Introduction
DEAH box helicase 15 (DHX15), which is a member of
the DEAH box RNA helicase family (1), is involved in several cellular
processes, such as splicing and ribosome biogenesis and facilitates
remodeling of large RNA-protein complexes (2–4). DHX15
was shown to carry out a role in antiviral immune response in
vivo (5). In RNA virus-induced
intestinal inflammation, DHX15 carries out a pivotal role in
controlling inflammation by sensing IFN-β and other cytokines
produced by RNA viruses (6).
Moreover, DHX15 is reported to be involved in tumorigenesis as a
tumor-promoting factor in acute lymphoblastic leukemia and cancer
of the lung, prostate or breast (7–10), but
it also acts as a tumor suppressor in hepatocellular carcinoma and
gastric cancer (11,12). Using gain- and loss-of-function
analyses, we previously revealed that DHX15 suppressed the
proliferation of glioma cell lines (13).
Colorectal cancer (CRC) is the third most common
type of cancer, with ~2 million cases diagnosed worldwide in 2020,
and the second most common cause of cancer-related mortality,
according to the International Agency for Research on Cancer of the
World Health Organization (14).
Repeat analyses of public single-cell sequence and bulk
transcriptome data revealed consistent upregulation of DHX15
expression in patients with CRC (15–17).
Analysis of the clinical pathological features of CRC samples from
the Cancer Gene Atlas and the Clinical Proteomic Tumor Analysis
Consortium revealed that reduced expression of DHX15 mRNA
and DHX15 protein associated with poor CRC prognosis in terms of
survival, metastasis and recurrence (16). Furthermore, the overall survival
positively associated with DHX15 expression levels in CRC (18). Conversely, in a case report of a
patient with CRC harboring a KRAS p.G12D mutation, DHX15 was
implicated as a critical mediator of microbiota-driven pathogenesis
of CRC. Specifically, DHX15 was found to be highly expressed and
functioned as a receptor on tumor cells, facilitating the invasion
of Fusobacterium nucleatum into the nuclei of intestinal
epithelial cells (IECs), thereby promoting tumorigenesis (19). The present study examined the
effects of DHX15 overexpression using CRC cell lines with different
characteristics, including KRAS mutations (19).
Materials and methods
Human clinical samples
Human CRC samples and adjacent normal tissues were
obtained by surgical resection from ten patients with CRC; five
male, average age 60 years (51–65 years), five female, average age
75.6 years (60–91 years) at Juntendo University Hospital (Tokyo,
Japan) between April 2022-August 2025 and embedded in paraffin. The
diagnoses of the ten patients were based on clinical and
pathological examinations (Table
I). Cancer staging was carried out in accordance with the Union
for International Cancer Control TNM classification (20). All human tissues were obtained with
written informed consent and approval from the medical ethical
committee of Juntendo University Hospital (Tokyo, Japan; approval
no. E22-0079).
 | Table I.DHX15 expression in human CRC tissues
and clinicopathological factors of the patients with CRC. |
Table I.
DHX15 expression in human CRC tissues
and clinicopathological factors of the patients with CRC.
|
|
|
|
|
|
|
|
|
|
|
| DHX15
immuno-Histochemical staining |
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Patient | Age (years) | Sex | Location | Stage | T | N | M | Histological
type | V | Ly | Adjacent
tissue | Cancer |
|---|
| 1 | 75 | W | C | IIIc | 4a | 2b | 0 | por | 0 | 1c | 48 | 58 |
| 2 | 62 | M | A | IIIb | 3 | 1a | 0 | tub2 | 1c | 0 | 34 | 56 |
| 3 | 82 | W | A | IIIb | 3 | 2a | 0 | tub2 | 1a | 1c | 17 | 74 |
| 4 | 60 | W | T | IIIb | 3 | 1b | 0 | tub1 | 0 | 1a | 47 | 76 |
| 5 | 51 | M | T | II | 3 | 0 | 0 | tub1 | 0 | 1a | 35 | 49 |
| 6 | 91 | W | S | IIIb | 3 | 1a | 0 | tub2 | 1b | 1a | 36 | 77 |
| 7 | 65 | M | S | IIIc | 4a | 3 | 0 | tub2 | 0 | 1a | 21 | 59 |
| 8 | 60 | M | S | II | 3 | 0 | 0 | tub1 | 1a | 0 | 14 | 48 |
| 9 | 62 | M | RS | IIa | 3 | 0 | 0 | tub1 | 0 | 0 | 29 | 57 |
| 10 | 70 | W | RS | II | 3 | 0 | 0 | tub2 | 1c | 1a | 49 | 69 |
Reagents, CRC cell lines, transfection
and cell sorting
Porcupine inhibitor C59 (cat. no. S7037) was
obtained from Selleck Chemicals and the NF-κB inhibitor IMD-0354
(cat. no. HY-10172) was obtained from MedChemExpress. The human CRC
cell lines HCT116 (21), SW480
(22) and Caco2 (23) (provided by Dr Akira Orimo, Juntendo
University, Tokyo, Japan) were cultured in low glucose DMEM
(Nacalai Tesque, Inc.) and DLD1 cells (24) (provided by Dr Akira Orimo, Juntendo
University, Tokyo, Japan) were cultured in RPMI1640 (Nacalai
Tesque, Inc.); both culture media were supplemented with 10% FBS
(MilliporeSigma) and penicillin-streptomycin (MilliporeSigma). The
characteristics of the four CRC cell lines are summarized in
(Table SI) (25).
HCT116, SW480, Caco2 and DLD1 cells
(1.5×106 cells) were seeded on 10-cm plates (Nippon Gene
Co., Ltd.) and transfected using GeneJuice Transfection Reagent
(Cat. no. 70967-3CN; Merck KGaA) with a combination of the plasmids
pMXs-IP (control vector) or pMX-DHX15 and EGFP expression
(pCAG-EGFP), at DNA amounts of 0.03 µg for pCAG-EGFP and 0.21 µg
for pMXs-IP or pMX-DHX15 per well. The cells were harvested at 37°C
after 12 h of culture and EGFP-positive cells were collected using
a cell sorter (FACS ARIA III, BD Biosciences). Total RNA was
purified using Trizol® reagent (Thermo Fisher
Scientific, Inc.).
Quantitative reverse transcription PCR
(RT-qPCR)
RT-qPCR was carried out as previously described
(26). The expression levels of the
transcripts of interest were normalized using GAPDH and
ACTB values. The respective forward and reverse primer
sequences were as follows: human ACTB:
5′-GAAGGAGATCACTGCCCTGG-3′ and 5′-ACTCCTGCTTGCTGATCCAC-3′;
GAPDH: 5′-ATTGCCCTCAACGACCACTT-3′ and
5′-TGGTCCAGGGGTCTTACTCC-3′; DHX15:
5′-TGCTGAACGTCTACCATGCTT-3′ and 5′-ATTCGAGATAGCTGCTGGCG-3′;
IRF1: 5′-AAGGGGTGTGGCCTTTTTAGA-3′ and
5′-TGTCCCTGTTCACCCCAAAG-3′; IRF3: 5′-CCTGCACATTTCCAACAGCC-3′
and 5′-AATCCATGCCCTCCACCAAG-3′: IRF7:
5′-GCTCCCCACGCTATACCATC-3′ and 5′-CAGGGAAGACACACCCTCAC-3′;
NFKB1: 5′-GTGAAGACCACCTCTCAGGC-3′ and
5′-CTGTCGCAGACACTGTCACT-3′; NFKB2:
5′-ACGCCTCTTGACCTCACTTG-3′ and 5′-GTGGCTCCATGGTGTTCTGA-3′;
RELA: 5′-GGACATGGACTTCTCAGCCC-3′ and
5′-AAAGTTGGGGGCAGTTGGAA-3′; and REL:
5′-TCCTTAGCCCAGCCATCTCT-3′ and 5′-GGCAGTCTCCGCTCATCTTT-3′.
Immunohistochemistry
DHX15 protein expression in human clinical samples
was examined by immunohistochemistry using an anti-DDX15 antibody
(H-4, 1:500; Santa Cruz Biotechnology; cat. no. sc-271686).
Paraffin-embedded 3-µm sections were first deparaffinized with EZ
Prep (cat. no. 950-102) and subjected to antigen retrieval using
Cell Conditioner #2 (pH 9.0; cat. no. 950-223) on a BenchMark Ultra
automated stainer (Roche Tissue Diagnostics). After antigen
retrieval, the sections were incubated with the primary anti-DDX15
antibody at 37°C for 1 h.
Primary antibody signals were then detected using
the UltraView Universal DAB Detection Kit (Roche Tissue
Diagnostics, cat. no. 760-500) under standard BenchMark Ultra
settings, including an 8-min incubation with the HRP-conjugated
multimer reagent, followed by DAB chromogen visualization.
Counterstaining was performed with Hematoxylin II for 8 min and
Bluing Reagent for 4 min at 37°C. Stained sections were observed
using a Zeiss Axio Imager M2 fluorescence microscope (Zeiss GmbH),
and images were acquired using ZEN software (version 3.12; Zeiss
GmbH).
Stained samples were observed using Zeiss Axio
Imager M2 fluorescence microscope (Zeiss GmbH) and images were
acquired using ZEN software (ZEN3.12; Zeiss GmbH). For
quantification of DHX15 positive signals, we used ImageJ (version
1.54p; National Institutes of Health). The images were imported to
ImageJ and DAB signals were isolated using the color deconvolution
command of ImageJ. DAB images were converted to gray scale binary
images, and doublet cells were separated by the Watershed command
and positive and negative cells were counted. To quantify intensity
of the signals, the log10 value of mean of sample ROI vs. mean of
background ROI by using 8-bit index color was calculated.
Immunohistochemistry of the cultured cells was
carried out as described previously (27). The primary antibodies used were
mouse monoclonal antibodies against Ki67 (1:250; cat. no. 55060;
BD); LC3 (1:2,000; cat. no. M152-3; MEDICAL & BIOLOGICAL
LABORATORIES CO., LTD.); rabbit polyclonal antibody against active
Caspase-3 (AC3; 1:250; cat. no. 9664S; Promega Corporation); and
chicken polyclonal antibody against GFP (1:2,000; cat. no. ab13970;
Abcam). Appropriate secondary antibodies conjugated with Alexa
Fluor 488 (1:500; cat. no. A11039; Thermo Fisher Scientific, Inc.)
or Alexa Fluor 594 (1:500; cat. no. C10639; Thermo Fisher
Scientific, Inc.) were used. The nuclei were visualized using DAPI.
Stained cells were observed using Zeiss Axio Imager I fluorescence
microscope (Zeiss GmbH) and images were acquired using ZEN software
(version, 3.12; Zeiss GmbH).
Luciferase reporter assay for
Wnt/β-catenin and NF-κB signaling pathways
Luciferase reporter assays for the Wnt/β-catenin and
NF-κB signaling pathways were performed using TCF-luc (IFN minimal
promoter with 7× TCF-binding sites driving firefly luciferase) and
NFκB-luc (SV40 early promoter with 8× NF-κB-binding sites driving
firefly luciferase) (28). These
plasmids were kindly provided by Dr. K. Shimotono (Kyoto
University, Kyoto, Japan) and Dr. R. Tsuruta (University of Tokyo,
Tokyo, Japan), respectively. No 3′ UTR reporter constructs and no
miRNA mimics or inhibitors were used in this study. Cells were
transfected with the reporter plasmids using GeneJuice Transfection
Reagent (cat. no. 70967-3CN; MilliporeSigma) according to the
manufacturer's instructions. Luciferase activity was measured 18 h
after transfection using a luminometer (Lumat LB9507;
Titertek-Berthold) and the Luciferase Assay System (Promega, cat.
no. E1500). Because only firefly luciferase reporters were used,
luciferase activity was analyzed without Renilla luciferase
normalization.
Cell proliferation assay
SW480 (1×104cell/well), Caco2
(1×104 cell/well), HCT116 (2×104 cells) and
DLD1 (2×104 cells) cells were plated on 48-well plates
(Corning, Inc.) and then transfected with a combination of plasmids
pMXs-IP (control vector) or pMX-DHX15 and pCAG-EGFP using GeneJuice
(cat. no. 70967-3CN; MilliporeSigma). After 1 h of incubation, the
plate was set on IncuCyte ZOOM (Sartorius AG). Live-cell images
were captured every 8 h and analyzed using IncuCyte Live-Cell
Imaging Software (v2024B).
To evaluate the time-dependent changes in the number
of GFP-positive cells after transfection, cells were monitored
using an Incucyte live-cell imaging system under standard culture
conditions (37°C; 5% CO2) at 8-h intervals immediately
after plating.
Cell proliferation was examined by incorporating
5-ethynyl 2′-deoxyuridine (EdU) using Click-iT EdU Alexa Fluor 594
Imaging kit (Thermo Fisher Scientific, Inc.). After transfecting
the cells as aforementioned, EdU was added to the culture at a
final concentration of 10 µM and incubated for 2 h at 37°C. For
inhibitor experiments, cells were treated with the Wnt inhibitor
C59 (10 µM) or the NF-κB inhibitor IMD-0354 (5 µM) at 37°C for 2 h
prior to EdU addition. The incorporated EdU signals were visualized
by immunostaining with a chicken polyclonal antibody against GFP
(cat. no. ab13970; 1:2,000; Abcam) at room temperature overnight
and a secondary antibody conjugated with Alexa Fluor 488 at room
temperature for 30 min (1:500; Thermo Fisher Scientific, Inc.).
EGFP was immunostained with an anti-GFP antibody (1:2,000; cat. no.
ab13970; Abcam). Nuclei were stained with DAPI. Stained cells were
observed using Zeiss Axio Imager I (Zeiss GmbH) and images were
acquired using ZEN software (Zeiss GmbH).
Western blotting
Cell lysates were collected from cell lines
(2×106 cells) and analyzed by western blotting, as
previously described (29). Protein
was extracted using RIPA buffer (Nacalai Tesque, Inc.), containing
50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium
deoxycholate and 0.1% SDS, supplemented with protease inhibitor
cocktail (Nacalai Tesque, Inc.).
Protein concentration was determined using the BCA
assay (Nacalai Tesque, Inc..) according to the manufacturer's
protocol. Equal amounts of protein (3 µg per lane) were mixed with
4× Laemmli sample buffer (Nacalai Tesque, Inc.) and boiled at 98°C
for 5 min before electrophoresis. Proteins were separated on 10%
SDS-PAGE gels. Proteins were transferred onto PVDF membranes.
Membranes were blocked in 5% non-fat dry milk (Bio-Rad) in
TBS-Tween (0.1%) at room temperature for 1 h. Membranes were
incubated with primary antibodies at 4°C overnight, followed by
incubation with HRP-conjugated secondary antibodies for 1 h at room
temperature. Signals were detected using Chemi-Lumi One Super
(Nacalai Tesque, Inc.). The primary antibodies used were mouse
monoclonal antibodies against ACTIN (1:1,000; cat. no. A4700;
MilliporeSigma) and DDX15 (1:1,000; cat. no. sc-271686; Santa Cruz
Biotechnology, Inc.). The secondary antibody used was HRP-linked
secondary antibodies (1:5,000; cat. no. NA931; Cytiva).
Statistical analysis
Statistical significance was calculated by an
unpaired Student's t test (two tailed). P<0.05 was considered to
indicate a statistically significant difference; *P<0.05,
**P<0.01. Experiments were conducted in triplicate. Data are
shown as mean ± standard deviation)
Results
DHX15 expression in human CRC
tissues
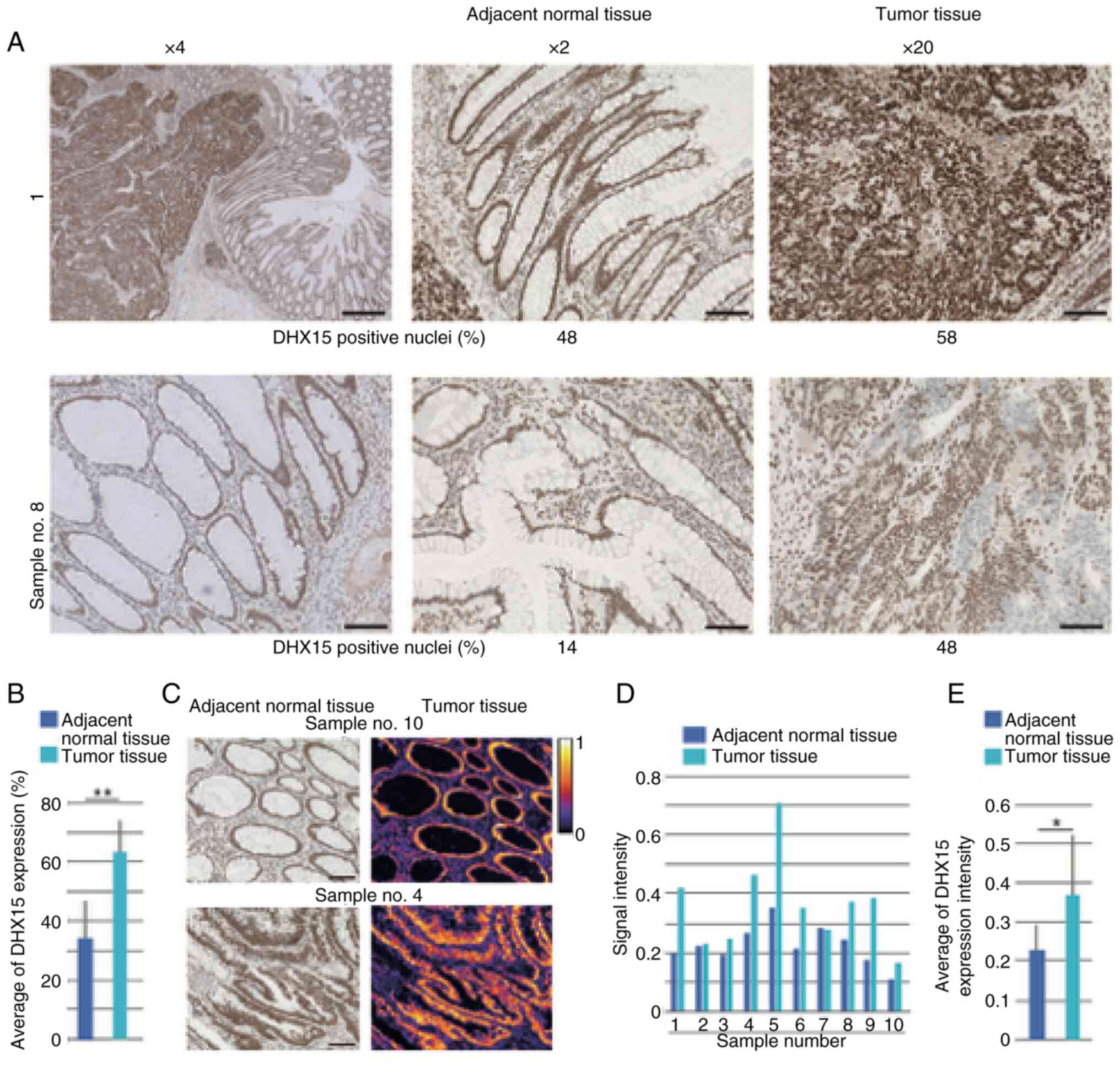
On examination of the clinical samples, DHX15
protein was detectable in the cell nuclei in both tumor and
adjacent normal tissues of all 10 patients (Fig. 1A). The evaluation values of DHX15
expression strength for all 10 samples are summarized (Table I). The average positive nuclei
staining of DHX15 in the ten tumor samples was increased compared
with that of adjacent tissue (Fig.
1B). Among the CRC tissues, four showed >60% of DHX15
positive signals in tumor tissues, but there was no correlation
between clinicopathological factors and DHX15 staining intensity
(Table I). Heat-map (Fig. 1C) represents intensity of DHX15
immunostaining of adjacent normal tissue and tumor tissue (Fig. 1D). Again, clear association between
pathological observation and signal intensity was not found, but
the average DHX15 expression intensity in the tumor region was
increased compared with that in adjacent tissue (Fig. 1E).
Effects of DHX15 overexpression on the
proliferation of CRC cell lines
DHX15 protein expression levels were first examined
in the CRC cell lines HCT116, SW480, Caco2 and DLD1 using western
blotting of whole protein. All cell lines express DHX15 with
expected size (Fig. S1A; upper
bands). Subsequently, the cells were co-transfected with
DHX15-expression vector or empty control vector together with
EGFP-expression plasmid and EGFP positive cells were purified by a
cell sorter after 12 h of culture. Overexpression of DHX15
transcripts was confirmed by RT-qPCR in all four cell lines
(Fig. S1B).
To examine the effects of DHX15 overexpression on
proliferation, the cells were cotransfected with DHX15-expressing
vector or empty control vector with EGFP-expression plasmid. The
transfected cells were cultured for 2 days, and the number of
EGFP-positive cells was counted over time using IncuCyte live-cell
imaging. The number of EGFP-positive cells is shown on Fig. 2A and B. Compared with the control
samples, samples with DHX15 overexpression had a significantly
reduced cell number in all the examined CRC cell lines (Fig. 2B). These results suggested that
DHX15 suppressed cell proliferation.
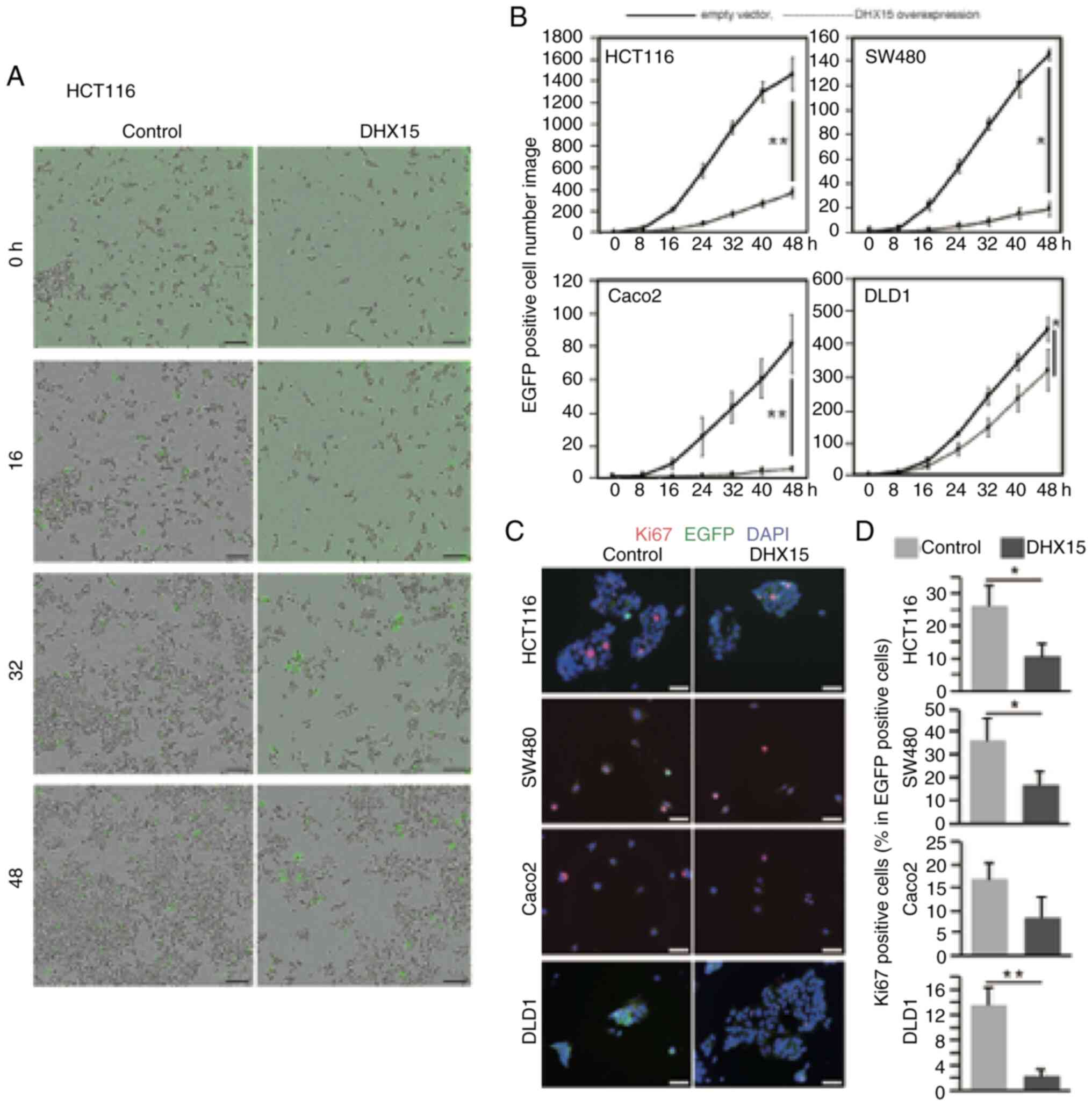 | Figure 2.Effects of DHX15 overexpression on
the proliferation of various CRC cell lines. The CRC cell lines
HCT116, SW480, Caco2 and DLD1 were transfected with
DHX15-expressing or empty vector with EGFP-expressing vector. The
cells were incubated in IncuCyte ZOOM, and live-cell images were
captured every 8 h. (A) The population of EGFP positive cells was
analyzed using IncuCyte Live-Cell Imaging Software. The captured
images of HCT116 of control and DHX15 overexpressed cells at 0, 16,
32 and 48 h after transfection. (B) Transition of number of EGFP
positive cells from 0 to 48 h after transfection of empty vector or
DHX15 overexpression vector. Cell proliferation was examined by
Ki67 immunostaining. Transfected cells were cultured for 24 h,
followed by immunostaining with an anti-Ki67 antibody. (C)
Representative images, DAPI (blue) shows nucleus. (D) Population of
Ki67 positive cells in total EGFP positive cells in the 4 cell
lines. Scale bar, 100 µm in A and C. Data are average of 3
independent samples with SEM. Statistical significance was
calculated by Student's t test (two tailed). *P<0.01;
**P<0.01. DHX15, DEAH (Asp-Glu-Ala-His) box helicase 15; CRC,
colorectal cancer. |
The number of Ki67-positive proliferating HCT116,
SW480 and DLD1 cells was significantly reduced after transfection
with DHX15 and EGFP-expression plasmid (Fig. 2C and D). The EdU incorporation assay
revealed that DHX15 overexpression significantly reduced the number
of EdU-positive cells in all four CRC cell lines (Fig. S2A and B). These results suggested
that DHX15 suppressed the proliferation of CRC cell lines.
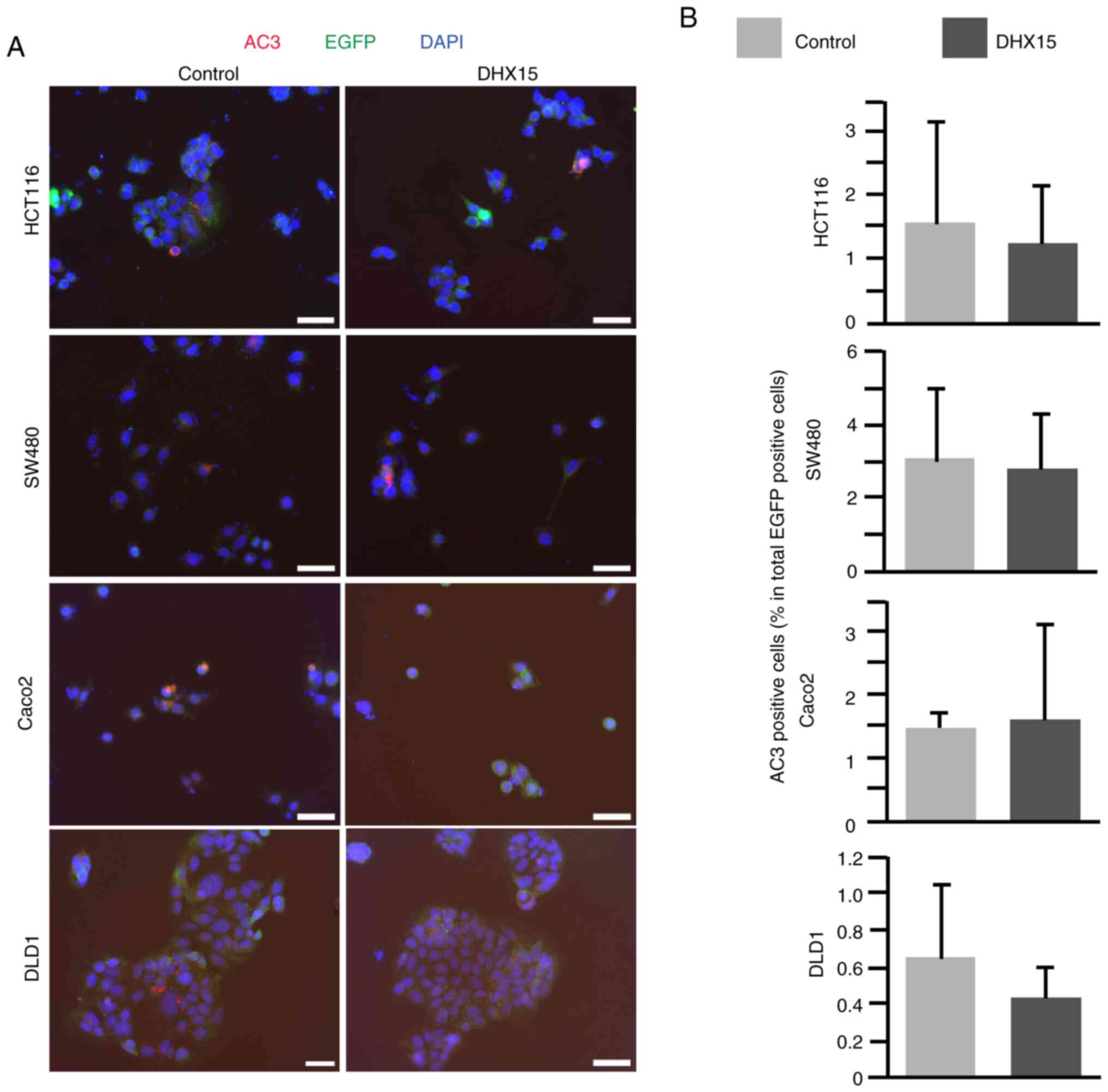
Next, the DHX15-overexpressing CRC cell lines were
investigated using the immunohistochemistry apoptosis marker AC3.
The cells were cotransfected with either the empty control or
DHX15-expressing vector with EGFP-expression plasmid and
immunostaining was carried out after 2 days of culture. The number
of AC3-positive apoptotic cells was comparable between the control
and DHX15-transfected cells in the four CRC cell lines, except
HCT116, which showed slight decrease in AC3-positive cells in the
DHX15 overexpression group, although this was not significant
(Fig. 3A and B). These results
indicated that DHX15 overexpression may not affect apoptosis in the
CRC cell lines.
Examination of the possible
involvement of Wnt, NF-κB and autophagy in DHX15-overexpressing
cells
To investigate the potential mechanisms underlying
this finding, the involvement of the Wnt and NF-κB signaling
pathways were examined, as well as autophagy, in
DHX15-overexpressing cells. These pathways have previously been
reported to play key roles in the regulation of cell proliferation
(11,30,31).
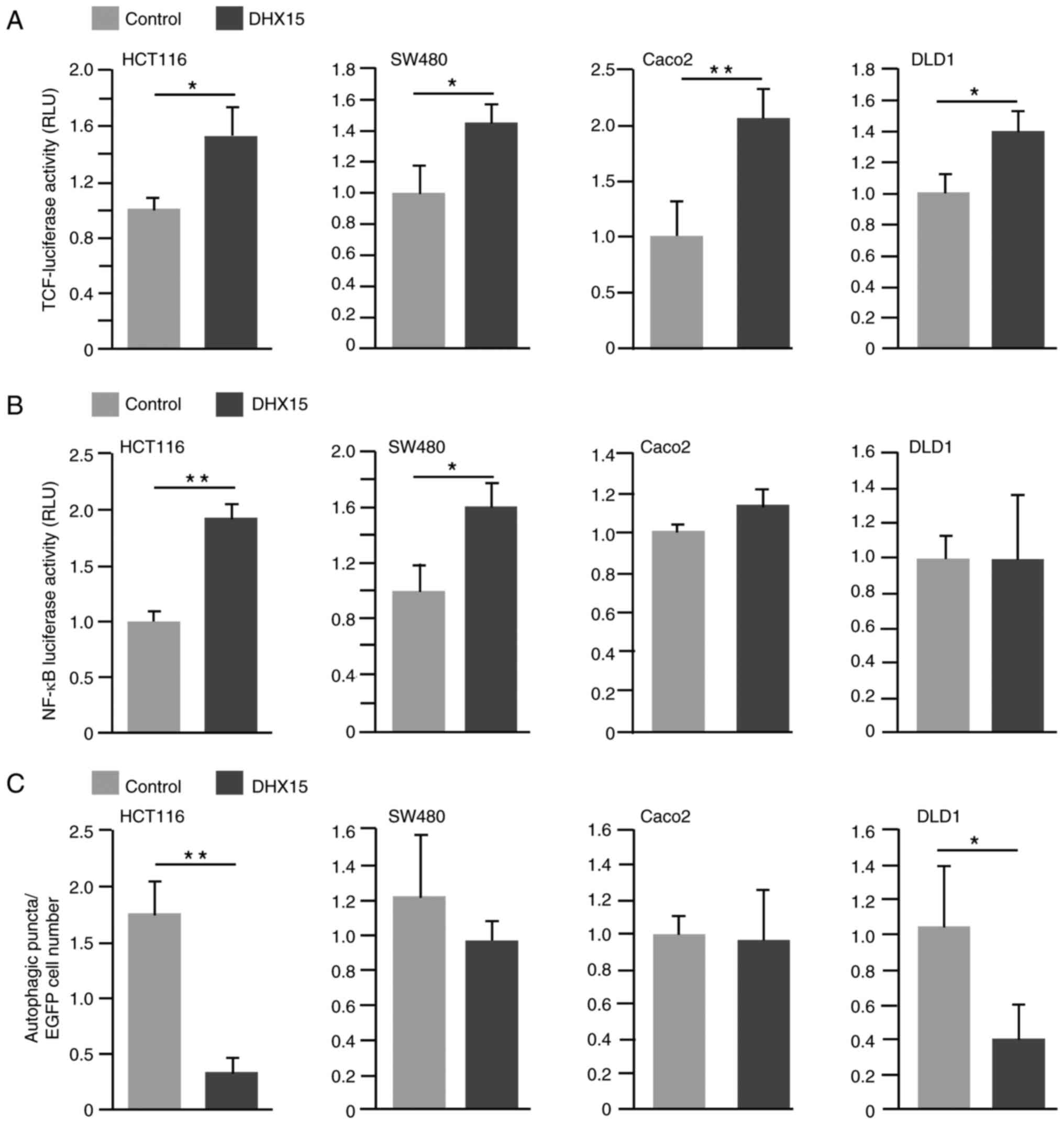
Analysis revealed that DHX15 overexpression significantly induced
luciferase activities that were dependent on the TCF-binding motif
in all examined CRC cells (Fig.
4A). The effects of the Wnt signaling inhibitor C59 on the
proliferation of CRC cell lines with overexpressed DHX15 were
investigated using EdU incorporation assay. Analysis revealed a
tendency for higher EdU incorporation in HCT116 and SW480 cells
when compared with control cells (Fig.
S3A and B).
On examination of NF-κB signaling using a NF-κB
target site-dependent luciferase assay, DHX15 overexpression
enhanced luciferase activity in HCT116 and SW480 cells; however, in
Caco2 and DLD1 cells, luciferase activity was similar between the
control and DHX15-overexpressing cells (Fig. 4B). On examination of NF-κB-related
gene expression by RT-qPCR in HCT116 and SW480 cells, the control
and DHX15-overexpressing cells revealed similar transcript levels
of genes, except for IRF3 and REL in SW480 cells
(Fig. S4). Examination of the
effects of the NF-κB inhibitor IMD-0354 revealed that the number of
EdU-positive cells was not significantly different between
DHX15-transfected and IMD-0354 treated cells in all four CRC cell
lines (Fig. S5A and B).
The expression pattern of LC3 puncta, which is an
autophagy marker (32), was
subsequently examined. Upon counting the LC3-positive puncta in the
nuclei of control and DHX15-overexpressing cells, DHX15
overexpression led to a reduced number of LC3-positive signals in
HCT116 and DLD1 cells but not in SW480 and Caco2 cells (Figs. 4C and S6).
Discussion
The present study investigated the role of DHX15 in
CRC by examining DHX15 expression levels in clinical samples and
changes in DHX15 levels in CRC cell lines. In the clinical samples
from 10 patients with CRC, DHX15 expression levels varied. The
absence of a clear association between clinicopathological factors
and intensity of DHX15 expression was likely attributable to the
small sample size (n=10 patients). Further analysis of DHX15
expression using additional clinical samples may help clarify its
relationship with clinicopathological factors. Although the
relationship between DHX15 expression levels and tumor malignancy
remains inconclusive, the quantification of its expression may
provide a potential prognostic indicator, such as recurrence or
reduced survival in cases with low DHX15 expression. At present, to
the best of our knowledge, no association has been observed between
DHX15 expression and tumor stage. In addition, we hypothesize that
there is a possibility that the DHX15 expression patterns and
levels are associated with disease progression which may be
revealed by monitoring the condition of patients continuously and
carefully. Conversely, analysis revealed that DHX15 upregulation
hampered the proliferation activity of all examined CRC cell lines.
This result is consistent with a recent study which revealed that
DHX15 inhibits CRC progression, invasion and metastasis (18). In the present study, all CRC cell
lines examined exhibited endogenous DHX15 expression. Therefore,
further forced overexpression of DHX15 may have exceeded the
physiological threshold, leading to cytotoxicity. Such
supraphysiological expression levels could disrupt essential
cellular processes, ultimately resulting in growth suppression.
Mutation in the well-known protooncogene KRAS is found in
HCT116, SW480 and DLD1 cell lines but not in Caco2 cells;
therefore, the DHX15 effects observed in the present study were
likely not associated with KRAS mutation. However,
evaluation of DHX15 protein expression patterns and their
relationship with clinicopathological factors in hepatocellular
carcinoma revealed that DHX15 was markedly upregulated, and its
high expression associated with poor prognosis (33). Taken together with the reports of
other types of cancer, we hypothesize that the effects on DHX15
expression levels to cancer prognosis depend on the types of
cancer.
Among the various mechanisms of DHX15 activity, Wnt
involvement has been reported in different model systems. In one
study that analyzed the role of DHX15 in antibacterial responses in
IECs, mice with DHX15 deletion in IECs were susceptible to
infection with enteric bacteria because of low levels of α
defensin, which is an antimicrobial peptide; considering that α
defensin is induced by the TLR/Wnt pathways this study suggested
that DHX15 regulates TLR/Wnt signals (34). Another study revealed that DHX15
regulates zebrafish intestinal development through the Wnt
signaling pathway (35). The
present study revealed that DHX15 overexpression significantly
induced Wnt signaling in the four CRC cell lines; however,
inhibition of Wnt signaling did not prevent the suppressed
proliferation of DHX15-overexpressing cells. Furthermore, given its
well-known contribution to the development and progression of
various tumors (36,37), Wnt signaling was less likely to
participate in the suppression of cell proliferation by DHX15.
However, other physiological processes, such as metastasis, were
not analyzed in the present study and requires in vivo
clarification in future research.
DHX15 acts as an immune modulator through regulation
of NF-κB. Human DHX15 contributes to activation of the NF-κB, JNK
and p38 MAPK pathways in HeLa cells in response to the synthetic
double-stranded RNA analog poly(I:C) (5). In acute myeloid leukemia, DHX15 is
downregulated in disease remission or cell line differentiation
(7). Furthermore, knockdown of
DHX15 inhibits nuclear translocation and activation of the
NF-κB subunit P65 in leukemia cells (7). Positive feedback of NF-κB and DHX15
was reported in breast cancer cells (10). The present study revealed that DHX15
overexpression induced NF-κB signaling in HCT116 and SW480 cells,
but the expression levels of known downstream target genes of the
NF-κB pathway were not upregulated. Furthermore, NF-κB inhibition
by the IMD-0354 did not affect cell proliferation, suggesting the
need to clarify the signaling pathways and biological roles of
DHX15 using cell lines in the future.
Involvement of autophagy in cancer progression has
been reported in various types of cancer (38–41).
DHX15 negatively regulates autophagy in association with mTORC1
activation in hepatoma cells (11).
There have been several reports on KRAS-activating mutations
and autophagy. KRAS-activating mutations increase autophagy
and contribute to the survival of CRC cells under starvation
conditions (42). In addition,
autophagy was induced to different degrees, depending on mutations
of the KRAS allele (42). In
the present study, given the decreased number of LC3 puncta in
HCT116 and DLD1 cells, which have a KRASG13D/−
mutation, a relationship between KRAS mutation and autophagy
was suspected. However, the contribution of this pathway to cell
proliferation remains unclear. The effects of other signaling
pathway inhibitors were also explored using this experimental model
(data not shown). Preliminary experiments were performed with
various inhibitor concentrations, and cell proliferation was
assessed by EdU immunostaining. No significant differences were
observed between the groups. However, because the efficacy of the
inhibitor itself was not fully confirmed, these data are not
included in the present study. Experiments in which knockdown of
DHX15 is achieved by siRNA oligonucleotide are planned as future
work.
Although the present study did not yield fully
conclusive mechanistic results, it provides novel evidence that
DHX15 overexpression suppresses colorectal cancer cell
proliferation and highlights DHX15 as a previously under-recognized
regulator of tumor biology. These findings offer a foundation for
future mechanistic studies and suggest that DHX15 may serve as a
potential biomarker or therapeutic target in colorectal cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr. Kanji Uchida
(Department of Anesthesiology, University of Tokyo Hospital, Tokyo,
Japan) for discussions and allowing the use of IncuCyte in their
department.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
YI and KoS performed the experiments, TI, SI, KoS,
KiS, KaS and SW designed the research, YI and SW wrote the
manuscript. SW contributed to the study conception, experimental
design, data interpretation, and provided critical revisions of the
manuscript. YI and SW confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All human tissues were obtained with written
informed consent and approval from the medical ethical committee of
Juntendo University Hospital, Tokyo, Japan (approval no.
E22-0079).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DHX15
|
DEAH (Asp-Glu-Ala-His) box helicase
15
|
|
CRC
|
colorectal cancer
|
|
IEC
|
epithelial cell
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
TCF-luc
|
TCF-binding site luciferase
|
|
NFκB-luc
|
NF-κB binding sites luciferase
|
References
|
1
|
Linder P: Dead-box proteins: A family
affair-active and passive players in RNP-remodeling. Nucleic Acids
Res. 34:4168–4180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arenas JE and Abelson JN: Prp43: An RNA
helicase-like factor involved in spliceosome disassembly. Proc Natl
Acad Sci USA. 94:11798–11802. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Combs DJ, Nagel RJ, Ares M Jr and Stevens
SW: Prp43p is a DEAH-box spliceosome disassembly factor essential
for ribosome biogenesis. Mol Cell Biol. 26:523–534. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tanaka N, Aronova A and Schwer B: Ntr1
activates the Prp43 helicase to trigger release of lariat-intron
from the spliceosome. Genes Dev. 21:2312–2325. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mosallanejad K, Sekine Y,
Ishikura-Kinoshita S, Kumagai K, Nagano T, Matsuzawa A, Takeda K,
Naguro I and Ichijo H: The DEAH-box RNA helicase DHX15 activates
NF-κB and MAPK signaling downstream of MAVS during antiviral
responses. Sci Signal. 7:ra402014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xing J, Zhou X, Fang M, Zhang E, Minze LJ
and Zhang Z: DHX15 is required to control RNA virus-induced
intestinal inflammation. Cell Rep. 35:1092052021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan L, Li Y, Zhang HY, Zheng Y, Liu XL, Hu
Z, Wang Y, Wang J, Cai YH, Liu Q, et al: DHX15 is associated with
poor prognosis in acute myeloid leukemia (AML) and regulates cell
apoptosis via the NF-kB signaling pathway. Oncotarget.
8:89643–89654. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao G, Chen K, Qin Y, Niu Y, Zhang X, Xu
S, Zhang C, Feng M and Wang K: Long non-coding RNA JHDM1D-AS1
interacts with DHX15 protein to enhance non-small-cell lung cancer
growth and metastasis. Mol Ther Nucleic Acids. 18:831–840. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing Y, Nguyen MM, Wang D, Pascal LE, Guo
W, Xu Y, Ai J, Deng FM, Masoodi KZ, Yu X, et al: DHX15 promotes
prostate cancer progression by stimulating Siah2-mediated
ubiquitination of androgen receptor. Oncogene. 37:638–650. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng W, Wang X, Yu Y, Ji C and Fang L:
CircRNF10-DHX15 interaction suppressed breast cancer progression by
antagonizing DHX15-NF-κB p65 positive feedback loop. Cell Mol Biol
Lett. 28:342023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao M, Ying L, Wang R, Yao J, Zhu L,
Zheng M, Chen Z and Yang Z: DHX15 inhibits autophagy and the
proliferation of hepatoma cells. Front Med (Lausanne).
7:5917362020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zong Z, Li H, Ning Z, Hu C, Tang F, Zhu X,
Tian H, Zhou T and Wang H: Integrative bioinformatics analysis of
prognostic alternative splicing signatures in gastric cancer. J
Gastrointest Oncol. 11:685–694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ito S, Koso H, Sakamoto K and Watanabe S:
RNA helicase DHX15 acts as a tumour suppressor in glioma. Br J
Cancer. 117:1349–1359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
15
|
Lv X, Ma W, Miao X, Hu S and Xie H:
Navigating colorectal cancer prognosis: A Treg-related signature
discovered through single-cell and bulk transcriptomic approaches.
Environ Toxicol. 39:3512–3522. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu P, Zhang Y, Cui Y, Liao Y, Liu Z, Cao
ZJ, Liu JE, Wen L, Zhou X, Fu W and Tang F: Systematic
characterization of full-length RNA isoforms in human colorectal
cancer at single-cell resolution. Protein Cell. 16:873–895. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tao Y, Li J, Pan J, Wang Q, Ke RW, Yuan D,
Wu H, Cao Y and Zhao L: Integration of scRNA-Seq and bulk RNA-Seq
identifies circadian rhythm disruption-related genes associated
with prognosis and drug resistance in colorectal cancer patients.
Immunotargets Ther. 14:475–489. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan L, Guo X, Zhang J, Wang Y, Wang J and
Li Y: Relationship between DHX15 expression and survival in
colorectal cancer. Rev Esp Enferm Dig. 115:234–240. 2023.PubMed/NCBI
|
|
19
|
Zhu H, Li M, Bi D, Yang H, Gao Y, Song F,
Zheng J, Xie R, Zhang Y, Liu H, et al: Fusobacterium nucleatum
promotes tumor progression in KRAS p.G12D-mutant colorectal cancer
by binding to DHX15. Nat Commun. 15:16882024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang SH and O'Sullivan B: Overview of the
8th edition TNM classification for head and neck cancer. Curr Treat
Options Oncol. 18:402017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rajput A, Martin ID, Rose R, Beko A, Levea
C, Sharratt E, Mazurchuk R, Hoffman RM, Brattain MG and Wang J:
Characterization of HCT116 human colon cancer cells in an
orthotopic model. J Surg Res. 147:276–281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Verhagen MP, Xu T, Stabile R, Joosten R,
Tucci FA, van Royen M, Trerotola M, Alberti S, Sacchetti A and
Fodde R: The SW480 cell line as a model of resident and migrating
colon cancer stem cells. iScience. 27:1106582024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sambuy Y, De Angelis I, Ranaldi G, Scarino
ML, Stammati A and Zucco F: The Caco-2 cell line as a model of the
intestinal barrier: Influence of cell and culture-related factors
on Caco-2 cell functional characteristics. Cell Biol Toxicol.
21:1–26. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dexter DL, Spremulli EN, Fligiel Z,
Barbosa JA, Vogel R, VanVoorhees A and Calabresi P: Heterogeneity
of cancer cells from a single human colon carcinoma. Am J Med.
71:949–956. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahmed D, Eide PW, Eilertsen IA, Danielsen
SA, Eknæs M, Hektoen M, Lind GE and Lothe RA: Epigenetic and
genetic features of 24 colon cancer cell lines. Oncogenesis.
2:e712013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saita K, Moriuchi Y, Iwagawa T, Aihara M,
Takai Y, Uchida K and Watanabe S: Roles of CSF2 as a modulator of
inflammation during retinal degeneration. Cytokine. 158:1559962022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Koso H, Yi H, Sheridan P, Miyano S, Ino Y,
Todo T and Watanabe S: Identification of RNA-binding protein LARP4B
as a tumor suppressor in glioma. Cancer Res. 76:2254–2264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsuruta L, Lee HJ, Masuda ES, Yokota T,
Arai N and Arai K: Regulation of expression of the IL-2 and IL-5
genes and the role of proteins related to nuclear factor of
activated T cells. J Allergy Clin Immunol. 96:1126–1135. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuribayashi H, Iwagawa T, Murakami A,
Kawamura T, Suzuki Y and Watanabe S: NMNAT1 is essential for human
iPS cell differentiation to the retinal lineage. Invest Ophthalmol
Vis Sci. 65:372024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kabiri Z, Greicius G, Zaribafzadeh H,
Hemmerich A, Counter CM and Virshup DM: Wnt signaling suppresses
MAPK-driven proliferation of intestinal stem cells. J Clin Invest.
128:3806–3812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiao L, Jiang M, Liu J, Wei L and Wu M:
Nuclear factor-kappa B activation inhibits proliferation and
promotes apoptosis of vascular smooth muscle cells. Vascular.
26:634–640. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Levine B and Kroemer G: Biological
functions of autophagy genes: A disease perspective. Cell.
176:11–42. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xie C, Liao H, Zhang C and Zhang S:
Overexpression and clinical relevance of the RNA helicase DHX15 in
hepatocellular carcinoma. Hum Pathol. 84:213–220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, He K, Sheng B, Lei X, Tao W, Zhu
X, Wei Z, Fu R, Wang A, Bai S, et al: The RNA helicase Dhx15
mediates Wnt-induced antimicrobial protein expression in Paneth
cells. Proc Natl Acad Sci USA. 118:e20174321182021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yao J, Cai Y, Chen Z, Wang X, Lai X, Pan
L, Li Y and Wang S: Dhx15 regulates zebrafish intestinal
development through the Wnt signaling pathway. Genomics.
115:1105782023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu F, Yu C, Li F, Zuo Y, Wang Y, Yao L, Wu
C, Wang C and Ye L: Wnt/β-catenin signaling in cancers and targeted
therapies. Signal Transduct Target Ther. 6:3072021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ge X and Wang X: Role of Wnt canonical
pathway in hematological malignancies. J Hematol Oncol. 3:332010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chang SH, Huang SW, Wang ST, Chung KC,
Hsieh CW, Kao JK, Chen YJ, Wu CY and Shieh JJ: Imiquimod-induced
autophagy is regulated by ER stress-mediated PKR activation in
cancer cells. J Dermatol Sci. 87:138–148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Russell RC and Guan KL: The multifaceted
role of autophagy in cancer. EMBO J. 41:e1100312022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jalali P, Shahmoradi A, Samii A,
Mazloomnejad R, Hatamnejad MR, Saeed A, Namdar A and Salehi Z: The
role of autophagy in cancer: From molecular mechanism to
therapeutic window. Front Immunol. 16:15282302025. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Niu X, You Q, Hou K, Tian Y, Wei P, Zhu Y,
Gao B, Ashrafizadeh M, Aref AR, Kalbasi A, et al: Autophagy in
cancer development, immune evasion, and drug resistance. Drug
Resist Updat. 78:1011702025. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alves S, Castro L, Fernandes MS, Francisco
R, Castro P, Priault M, Chaves SR, Moyer MP, Oliveira C, Seruca R,
et al: Colorectal cancer-related mutant KRAS alleles function as
positive regulators of autophagy. Oncotarget. 6:30787–30802. 2015.
View Article : Google Scholar : PubMed/NCBI
|