Introduction
Epidermal growth factor receptor (EGFR)
mutations are major oncogenic drivers in non-small cell lung cancer
(NSCLC). In the Chinese NSCLC population, ~28.2% of patients harbor
EGFR mutations, with their prevalence in lung adenocarcinoma
exceeding 50% (1). The most common
EGFR mutations, exon 19 deletion (19del) and the L858R point
mutation, account for ~85% of cases, while the remaining 15% are
rare, highly heterogeneous mutations occurring at low frequencies
(2). Rare mutations frequently
respond poorly to standard targeted therapies, and evidence guiding
treatment for patients with concurrent common and rare EGFR
mutations remains limited, posing notable clinical challenges.
Among the rare EGFR mutations, the L861Q
mutation accounts for 1–2% of cases (2). The L861Q mutation is typically
resistant to first-generation EGFR tyrosine kinase inhibitors
(TKIs) but demonstrates sensitivity to second- and third-generation
TKIs (3,4). Epidemiologically, EGFR
mutations are detected in 30–50% of Asian patients with NSCLC,
compared with 10–20% in Western populations (5,6). While
most EGFR alterations are classical sensitizing mutations
(19del and L858R, 80–90% of cases), uncommon mutations such as
L861Q, G719X and S768I comprise 10–15%, presenting unique
therapeutic challenges due to variable drug sensitivity (7,8).
Compound EGFR mutations, defined as the coexistence of two
or more alterations within the same tumor, are rare. Among these,
the coexistence of 19del with L861Q is notably uncommon, and its
clinical significance and optimal management strategies remain
poorly defined, to the best of our knowledge (9).
Almonertinib, a third-generation EGFR TKI, has
demonstrated promising efficacy, achieving a progression-free
survival (PFS) time of up to 19.3 months as first-line therapy in
advanced EGFR-mutant NSCLC (10). Several novel agents have emerged for
subsequent-line treatment, including antibody-drug conjugates
(ADCs), which combine monoclonal antibodies with cytotoxic agents
and have garnered considerable attention. In the OptiTROP-Lung03
study, the tumor-associated calcium signal transducer 2
(TROP2)-targeted ADC, sacituzumab tirumotecan, demonstrated notable
efficacy in patients resistant to EGFR TKIs, achieving a median PFS
time of 11.5 months (11). Despite
these advances, no standardized treatment exists for advanced NSCLC
harboring both common and rare EGFR mutations, leaving
clinicians without clear guidance.
According to the latest National Comprehensive
Cancer Network (NCCN) and Chinese Society of Clinical Oncology
(CSCO) guidelines (12,13), targeted therapy remains the
recommended first-line approach for EGFR-mutant NSCLC.
However, to the best of our knowledge, evidence for rare compound
mutations, such as 19del + L861Q, is limited. Furthermore, tissue
samples for mutation analysis are often insufficient in advanced or
metastatic disease (14). Plasma
circulating tumor DNA (ctDNA) has emerged as a non-invasive
alternative for molecular profiling, providing real-time insights
into tumor mutation status (15).
Dynamic ctDNA monitoring is increasingly recognized as a valuable
tool in evaluating treatment efficacy and guiding therapy
adjustments.
The present case report presents the treatment and
ctDNA-guided management of a patient with advanced metastatic lung
adenocarcinoma harboring both EGFR 19del and L861Q
mutations. It highlights the integration of ctDNA monitoring with
conventional diagnostic tools, including imaging to inform
individualized therapeutic strategies for complex
EGFR-mutant NSCLC.
Case report
A 63-year-old Chinese man, with no smoking history,
was admitted to Ruijin Hospital, Shanghai Jiao Tong University
School of Medicine| (Shanghai, China), in June 2023 after a routine
health examination detected a pulmonary lesion in the right lung.
PET-CT revealed suspected metastases in the mediastinal and
bilateral supraclavicular lymph nodes, right cardiophrenic angle
lymph nodes, pleura, bones and pancreatic head. Lung biopsy
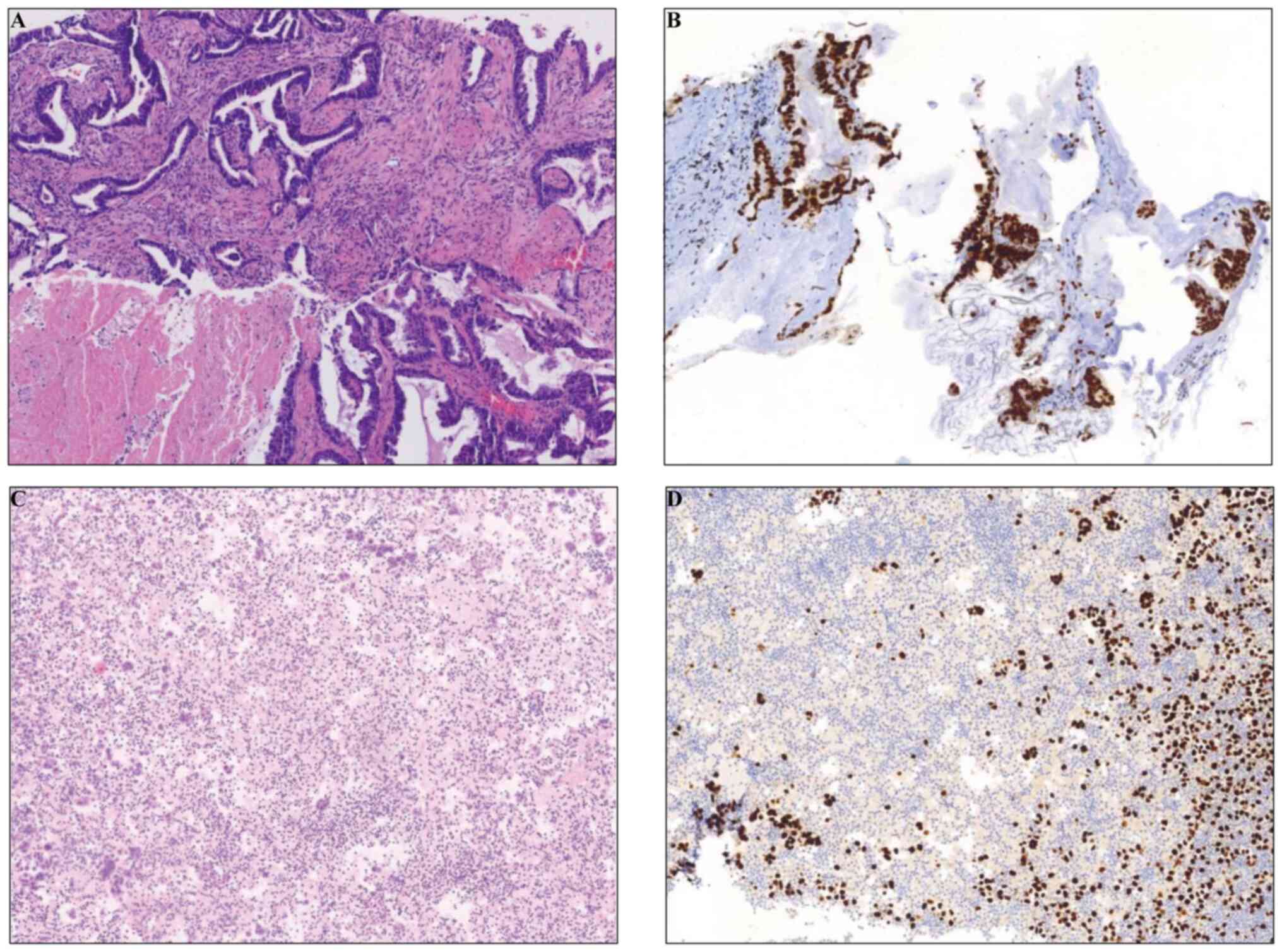
confirmed adenocarcinoma (Fig. 1A).
Immunohistochemical analysis demonstrated positive thyroid
transcription factor-1 (TTF-1) expression (Fig. 1B), using a standard HRP/DAB method
as described in Appendix S1. The
patient was staged as cT4N3M1c (stage IVb) according to the 8th
edition of the TNM classification for lung cancer (16). As recorded in the medical records,
initial tissue analysis identified an EGFR exon 21 L861Q
mutation using an amplification refractory mutation system PCR
assay (AmoyDX) and programmed death-ligand 1 tumor proportion score
<1% assessed by immunohistochemistry (IHC) using the 22C3
pharmDx kit (Agilent Technologies, Inc.), performed according to
the manufacturers' instructions. Repeat tissue-based
next-generation sequencing (NGS) was not performed due to the
invasive nature of re-biopsy. In line with the NCCN and CSCO NSCLC
guidelines (12,13), plasma-based NGS using ultra-deep
sequencing of a 2,365-gene panel was performed at Nanjing Geneseeq
Technology Inc. for ctDNA testing. Detailed procedures for
cell-free DNA extraction, library preparation, sequencing and
bioinformatics analysis are provided in Appendix S1. The analysis identified
EGFR 19del and L861Q as driver mutations (Table I). Variant allele frequency (VAF)
analysis indicated L861Q (30.53%) as the dominant clone compared
with exon 19del (0.45%).
 | Table I.Longitudinal monitoring of variant
allele frequencies of key genes in serial plasma ctDNA samples. |
Table I.
Longitudinal monitoring of variant
allele frequencies of key genes in serial plasma ctDNA samples.
|
|
|
Serial
monitoring timepoints |
|---|
|
|
|
|
|---|
| Gene | Variant | 2023.07 | 2023.08 | 2024.03 | 2024.04 | 2024.05 | 2024.06 | 2024.07 | 2024.08 | 2024.09 | 2024.10 | 2024.10 | 2024.11 | 2024.12 | 2025.02 |
|---|
| EGFR | p.L861Q | 30.53 | 0.39 | 1.11 | 2.85 | 6.93 | 1.11 | 0.69 | 15.62 | 1.69 | 3.96 | 4.59 | 0.29 | 0.00 | 0.00 |
| EGFR | p.E746_A750 del
(19del) | 0.45 | 0.00 | 0.34 | 1.37 | 0.93 | 2.30 | 29.81 | 1.33 | 9.53 | 15.32 | 6.34 | 1.97 | 1.66 | 1.83 |
| EGFR |
p.C797Sa | 0.00 | 0.00 | 0.09 | 0.08 | 0.07 | 0.30 | 0.34 | 0.25 | 0.68 | 1.01 | 0.80 | 0.28 | 0.83 | 0.12 |
| PIK3CA | p.R88Q | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.27 | 17.08 | 0.71 | 4.74 | 11.89 | 2.95 | 0.99 | 0.09 | 1.07 |
| TP53 | p.R337L | 0.39 | 0.00 | 0.11 | 0.42 | 0.37 | 1.13 | 23.39 | 0.93 | 5.97 | 13.88 | 3.79 | 1.00 | 0.78 | 1.05 |
| TP53 | p.S241C | 2.31 | 0.05 | 0.00 | 0.07 | 0.25 | 0.09 | 0.00 | 0.21 | 0.13 | 0.32 | 0.30 | 0.00 | 0.00 | 0.00 |
| RB1 | c.1498+2T>C | 2.19 | 0.00 | 0.00 | 0.00 | 0.42 | 0.00 | 0.00 | 0.00 | 0.00 | 0.16 | 0.35 | 0.00 | 0.00 | 0.00 |
Following multidisciplinary team (MDT) consultation,
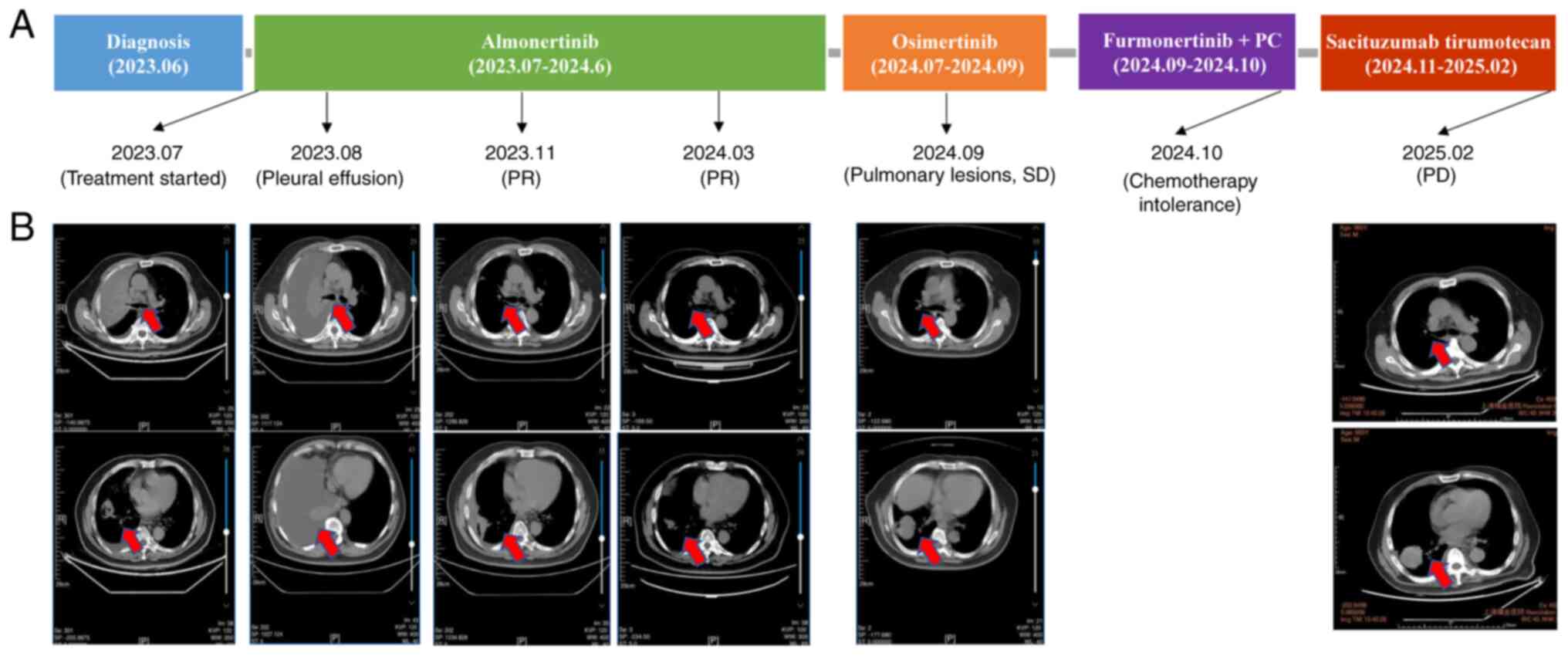
the patient initiated first-line almonertinib (165 mg, orally
administered once daily) in July 2023 (Fig. 2A). Bone metastases were managed
concurrently with denosumab (120 mg subcutaneously monthly). This
approach aligned with guideline recommendations from the NCCN and
CSCO (12,13), supporting the use of
third-generation EGFR TKIs, even for rare compound mutations.
Serial plasma ctDNA monitoring facilitated dynamic assessment of
clonal evolution and guided subsequent therapy adjustments,
highlighting the complementary role of liquid biopsy alongside
conventional imaging and tumor markers (17).
Subsequently, 1 month later, follow-up lung CT
revealed notable right pleural effusion (Fig. 2B). Pleural fluid drainage alleviated
symptoms and cytology confirmed malignancy (H&E staining and
TTF-1 IHC; Fig. 1C and D). Detailed
staining procedures are described in Appendix S1. The ctDNA equivalent,
expressed as haploid genome equivalents (hGE)/ml, quantitatively
reflects ctDNA levels and serves as a surrogate marker of tumor
burden (18). Reassessment of
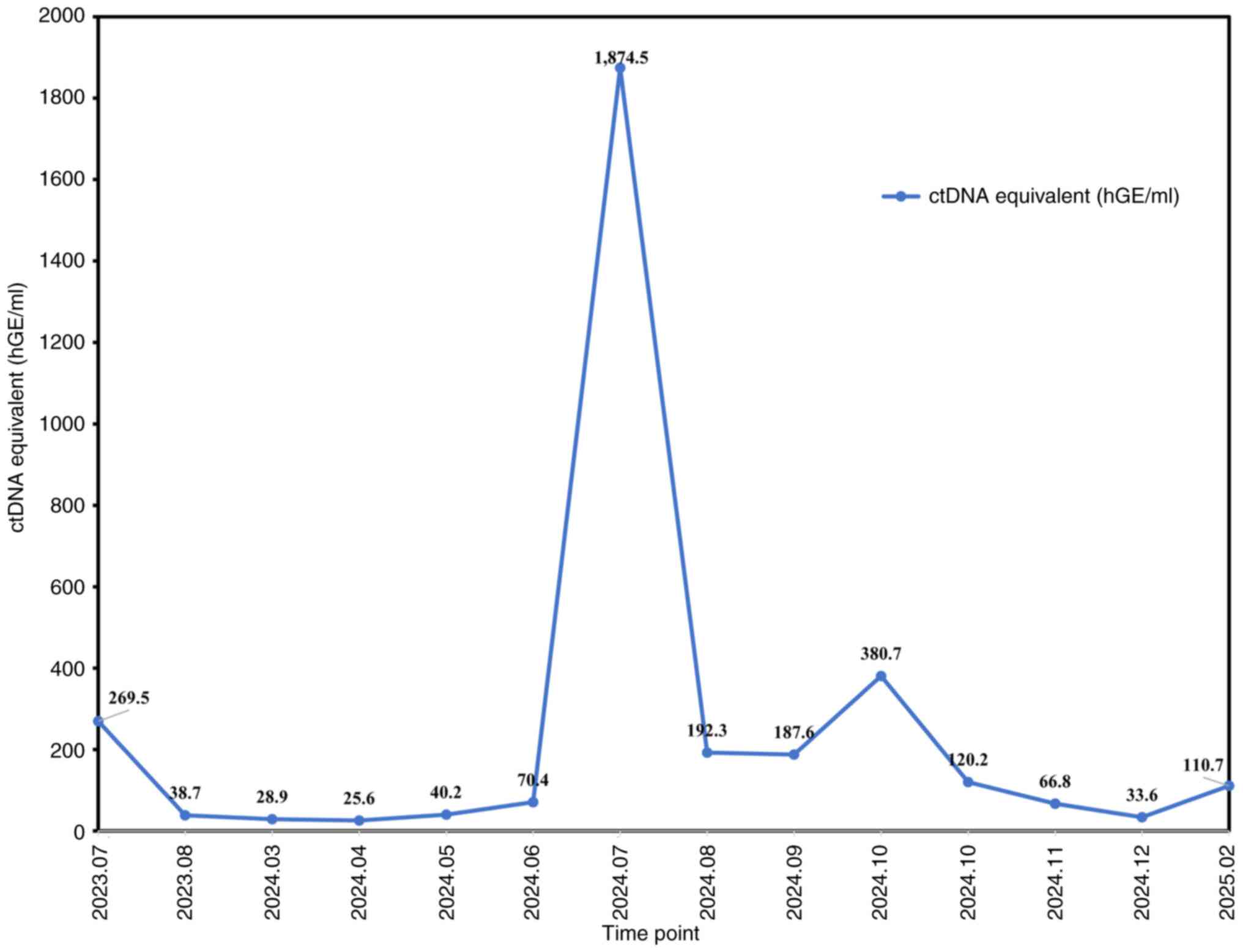
plasma ctDNA indicated a notable decline from 269.5 to 38.7 hGE/ml
(Fig. 3). Based on clinical
improvement and early molecular response, consistent with evidence
that rapid ctDNA decline is associated with treatment efficacy
(19,20), the MDT recommended continuation of
almonertinib.
In November 2023, imaging demonstrated a partial
response according to Response Evaluation Criteria in Solid Tumors
(RECIST) 1.1 criteria (21)
(Fig. 2B). In March 2024, ctDNA
remained low (28.9 hGE/ml; Fig. 3);
however, a low-abundance EGFR C797S mutation was detected by
ctDNA testing, with a VAF of 0.09% (Table I). An EGFR C797S mutation is
a known resistance mechanism to third-generation TKIs (22,23);
therefore, serial ctDNA surveillance was implemented to monitor
early progression.
In June 2024, ctDNA increased markedly to 70.4
hGE/ml (Fig. 3), with the
EGFR C797S allele fraction increasing to 0.30% (Table I), suggesting molecular progression
despite stable imaging. The patient achieved a PFS of 11 months on
almonertinib (Fig. 2A).
Subsequently, therapy was switched to osimertinib (80 mg,
administered orally once daily) as an individualized bridging
strategy, despite lateral third-generation TKI switching not being
the standard guideline practice (12,13).
In addition, in August 2024, ctDNA decreased from 1,874.5 hGE/ml to
192.3 hGE/ml (Fig. 3), but levels
did not continue to decline.
By September 2024, imaging revealed stable pulmonary
lesions (Fig. 2B) but progression
in bone lesions (pelvis, spine and femur) (Fig. S1A). Therefore, in September 2024,
the treatment regimen was modified to furmonertinib (80 mg,
administered orally once daily) combined with pemetrexed (800 mg)
and carboplatin (450 mg), both administered intravenously every 3
weeks (PC regimen), the standard of care following EGFR-TKI
resistance (13). ctDNA declined
from 380.7 to 120.2 hGE/ml; however, the patient discontinued
chemotherapy after 1 month due to intolerance. Following a third
MDT consultation, due to the chemotherapy intolerance of the
patient, disease progression, and the strong preference of the
patient and their family to avoid further cytotoxic therapy,
treatment with the TROP2-targeted ADC sacituzumab tirumotecan (300
mg, administered intravenously every 2 weeks) was initiated in
November 2024. Although the drug was not yet approved at the time
of administration, it subsequently received approval on March 10,
2025, by the National Medical Products Administration for adults
with locally advanced or metastatic EGFR-mutated NSCLC who
had progressed after EGFR-TKI and platinum-based chemotherapy
(24), which further supports its
use as an individualized option in the present case. Subsequently,
in December 2024, ctDNA decreased to 33.6 hGE/ml, indicating an
early molecular response.
In February 2025, plasma ctDNA monitoring detected a
slight ctDNA increase, and imaging indicated pulmonary lesion
shrinkage (Fig. 2B), whereas
earlier imaging in January 2025 revealed notable intracranial
progression (Fig. S1B). Central
nervous system-directed therapy was considered, in line with
European Association of Neuro-Oncology-European Society for Medical
Oncology and NCCN recommendations (12,25).
During plasma ctDNA monitoring, TP53 and RB1
mutations were identified, both of which have been reported to be
associated with unfavorable prognosis in lung cancer (26). Follow-up was conducted approximately
monthly during hospital visits for blood sampling, with the last
follow-up in February 2025. All mutations detected using plasma
ctDNA monitoring are summarized in Table SI.
Discussion
The present case highlighted the clinical complexity
of managing advanced NSCLC with a compound EGFR mutation
(19del + L861Q), a rare scenario with limited treatment evidence.
Co-occurring RB1 and TP53 mutations may partly
explain the shorter PFS of 11 months observed in this case,
compared with the median PFS of 19.3 months reported for
almonertinib in the AENEAS trial (10). This observation is consistent with
prior reports showing that baseline concurrent
TP53/RB1 alterations are associated with inferior PFS
and an increased risk of histologic transformation on first-line
third-generation EGFR TKIs (26,27).
In terms of prevalence, EGFR L861Q occurs in 1–2% of
patients with EGFR+ NSCLC, while compound
EGFR mutations account for 3–14% of EGFR+
cases (28). To the best of our
knowledge, Zhao et al (9)
reported the largest cohort of EGFR compound mutations to
date, including 1,025 patients, in which compound variants
comprised 12.7% of EGFR-mutated NSCLC. Within this cohort,
the 19del + L861Q combination was categorized as a ‘common + rare’
subtype and occurred less frequently compared with L858R combined
with uncommon variants. By contrast, common mutations such as 19del
or L858R alone represent 85–90% of EGFR-mutated NSCLC
(28). Clinically, patients with
rare or compound EGFR mutations typically exhibit reduced
sensitivity to first-generation TKIs and a shorter PFS compared
with patients with common mutations (9). Second- and third-generation TKIs may
still offer benefit in certain patients (28), as illustrated in the present case in
which almonertinib achieved an 11-month PFS with manageable
toxicity and sustained disease control. However, to the best of our
knowledge, clinical data on metastatic patterns and other features
of 19del + L861Q subtype remain limited due to its rarity. The
present case report adds to current evidence by documenting
longitudinal clinical outcomes and molecular monitoring in a
patient with this uncommon compound mutation. Although clinical
evidence for ctDNA-guided therapy in compound EGFR mutations
is limited, emerging evidence supports the utility of serial plasma
ctDNA monitoring to inform dynamic treatment decisions in such rare
cases (29).
In NSCLC tumors harboring compound EGFR
mutations, variants of unknown significance (VUSs) within the
kinase domain of EGFR may occur more frequently (9). The kinase domain, which mediates the
tyrosine kinase activity of EGFR, is the primary binding site for
TKIs. VUSs are genetic alterations whose functional or clinical
relevance has not yet been clearly established. When multiple
mutations coexist, certain VUSs may alter the three-dimensional
structure of the kinase domain, particularly the regions targeted
by TKIs. Such structural changes can reduce TKI binding efficiency,
potentially decreasing drug efficacy or promoting resistance
(9). Therefore, compound mutations
are not simply the sum of two or more known driver mutations; they
may also involve additional, previously uncharacterized variants
that influence drug binding and treatment response. This complexity
underscores the need for individualized monitoring and adaptive
treatment strategies, such as serial ctDNA analysis.
In the present case, early ctDNA dynamics provided
actionable insights. The detection of low-abundance EGFR
C797S mutation in March 2024 prompted increased surveillance and
therapy adjustments. Subsequent ADC therapy with sacituzumab
tirumotecan induced a rapid molecular response despite prior TKI
and chemotherapy exposure. The rationale for using an ADC in this
context lies in its ability to circumvent conventional TKI
resistance by delivering cytotoxic agents directly to tumor cells,
independent of EGFR mutation status. Beyond sacituzumab
tirumotecan, other ADCs, such as patritumab deruxtecan (HER3-DXd)
and datopotamab deruxtecan (Dato-DXd, a TROP2-directed ADC), are
under investigation for EGFR-mutant NSCLC post-TKI
resistance. Ongoing trials, including TROPION-Lung14
(ClinicalTrials.gov identifier: NCT06350097), TROPION-Lung15
(ClinicalTrials.gov identifier: NCT06417814) and HERTHENA-Lung02
(30) aim to further evaluate their
safety and efficacy.
Several previous studies have demonstrated the
clinical utility of ctDNA in advanced NSCLC. In the AURA trial,
plasma detection of T790M predicted outcomes with osimertinib
similar to tissue-based analysis (31). Similarly, the SWOG S1403 study
reported that early clearance of mutant EGFR ctDNA in plasma
markedly predicted PFS and overall survival, outperforming RECIST
in anticipating long-term benefit (32). Another previous study confirmed that
decreasing ctDNA levels during EGFR TKI therapy are associated with
longer survival (33). In addition
to its applications in advanced disease, ctDNA has also
demonstrated value in assessing minimal residual disease (MRD) in
early-stage NSCLC. The TRACERx study demonstrated that ctDNA
detected relapse earlier compared with imaging (34) and subsequent trials validated that
MRD positivity predicts recurrence risk (35,36).
Collectively, these findings highlighted the broad clinical utility
of ctDNA, from monitoring treatment response in advanced NSCLC to
guiding risk stratification in curative settings. The present case
further illustrated the potential of serial ctDNA surveillance to
inform clinical decisions, emphasizing its value as a non-invasive
biomarker to guide individualized therapy for rare compound
EGFR mutations. Combining ctDNA with imaging and tumor
markers may improve assessment in complex scenarios such as
pseudo-progression or discordant radiographic findings (37). As a single-patient report, the
present case findings require validation in larger cohorts to
establish the potential effectiveness of ctDNA-guided therapy in
compound EGFR-mutated NSCLC in the future.
In conclusion, the present case highlighted the
challenges of managing advanced NSCLC with rare EGFR
compound mutations. Dynamic plasma ctDNA monitoring complemented
conventional diagnostics, enabling real-time assessment of tumor
evolution and therapy response. Personalized therapy guided by
ctDNA, imaging and tumor markers facilitated timely therapy
adjustments, including ADC introduction after TKI and chemotherapy
resistance. With ongoing research and emerging treatments,
integrating ctDNA into routine clinical practice may potentially
improve outcomes for patients with complex EGFR-mutant NSCLC
in the future.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present case report was supported by Wuxi Taihu Talent
Plan-High-level Medical Talents Project (grant no. RJWX-001).
Availability of data and materials
The sequencing data generated in the present study
are deposited in the Genome Sequence Archive for Human (GSA-Human)
under accession number HRA013443 or at the following URL:
https://ngdc.cncb.ac.cn/gsa-human/s/0ee9t310. The
other data generated in the present study may be requested from the
corresponding author.
Authors' contributions
JZ conceived and designed the present case report,
and supervised the study. XBZ, YS and XC collected clinical data
and performed patient follow-ups. CL contributed to data
interpretation and critically revised the manuscript. RC, XWZ and
WZ contributed to data analysis and interpretation, and assisted
with manuscript revision. AS contributed to study design, data
interpretation and manuscript editing. XBZ, YS, CL, RC, WZ, XWZ,
AS, and JZ drafted and critically revised the manuscript. XBZ and
JZ confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Written informed consent was obtained from the
patient for the publication of the present case report and any
accompanying images.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ADC
|
antibody-drug conjugate
|
|
CSCO
|
Chinese Society of Clinical
Oncology
|
|
ctDNA
|
circulating tumor DNA
|
|
EGFR
|
epidermal growth factor receptor
|
|
NSCLC
|
non-small cell lung cancer
|
|
MRD
|
minimal residual disease
|
|
NCCN
|
National Comprehensive Cancer
Network
|
|
NGS
|
next-generation sequencing
|
|
PFS
|
progression-free survival
|
|
RECIST
|
Response Evaluation Criteria in Solid
Tumors
|
|
TKI
|
tyrosine kinase inhibitor
|
|
TROP2
|
tumor-associated calcium signal
transducer 2
|
|
TTF-1
|
thyroid transcription factor-1
|
|
VUS
|
variant of unknown significance
|
|
19del
|
exon 19 deletion
|
References
|
1
|
Chen P, Liu Y, Wen Y and Zhou C: Non-small
cell lung cancer in China. Cancer Commun (Lond). 42:937–970. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fukuda A and Okuma Y: From rarity to
reality: Osimertinib's promising horizon in treating uncommon EGFR
mutations in Non-Small cell lung cancer. Clin Cancer Res.
30:3128–3136. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okuma Y, Kubota K, Shimokawa M, Hashimoto
K, Kawashima Y, Sakamoto T, Wakui H, Murakami S, Okishio K,
Hayashihara K, et al: First-line osimertinib for previously
untreated patients with NSCLC and uncommon EGFR mutations: The
UNICORN phase 2 nonrandomized clinical trial. JAMA Oncol. 10:43–51.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang JC, Sequist LV, Geater SL, Tsai CM,
Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SH, Zhou C, et al:
Clinical activity of afatinib in patients with advanced
non-small-cell lung cancer harbouring uncommon EGFR mutations: A
combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung
6. Lancet Oncol. 16:830–838. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Miao J, Wang Y, Xing W, Xu X and
Wu R: Comparison of the efficacy, safety of first-line treatments
for of advanced EGFR mutation-positive non-small-cell lung cancer
in Asian populations: A systematic review and network
meta-analysis. Front Pharmacol. 14:12123132023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hofman P, Calabrese F, Kern I, Adam J,
Alarcão A, Alborelli I, Anton NT, Arndt A, Avdalyan A, Barberis M,
et al: Real-world EGFR testing practices for non-small-cell lung
cancer by thoracic pathology laboratories across Europe. ESMO Open.
8:1016282023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Borgeaud M, Parikh K, Banna GL, Kim F,
Olivier T, Le X and Addeo A: Unveiling the landscape of uncommon
EGFR mutations in NSCLC-A systematic review. J Thorac Oncol.
19:973–983. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu Y, Yao F, Wang J, Huang Z, Hu T, Zhu C
and Lu S: An in-depth exploration of four heterogeneity
structure-based EGFR mutation subgroups in Chinese non-small cell
lung cancer. BMC Pulm Med. 25:3162025. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao W, Song A, Xu Y, Wu Q, Liu C, Yin JC,
Ou Q, Wu X, Shao Y and Zhao X: Rare mutation-dominant compound
EGFR-positive NSCLC is associated with enriched kinase
domain-resided variants of uncertain significance and poor clinical
outcomes. BMC Med. 21:732023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu S, Dong X, Jian H, Chen J, Chen G, Sun
Y, Ji Y, Wang Z, Shi J, Lu J, et al: AENEAS: A randomized phase III
trial of aumolertinib versus gefitinib as first-line therapy for
locally advanced or metastatic non-small-cell lung cancer with EGFR
Exon 19 deletion or L858R mutations. J Clin Oncol. 40:3162–3171.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang W, Cheng Y, Chen Z, Wang W, Li Y, Yin
Y, Li X, Xu X, Yu G, Mi Y, et al: Abstract CT247: Updated efficacy
and safety of anti-TROP2 ADC SKB264 (MK-2870) for previously
treated advanced NSCLC in Phase 2 study. Cancer Res. 84:CT2472024.
View Article : Google Scholar
|
|
12
|
Riely GJ, Wood DE, Ettinger DS, Aisner DL,
Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, et
al: Non-small cell lung cancer, version 4.2024, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
22:249–274. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang J, Qiu T and Ren S: Updates to the
2024 CSCO advanced non-small cell lung cancer guidelines. Cancer
Biol Med. 22:77–82. 2025.PubMed/NCBI
|
|
14
|
Brice K, Arline C, Raez LE, Dumais K and
Block M: Molecular profiling using Next-generation sequencing of
sufficient endobronchial ultrasound-guided transbronchial needle
aspiration and liquid biopsy samples in patients with advanced lung
cancer. J Clin Transl Pathol. 4:110–114. 2024.
|
|
15
|
Zhang YC, Zhou Q and Wu YL: The emerging
roles of NGS-based liquid biopsy in non-small cell lung cancer. J
Hematol Oncol. 10:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (Eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moding EJ, Nabet BY, Alizadeh AA and Diehn
M: Detecting liquid remnants of solid tumors: Circulating tumor DNA
minimal residual disease. Cancer Discov. 11:2968–2986. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chaudhuri AA, Chabon JJ, Lovejoy AF,
Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL,
Zhou L, et al: Early detection of molecular residual disease in
localized lung cancer by circulating tumor DNA profiling. Cancer
Discov. 7:1394–1403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng ML, Lau CJ, Milan MSD, Supplee JG,
Riess JW, Bradbury PA, Jänne PA, Oxnard GR and Paweletz CP: Plasma
ctDNA response is an early marker of treatment effect in advanced
NSCLC. JCO Precis Oncol. 5:PO.20.00419, 2021.
|
|
20
|
Leite da Silva LF, Saldanha EF, de Menezes
JSA, Halamy Pereira L, de Bragança Dos Santos JAR, Buonopane IR, de
Souza EM, de Menezes CUG and Lopes G: Plasma ctDNA kinetics as a
predictor of systemic therapy response for advanced non-small cell
lung cancer: A systematic review and meta-analysis. Oncologist.
30:oyae3442025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi K, Wang G, Pei J, Zhang J, Wang J,
Ouyang L, Wang Y and Li W: Emerging strategies to overcome
resistance to third-generation EGFR inhibitors. J Hematol Oncol.
15:942022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He J, Huang Z, Han L, Gong Y and Xie C:
Mechanisms and management of 3rd-generation EGFR-TKI resistance in
advanced non-small cell lung cancer (review). Int J Oncol.
59:902021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sichuan Kelun-Biotech Biopharmaceutical
Co.Ltd, . Kelun-Biotech's TROP2 ADC Sacituzumab Tirumotecan
(sac-TMT) Approved for Marketing in Second Indication by NMPA for
EGFRm NSCLC. Mar 10–2025.
|
|
25
|
Le Rhun E, Guckenberger M, Smits M, Dummer
R, Bachelot T, Sahm F, Galldiks N, de Azambuja E, Berghoff AS,
Metellus P, et al: EANO-ESMO Clinical Practice Guidelines for
diagnosis, treatment and follow-up of patients with brain
metastasis from solid tumours. Ann Oncol. 32:1332–1347. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Choudhury NJ, Marra A, Sui JSY, Flynn J,
Yang SR, Falcon CJ, Selenica P, Schoenfeld AJ, Rekhtman N, Gomez D,
et al: Molecular biomarkers of disease outcomes and mechanisms of
acquired resistance to First-Line osimertinib in advanced
EGFR-mutant lung cancers. J Thorac Oncol. 18:463–475. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Offin M, Chan JM, Tenet M, Rizvi HA, Shen
R, Riely GJ, Rekhtman N, Daneshbod Y, Quintanal-Villalonga A,
Penson A, et al: Concurrent RB1 and TP53 alterations define a
subset of EGFR-mutant lung cancers at risk for histologic
transformation and inferior clinical outcomes. J Thorac Oncol.
14:1784–1793. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang T, Wan B, Zhao Y, Li C, Liu H, Lv T,
Zhan P and Song Y: Treatment of uncommon EGFR mutations in
non-small cell lung cancer: New evidence and treatment. Transl Lung
Cancer Res. 8:302–316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duffy MJ: Circulating tumor DNA (ctDNA) as
a biomarker for lung cancer: Early detection, monitoring and
therapy prediction. Tumour Biol. 46 (Suppl 1):S283–S295. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mok T, Janne PA, Nishio M, Novello S, Reck
M, Steuer C, Wu YL, Fougeray R, Fan PD, Meng J, et al:
HERTHENA-Lung02: Phase III study of patritumab deruxtecan in
advanced EGFR-mutated NSCLC after a third-generation EGFR TKI.
Future Oncol. 20:969–980. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oxnard GR, Thress KS, Alden RS, Lawrance
R, Paweletz CP, Cantarini M, Yang JC, Barrett JC and Jänne PA:
Association between plasma genotyping and outcomes of treatment
with osimertinib (AZD9291) in advanced Non-small-cell lung cancer.
J Clin Oncol. 34:3375–3382. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mack PC, Miao J, Redman MW, Moon J,
Goldberg SB, Herbst RS, Melnick MA, Walther Z, Hirsch FR, Politi K,
et al: Circulating tumor DNA kinetics predict Progression-free and
overall survival in EGFR TKI-Treated patients with EGFR-Mutant
NSCLC (SWOG S1403). Clin Cancer Res. 28:3752–3760. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Andrews HS, Zariffa N, Nishimura KK, Choi
SH, Deng S, Eisele M, Espenschied CR, Goren EM, Guha M, Hong S, et
al: ctDNA clearance as an early indicator of improved clinical
outcomes in advanced NSCLC treated with TKI: Findings from an
aggregate analysis of eight clinical trials. Clin Cancer Res.
31:2162–2172. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Abbosh C, Birkbak NJ, Wilson GA,
Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA,
Veeriah S, Rosenthal R, et al: Phylogenetic ctDNA analysis depicts
early-stage lung cancer evolution. Nature. 545:446–451. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hong TH, Hwang S, Dasgupta A, Abbosh C,
Hung T, Bredno J, Walker J, Shi X, Milenkova T, Horn L, et al:
Clinical utility of Tumor-naive presurgical circulating tumor DNA
detection in Early-stage NSCLC. J Thorac Oncol. 19:1512–1524. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhong R, Gao R, Fu W, Li C, Huo Z, Gao Y,
Lu Y, Li F, Ge F, Tu H, et al: Accuracy of minimal residual disease
detection by circulating tumor DNA profiling in lung cancer: A
meta-analysis. BMC Med. 21:1802023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee JH, Long GV, Menzies AM, Lo S,
Guminski A, Whitbourne K, Peranec M, Scolyer R, Kefford RF, Rizos H
and Carlino MS: Association between circulating tumor DNA and
pseudoprogression in patients with metastatic melanoma treated with
Anti-programmed cell death 1 antibodies. JAMA Oncol. 4:717–721.
2018. View Article : Google Scholar : PubMed/NCBI
|