared with
the cells treated by DMSO (BIC group, P=0.0160; ENZ group,
P=0.0120; Fig. 6F and G). Since
MYH11 was initially identified from LNCaP cells and these differ
biologically from C4-2 cells, the present study also performed
parallel analyses in LNCaP cells. In LNCaP cells, treatment with
BIC or ENZ significantly increased MYH11 expression and reduced
proliferation (BIC group, P=0.0001; ENZ group, P=0.0001; Fig. S2A-D). The shMYH11 lentivirus also
significantly decreased MYH11 expression in LNCaP cells (P=0.0001;
Fig. S2E), while shMYH11 knockdown
sensitized cells to drug treatment (BIC group, P=0.0180; ENZ group,
P=0.0160; Fig. S2F and G).
Together, these results indicate that MYH11 is key to determining
PCa cell sensitivity to antiandrogen drugs.
Discussion
The increasing life expectancy of men has
contributed to rising PCa morbidity (30). Improvements in health awareness and
the widespread use of prostate-specific antigen screening have
facilitated early detection (31).
Nonetheless, aggressive treatment is warranted to achieve improved
therapeutic effects. Among the available treatments, androgen
deprivation therapy (ADT), which lowers androgen levels or
decreases AR expression, has demonstrated therapeutic efficacy and
can prolong survival (6). However,
most patients (86%) develop resistance to ADT ≤2 years (32), highlighting the need to elucidate
the mechanisms underlying antiandrogen drug resistance.
BIC, ENZ and APN are three antiandrogen drugs with
confirmed therapeutic benefits in PCa (8). These agents markedly prolong median
survival in patients (33–35); however, resistance eventually
develops and the mechanisms remain to be elucidated. Genetic
alterations are considered to be a key contributing factor. For
example, loss of the chromodomain helicase DNA binding protein 1
gene has been associated with antiandrogen resistance (36). Furthermore, changes in neuregulin 1
expression may alter the tumor microenvironment and promote
resistance (37). Collectively,
these findings emphasize the importance of genetic alterations in
driving resistance to antiandrogen drugs.
Due to the role of genetic alterations in
resistance, the present study aimed to identify potential hub genes
involved in this process. RNA-sequencing analysis of the GSE211781
dataset identified nine hub genes among 54 ADRGs that may be key
for resistance. Further analysis indicated that several of these
genes also serve roles in PCa occurrence. MYH11 emerged as a
particularly key gene associated with PCa occurrence, progression
and prognosis. Two PCa cell lines, LNCaP and C4-2, were
subsequently used to examine whether MYH11 could influence
cellular sensitivity to antiandrogen drugs. Since C4-2 cells were
derived from bone metastasis of LNCaP in nude mice, they represent
a castration-resistant line with distinct biological features
(38). Here, the present study
found that although the rate of cell proliferation slowed down
slightly in the cells of MYH11 knockdown compared with the
normal control, there were still statistically significant
differences for C4-2 cells (BIC group, P=0.016; ENZ group, P=0.012)
and LNCaP cells (BIC group, P=0.018; ENZ group, P=0.016). So, the
present study identified that MYH11 modulated the
sensitivity of both LNCaP and C4-2 cells to BIC and ENZ, suggesting
that MYH11 can be an indicator for predicting PCa to
antiandrogen drugs resistance. However, the present study verified
the function of MYH11 only in vitro and did not investigate
underlying mechanisms. Due to the complexity of drug resistance,
including the possibility of acquired mechanisms, these results
require additional confirmation in future studies.
MYH11, encoded by the MYH11 gene, is a smooth
muscle myosin belonging to the myosin heavy chain family (39). Mutations in MYH11 have been
implicated in the development of several cancer types (40). For example, MYH11
hypermethylation promotes gastric cancer progression (41) and the gene is key in acute myeloid
leukemia (42,43). MYH11 also serves as a
biomarker for the prediction of lung cancer prognosis (39). Furthermore, MYH11 has been
associated with PCa development: Somatic mutations in MYH11
contribute to PCa occurrence and predict disease progression
(44,45).
In the present study however, some unexpected
phenomenon were observed: First, MYH11 was downregulated in
the tissues of patients with PCa but demonstrated increased
expression in PCa cells treated with antiandrogen drugs. Several
factors may explain this discrepancy. First, biological complexity
and interindividual variability can produce different results in
tissues compared with cell lines. Second, drug treatment can alter
gene expression patterns. For instance, previous studies have
reported that antiandrogen drugs initially suppress AR expression
but eventually increase it through cellular adaptation (5,46). A
similar mechanism may underlie the regulation of MYH11. Another
phenomenon noted in the present study is that the cells without
MYH11 knockdown would still proliferate after treatment with
BIC or ENZ but exhibited decreased proliferation after MYH11
knockdown. This could be because in PCa cells with MYH11
expression, proteins such as AR which affect the function of BIC or
ENZ still have normal expression but after MYH11 knockdown,
the expression of these proteins may decrease. After a protein such
as AR decreases, the cells would be more sensitive to antiandrogen
drugs and proliferate at a decreased rate. Although, to the best of
our knowledge, no direct studies have reported an association
between MYH11 and AR in PCa, a previous study in penile tissue
identified such a relationship (47), suggesting a possible association in
other tissues. In the present study, drugs that inhibited AR
expression also affected MYH11 expression, supporting the idea of a
regulatory association between MYH11 and AR in PCa. Cells with
MYH11 knockdown decreased after treated by BIC or ENZ which
further indicated that MYH11 may influence AR expression in PCa
cells. These findings suggest that MYH11 could be a therapeutic
target for drug-resistant PCa.
However, the present study had several limitations.
First, the present study identified 54 ADRGs as potentially key for
resistance to antiandrogen drugs in PCa and PPI network analysis
highlighted nine hub genes. The functional roles of the remaining
ADRGs, however, remain to be elucidated. Second, the present
examined gene expression using RNA-sequencing data from different
databases; however, these findings were not validated in clinical
samples. The mechanism of MYH11 in PCa therefore requires further
experimental investigation in the future. Third, although
MYH11 expression was generally lower in patients with PCa,
higher MYH11 expression patients were associated with
improved DFS compared with patients with low MYH11 level.
This apparent contradiction warrants additional study. Lastly, the
present functional analysis was performed using bioinformatics
approaches and the mechanisms by which these genes influence PCa
development and drug resistance remain to be elucidated.
Additional experiments should be carried out in the
future to investigate the mechanism of MYH11, including its
interactions with AR signaling. For example, one group of C4-2
cells may be treated with shMYH11 lentivirus and another group with
ENZ to make these cells resistant to ENZ. Subsequently,
RNA-sequencing could be performed and the differentially expressed
genes could be obtained. A pathway analysis could be performed to
identify the genes associated with both BIC and ENZ resistance.
After identifying these genes, basic experiments such as PCR,
western blotting should be performed to further identify the
association of potential genes with MYH11. Furthermore, validation
in additional PCa cell lines such as 22Rv1 and vertebral-cancer of
the prostate is necessary to strengthen the generalizability of the
findings in the future. Despite these limitations, the present
study suggests that MYH11 serves a key role in resistance to
antiandrogen drugs and may potentially serve as a predictive
indicator for PCa development and therapeutic response in the
future.
In summary, the present study identified nine hub
genes among 54 ADRGs from the GSE211781 dataset, with MYH11
emerging as a key indicator for the prediction of PCa occurrence,
progression and also key for PCa cells resistant to antiandrogen
drugs.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LP and JS designed the study and got the data from
public databases. LP and ZL analyzed the data from pubic databases
and wrote the paper. GX and CC conducted all the experiments
including cell culture, western blot and CCK-8 assay. CC
contributed to the statistical analysis and revised the manuscript.
CC and LP confirm the authenticity of all the raw data. All authors
reviewed the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI
|
|
2
|
Fu ZT, Guo XL, Zhang SW, Zheng RS, Zeng
HM, Chen R, Wang SM, Sun KX, Wei WW and He J: Statistical analysis
of incidence and mortality of prostate cancer in China, 2015.
Zhonghua Zhong Liu Za Zhi. 42:718–722. 2020.(In Chinese).
PubMed/NCBI
|
|
3
|
Sekhoacha M, Riet K, Motloung P, Gumenku
L, Adegoke A and Mashele S: Prostate cancer review: Genetics,
diagnosis, treatment options, and alternative approaches.
Molecules. 27:57302022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Debes JD and Tindall DJ: Mechanisms of
androgen-refractory prostate cancer. N Engl J Med. 351:1488–1490.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Watson PA, Arora VK and Sawyers CL:
Emerging mechanisms of resistance to androgen receptor inhibitors
in prostate cancer. Nat Rev Cancer. 15:701–711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ross RW, Xie W, Regan MM, Pomerantz M,
Nakabayashi M, Daskivich TJ, Sartor O, Taplin ME, Kantoff PW and Oh
WK: Efficacy of androgen deprivation therapy (ADT) in patients with
advanced prostate cancer: Association between Gleason score,
prostate-specific antigen level, and prior ADT exposure with
duration of ADT effect. Cancer. 112:1247–1253. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garje R, Chennamadhavuni A, Mott SL,
Chambers IM, Gellhaus P, Zakharia Y and Brown JA: Utilization and
outcomes of surgical castration in comparison to medical castration
in metastatic prostate cancer. Clin Genitourin Cancer.
18:e157–e166. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patke R, Harris AE, Woodcock CL, Thompson
R, Santos R, Kumari A, Allegrucci C, Archer N, Gudas LJ, Robinson
BD, et al: Epitranscriptomic mechanisms of androgen signalling and
prostate cancer. Neoplasia. 56:1010322024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kolvenbag GJ and Nash A: Bicalutamide
dosages used in the treatment of prostate cancer. Prostate.
39:47–53. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scott LJ: Enzalutamide: A review in
Castration-resistant prostate cancer. Drugs. 78:1913–1924. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clegg NJ, Wongvipat J, Joseph JD, Tran C,
Ouk S, Dilhas A, Chen Y, Grillot K, Bischoff ED, Cai L, et al:
ARN-509: A novel antiandrogen for prostate cancer treatment. Cancer
Res. 72:1494–1503. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Li H, Liu B, Wang X, Zhou W, Wu G
and Xu C: Identification and validation of MSMB as a critical gene
for prostate cancer development in obese people. Am J Cancer Res.
13:1582–1593. 2023.PubMed/NCBI
|
|
13
|
Chen X, Ma J, Wang X, Zi T, Qian D, Li C
and Xu C: CCNB1 and AURKA are critical genes for prostate cancer
progression and castration-resistant prostate cancer resistant to
vinblastine. Front Endocrinol (Lausanne). 13:11061752022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Wang X, Zhai M, Xu C and Chen X:
Exploration of the influence of GOLGA8B on prostate cancer
progression and the resistance of castration-resistant prostate
cancer to cabazitaxel. Discov Oncol. 15:1522024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma J, Qin X, Le W, Chen X, Wang X and Xu
C: Identification of BBC3 as a novel indicator for predicting
prostate cancer development and olaparib resistance. Discov Oncol.
15:4962024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Wu Y, Wang X, Xu C, Wang L, Jian
J, Wu D and Wu G: CDK6 is upregulated and may be a potential
therapeutic target in Enzalutamide-resistant castration-resistant
prostate cancer. Eur J Med Res. 27:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blum A, Wang P and Zenklusen JC: SnapShot:
TCGA-Analyzed tumors. Cell. 173:5302018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z,
Wang H, Yu Y, Yang C, Gao X, et al: A genomic and epigenomic atlas
of prostate cancer in Asian populations. Nature. 580:93–99. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chawla S, Rockstroh A, Lehman M, Ratther
E, Jain A, Anand A, Gupta A, Bhattacharya N, Poonia S, Rai P, et
al: Gene expression based inference of cancer drug sensitivity. Nat
Commun. 13:56802022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He Y, Wei T, Ye Z, Orme JJ, Lin D, Sheng
H, Fazli L, Jeffrey Karnes R, Jimenez R, Wang L, et al: A
noncanonical AR addiction drives enzalutamide resistance in
prostate cancer. Nat Commun. 12:15212021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shah S, Carriveau WJ, Li J, Campbell SL,
Kopinski PK, Lim HW, Daurio N, Trefely S, Won KJ, Wallace DC, et
al: Targeting ACLY sensitizes castration-resistant prostate cancer
cells to AR antagonism by impinging on an ACLY-AMPK-AR feedback
mechanism. Oncotarget. 7:43713–43730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled Protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen X, Ma J, Xu C, Wang L, Yao Y, Wang X,
Zi T, Bian C, Wu D and Wu G: Identification of hub genes predicting
the development of prostate cancer from benign prostate hyperplasia
and analyzing their clinical value in prostate cancer by
bioinformatic analysis. Discov Oncol. 13:542022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen X, Li H, Xu C, Wang X, Wu G, Li C and
Wu D: CYP19A1 is downregulated by BRD4 and suppresses
castration-resistant prostate cancer cell invasion and
proliferation by decreasing AR expression. Am J Cancer Res.
13:4003–4020. 2023.PubMed/NCBI
|
|
29
|
Paner GP, Stadler WM, Hansel DE, Montironi
R, Lin DW and Amin MB: Updates in the eighth edition of the
Tumor-Node-Metastasis staging classification for urologic cancers.
Eur Urol. 73:560–569. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
31
|
Etzioni R, Tsodikov A, Mariotto A, Szabo
A, Falcon S, Wegelin J, DiTommaso D, Karnofski K, Gulati R, Penson
DF and Feuer E: Quantifying the role of PSA screening in the US
prostate cancer mortality decline. Cancer Causes Control.
19:175–181. 2008.PubMed/NCBI
|
|
32
|
Arsov C, Winter C, Rabenalt R and Albers
P: Current second-line treatment options for patients with
castration resistant prostate cancer (CRPC) resistant to docetaxel.
Urol Oncol. 30:762–771. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fradet Y: Bicalutamide (Casodex) in the
treatment of prostate cancer. Expert Rev Anticancer Ther. 4:37–48.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Armstrong AJ, Azad AA, Iguchi T,
Szmulewitz RZ, Petrylak DP, Holzbeierlein J, Villers A, Alcaraz A,
Alekseev B, Shore ND, et al: Improved survival with enzalutamide in
patients with metastatic hormone-sensitive prostate cancer. J Clin
Oncol. 40:1616–1622. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Smith MR, Saad F, Chowdhury S, Oudard S,
Hadaschik BA, Graff JN, Olmos D, Mainwaring PN, Lee JY, Uemura H,
et al: Apalutamide treatment and Metastasis-free survival in
prostate cancer. N Engl J Med. 378:1408–1418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Z, Zhou C, Li X, Barnes SD, Deng S,
Hoover E, Chen CC, Lee YS, Zhang Y, Wang C, et al: Loss of CHD1
Promotes heterogeneous mechanisms of resistance to AR-Targeted
therapy via chromatin dysregulation. Cancer Cell. 37:584–598.e11.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Z, Karthaus WR, Lee YS, Gao VR, Wu
C, Russo JW, Liu M, Mota JM, Abida W, Linton E, et al: Tumor
Microenvironment-derived NRG1 promotes antiandrogen resistance in
prostate cancer. Cancer Cell. 38:279–296.e9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Spans L, Helsen C, Clinckemalie L, Van den
Broeck T, Prekovic S, Joniau S, Lerut E and Claessens F:
Comparative genomic and transcriptomic analyses of LNCaP and C4-2B
prostate cancer cell lines. PLoS One. 9:e900022014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nie MJ, Pan XT, Tao HY, Xu MJ, Liu SL, Sun
W, Wu J and Zou X: Clinical and prognostic significance of MYH11 in
lung cancer. Oncol Lett. 19:3899–3906. 2020.PubMed/NCBI
|
|
40
|
Islam T, Rahman R, Gov E, Turanli B,
Gulfidan G, Haque A, Arga KY and Haque Mollah N: Drug targeting and
biomarkers in head and neck cancers: Insights from systems biology
analyses. OMICS. 22:422–436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Xu P, Hao Y, Yu T, Liu L, Song Y
and Li Y: Interaction between DNMT3B and MYH11 via hypermethylation
regulates gastric cancer progression. BMC Cancer. 21:9142021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cho BS, Min GJ, Park SS, Park S, Jeon YW,
Shin SH, Yahng SA, Yoon JH, Lee SE, Eom KS, et al: Prognostic
values of D816V KIT mutation and Peri-transplant CBFB-MYH11 MRD
monitoring on acute myeloid leukemia with CBFB-MYH11. Bone Marrow
Transplant. 56:2682–2689. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ishikawa Y, Kawashima N, Atsuta Y, Sugiura
I, Sawa M, Dobashi N, Yokoyama H, Doki N, Tomita A, Kiguchi T, et
al: Prospective evaluation of prognostic impact of KIT mutations on
acute myeloid leukemia with RUNX1-RUNX1T1 and CBFB-MYH11. Blood
Adv. 4:66–75. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Alhopuro P, Karhu A, Winqvist R, Waltering
K, Visakorpi T and Aaltonen LA: Somatic mutation analysis of MYH11
in breast and prostate cancer. BMC Cancer. 8:2632008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang Y, Cao Q, Song Z, Ruan H, Wang K,
Chen K and Zhang X: The identification of key gene expression
signature in prostate cancer. Crit Rev Eukaryot Gene Expr.
30:153–168. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Aurilio G, Cimadamore A, Mazzucchelli R,
Lopez-Beltran A, Verri E, Scarpelli M, Massari F, Cheng L, Santoni
M and Montironi R: Androgen receptor signaling pathway in prostate
cancer: From genetics to clinical applications. Cells. 9:26532020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Okumu LA, Bruinton S, Braden TD, Simon L
and Goyal HO: Estrogen-induced maldevelopment of the penis involves
down-regulation of myosin heavy chain 11 (MYH11) expression, a
biomarker for smooth muscle cell differentiation. Biol Reprod.
87:1092012. View Article : Google Scholar : PubMed/NCBI
|