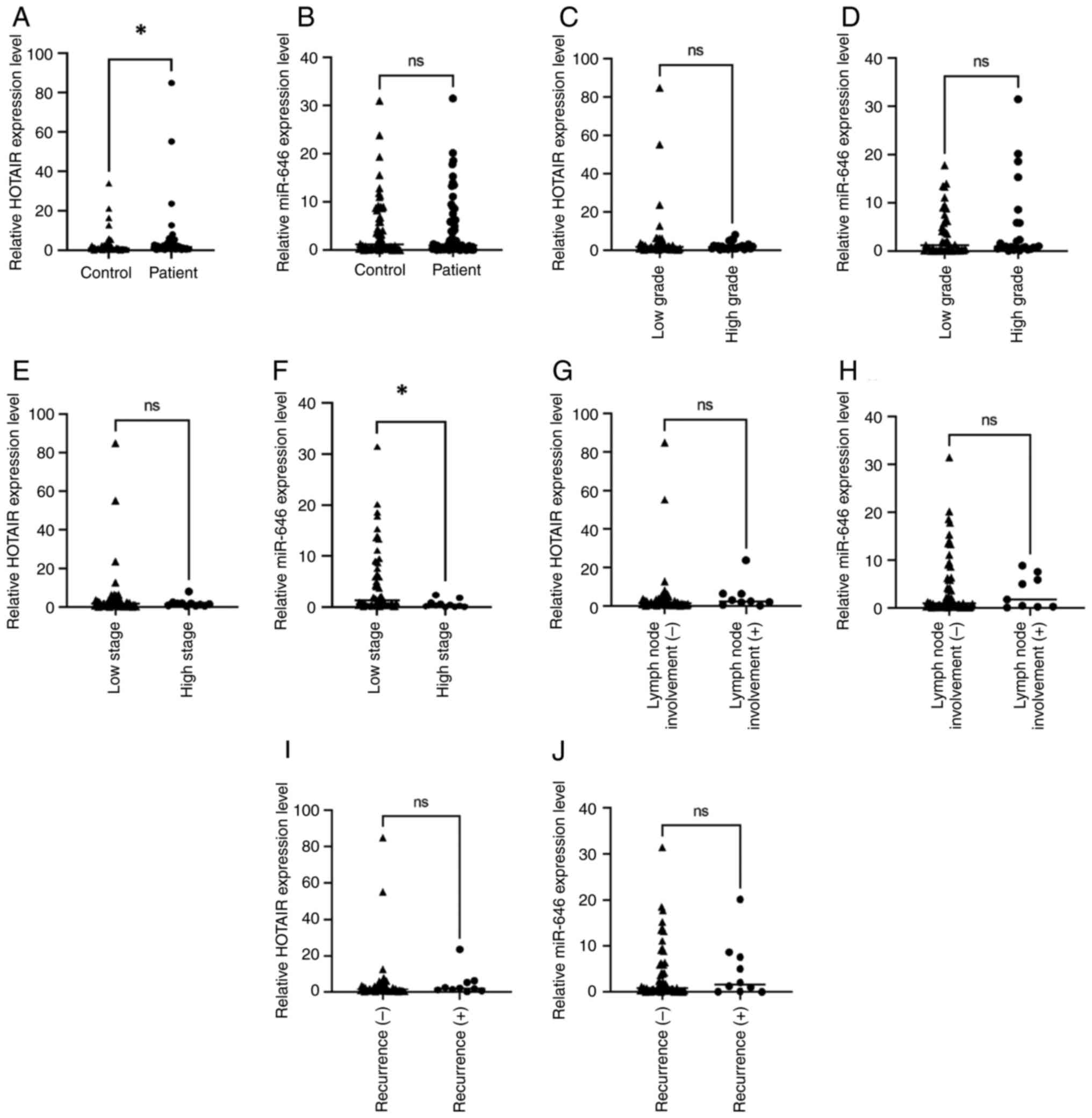
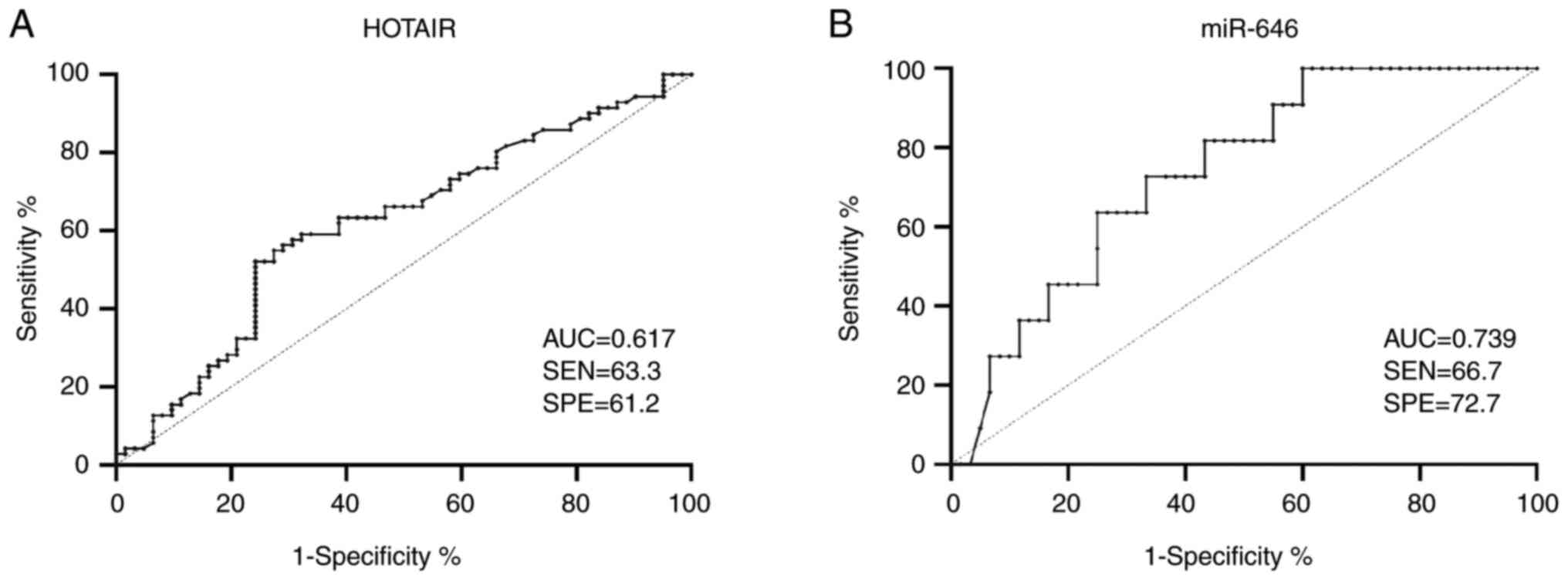
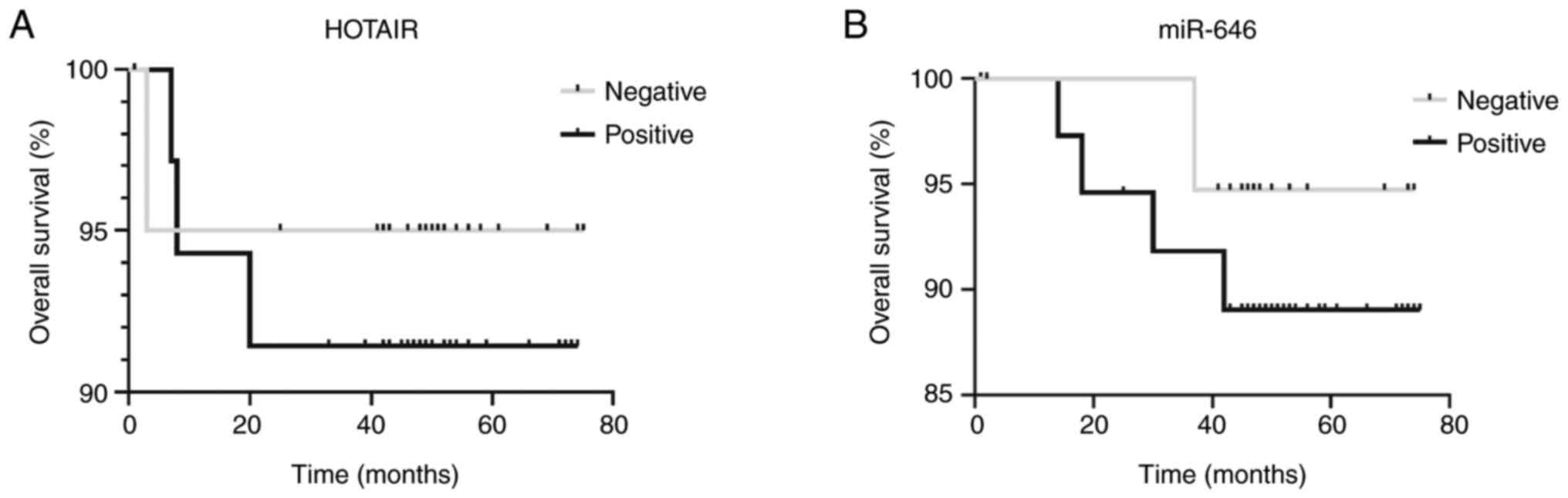
|
1
|
Baker-Rand H and Kitson SJ: Recent
advances in endometrial cancer prevention, early diagnosis and
treatment. Cancers (Basel). 16:10282024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zaino RJ: Introduction to endometrial
cancer. Molecular Pathology of Gynecologic Cancer. Giordano A,
Bovicelli A and Kurman RJ: Humana Press; Totowa, NJ: pp. 51–72.
2007, View Article : Google Scholar
|
|
3
|
Patni R: Current concepts in endometrial
cancer. Endometrial Carcinoma: Evolution and Overview. Patni R:
Springer; Singapore: pp. 1–10. 2017
|
|
4
|
Pokharna S: Epidemiology and prevention of
endometrial carcinoma. Current Concepts in Endometrial Cancer.
Patni R: Springer; Singapore: pp. 11–18. 2017, View Article : Google Scholar
|
|
5
|
Bokhman JV: Two pathogenetic types of
endometrial carcinoma. Gynecol Oncol. 15:10–17. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilczyński M, Danielska J and Wilczyński
JR: An update of the classical Bokhman's dualistic model of
endometrial cancer. Menopause Rev. 15:63–68. 2016. View Article : Google Scholar
|
|
7
|
Collins FS and Mansoura MK: The human
genome project. Cancer. 91:221–225. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu M, Fu P, Qu L, Liu J and Lin A: Long
noncoding RNAs: New critical regulators in cancer immunity. Front
Oncol. 10:5509872020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15(Spec No 1): R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhattacharjee R, Prabhakar N, Kumar L,
Bhattacharjee A, Kar S, Malik S, Kumar D, Ruokolainen J, Negi A,
Jha NK and Kesari KK: Crosstalk between long noncoding RNA and
microRNA in cancer. Cell Oncol (Dordr). 46:885–908. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slack FJ and Chinnaiyan AM: The role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Wang W, Zhu W, Dong J, Cheng Y,
Yin Z and Shen F: Mechanisms and functions of long non-coding RNAs
at multiple regulatory levels. Int J Mol Sci. 20:55732019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hajjari M and Salavaty A: HOTAIR: An
oncogenic long non-coding RNA in different cancers. Cancer Biol
Med. 12:1–9. 2015.PubMed/NCBI
|
|
17
|
Rajagopal T, Talluri S, Akshaya RL and
Dunna NR: HOTAIR lncRNA: A novel oncogenic propellant in human
cancer. Clin Chim Acta. 503:1–18. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu X, Alsager S, Zhuo Y and Shan B: HOX
transcript antisense RNA (HOTAIR) in cancer. Cancer Lett.
454:90–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F,
Yu W, Wang X, Zhang L, Yu J and Hao X: Long noncoding RNA HOTAIR
involvement in cancer. Tumour Biol. 35:9531–9538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mozdarani H, Ezzatizadeh V and Rahbar
Parvaneh R: The emerging role of the long non-coding RNA HOTAIR in
breast cancer development and treatment. J Transl Med. 18:1522020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vallone C, Rigon G, Gulia C, Baffa A,
Votino R, Morosetti G, Zaami S, Briganti V, Catania F, Gaffi M, et
al: Coding RNAs and endometrial cancer. Genes (Basel). 9:1872018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He X, Bao W, Li X, Chen Z, Che Q, Wang H
and Wan XP: The long non-coding RNA HOTAIR is upregulated in
endometrial carcinoma and correlates with poor prognosis. Int J Mol
Med. 33:325–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Romano G, Veneziano D, Acunzo M and Croce
C: Small non-coding RNA and cancer. Carcinogenesis. 38:485–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Fu LL, Wen X, Liu B, Huang J, Wang
JH and Wei YQ: Oncogenic and tumor suppressive roles of microRNAs
in apoptosis and autophagy. Apoptosis. 19:1177–1189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng Y and Croce CM: The role of microRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan Y, Chen Y, Ma D, Ji Z, Cao F, Chen Z,
Ning Y and Bai C: miR-646 is a key negative regulator of EGFR
pathway in lung cancer. Exp Lung Res. 42:286–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zang Y, Li J, Wan B and Tai Y: circRNA
circ-CCND1 promotes the proliferation of laryngeal squamous cell
carcinoma via elevating CCND1 expression by interacting with HuR
and miR-646. J Cell Mol Med. 24:2423–2433. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li W, Liu M, Feng Y, Xu YF, Huang YF, Che
JP, Wang GC, Yao XD and Zheng JH: Downregulated miR-646 in clear
cell renal carcinoma correlated with tumour metastasis by targeting
the nin one binding protein (NOB1). Br J Cancer. 111:1188–1200.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Wu J, Li Y, Jiang Y, Wang L, Chen
Y, Lv Y, Zou Y and Ding X: Circ_0000527 promotes the progression of
retinoblastoma by regulating miR-646/LRP6 axis. Cancer Cell Int.
20:3012020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang L, Liu G, Xiao S, Wang L, Liu X, Tan
Q and Li Z: Long noncoding MT1JP enhanced the inhibitory effects of
miR-646 on FGF2 in osteosarcoma. Cancer Biother Radiopharm.
35:371–376. 2020.PubMed/NCBI
|
|
31
|
Zhang P, Tang WM, Zhang H, Li YQ, Peng Y,
Wang J, Liu GN, Huang XT, Zhao JJ, Li G, et al: miR-646 inhibits
cell proliferation and EMT-induced metastasis by targeting FOXK1 in
gastric cancer. Br J Cancer. 117:525–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Chen S, Zong ZH, Guan X and Zhao Y:
CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the
development of endometrial cancer. J Cell Mol Med. 24:6898–6907.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou YX, Wang C, Mao LW, Wang YL, Xia LQ,
Zhao W, Shen J and Chen J: Long noncoding RNA HOTAIR mediates the
estrogen-induced metastasis of endometrial cancer cells via the
miR-646/NPM1 axis. Am J Physiol Cell Physiol. 314:C690–C701. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang WT, Zhao YN, Yan JX, Weng MY, Wang Y,
Chen YQ and Hong SJ: Differentially expressed microRNAs in the
serum of cervical squamous cell carcinoma patients before and after
surgery. J Hematol Oncol. 7:62014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Murali R, Davidson B, Fadare O, Carlson
JA, Crum CP, Gilks CB, Irving JA, Malpica A, Matias-Guiu X,
McCluggage WG, et al: High-grade endometrial carcinomas:
Morphologic and immunohistochemical features, diagnostic challenges
and recommendations. Int J Gynecol Pathol. 38:40–63. 2019.
View Article : Google Scholar
|
|
36
|
Zhang C and Zheng W: High-grade
endometrial carcinomas: Morphologic spectrum and molecular
classification. Semin Diagn Pathol. 39:176–186. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oliva E and Soslow RA: High-grade
endometrial carcinomas. Surg Pathol Clin. 4:199–241. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Concin N, Matias-Guiu X, Vergote I, Cibula
D, Mirza MR, Marnitz S, Lederman S, Bosse T, Chargari C, Fagotti A,
et al: ESGO/ESTRO/ESP guidelines for the management of patients
with endometrial carcinoma. Int J Gynecol Cancer. 31:12–39. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
NCCN Clinical Practice Guidelines in
Oncology (NCCN Guidelines®), . Uterine Neoplasms. Version 3.2025.
National Comprehensive Cancer Network; Plymouth Meeting, PA:
2025
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mohamad N, Khedr AMB, Shaker OG and Hassan
M: Expression of long noncoding RNA, HOTAIR, and MicroRNA-205 and
their relation to transforming growth factor β1 in patients with
alopecia Areata. Skin Appendage Disord. 9:111–120. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang B, Sun Q, Ye W, Li L and Jin P: Long
non-coding RNA CDKN2B-AS1 enhances LPS-induced apoptotic and
inflammatory damages in human lung epithelial cells via regulating
the miR-140-5p/TGFBR2/Smad3 signal network. BMC Pulm Med.
21:2002021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
İlhan B, Kuraş S, Kılıç B, Tilgen
Yasasever C, Oğuz Soydinç H, Alsaadoni H, Öztan G, Adamnejad
Ghafour A, Ucuncu M, Kunduz E and Bademler S: Exploratory analysis
of circulating serum miR-197-3p, miR-1236, and miR-1271 expression
in early breast cancer. Int J Mol Sci. 26:89442025. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Melton C, Reuter JA, Spacek DV and Snyder
M: Recurrent somatic mutations in regulatory regions of human
cancer genomes. Nat Genet. 47:710–716. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Egranova SD, Hua Q, Lina C and Yanga L:
lncRNAs as tumor cell intrinsic factors that affect cancer
immunotherapy. RNA Biol. 17:1625–1627. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kansara S, Pandey V, Lobie PE, Sethi G,
Garg M and Pandey AK: Mechanistic involvement of long noncoding
RNAs in oncotherapeutics resistance in triple-negative breast
cancer. Cells. 9:15112020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Basak P, Chatterjee S, Bhat V, Su A, Jin
H, Lee-Wing V, Liu Q, Hu P, Murphy LC and Raouf A: Long non-coding
RNA H19 acts as an estrogen receptor modulator that is required for
endocrine therapy resistance in ER+ breast cancer cells. Cell
Physiol Biochem. 51:1518–1532. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Whitfield ML, George LK, Grant GD and
Perou CM: Common markers of proliferation. Nat Rev Cancer.
6:99–106. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ding L, Wang R, Shen D, Cheng S, Wang H,
Lu Z, Zheng Q, Wang L, Xia L and Li G: Role of noncoding RNA in
drug resistance of prostate cancer. Cell Death Dis. 12:5902021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen N, Li Y and Li X: Dynamic role of
long noncoding RNA in liver diseases: Pathogenesis and diagnostic
aspects. Clin Exp Med. 25:1602025. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lin Y, Zhao W, Pu R, Lv Z, Xie H, Li Y and
Zhang Z: Long non coding RNAs as diagnostic and prognostic
biomarkers for colorectal cancer (Review). Oncol Lett. 28:4862024.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li S, Zhao Y and Chen X: Microarray
expression profile analysis of circular RNAs and their potential
regulatory role in bladder carcinoma. Oncol Rep. 45:239–253. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kawai M, Fukuda A, Otomo R, Obata S,
Minaga K, Asada M, Umemura A, Uenoyama Y, Hieda N, Morita T, et al:
Early detection of pancreatic cancer by comprehensive serum miRNA
sequencing with automated machine learning. Br J Cancer.
131:1158–1168. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wong NK, Huang CL, Islam R and Yip SP:
Long non-coding RNAs in hematological malignancies: Translating
basic techniques into diagnostic and therapeutic strategies. J
Hematol Oncol. 11:1312018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ludwig N, Leidinger P, Becker K, Backes C,
Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E
and Keller A: Distribution of miRNA expression across human
tissues. Nucleic Acids Res. 44:3865–3877. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Valihrach L, Androvic P and Kubista M:
Circulating miRNA analysis for cancer diagnostics and therapy. Mol
Aspects Med. 72:1008262020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chung YW, Bae HS, Song JY, Lee JK, Lee NW,
Kim T and Lee KW: Detection of microRNA as novel biomarkers of
epithelial ovarian cancer from the serum of ovarian cancer
patients. Int J Gynecol Cancer. 23:673–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Iorio MV and Croce CM: microRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Donkers H, Bekkers R and Galaal K:
Diagnostic value of microRNA panel in endometrial cancer: A
systematic review. Oncotarget. 11:2010–2023. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ayers D: Long non-coding RNAs: Novel
emergent biomarkers for cancer diagnostics. J Cancer Res Treat.
1:31–35. 2013.
|
|
65
|
Akman HB and Bensan AE: Noncoding RNAs and
cancer. Turk J Biol. 38:817–828. 2014. View Article : Google Scholar
|
|
66
|
Beckedorff FC, Amaral MS,
Deocesano-Pereira C and Verjovski-Almeida S: Long noncoding RNAs
and their implications in cancer epigenetics. Biosci Rep.
33:701–713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bhan A, Hussain I, Ansari KI, Kasiri S,
Bashyal A and Mandal SS: Antisense transcript long noncoding RNA
(lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol
Biol. 425:3707–3722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang J, Ke P, Guo L, Wang W, Tan H, Liang
Y and Yao S: Lentivirus-mediated RNA interference targeting the
long noncoding RNA HOTAIR inhibits proliferation and invasion of
endometrial carcinoma cells in vitro and in vivo. Int J Gynecol
Cancer. 24:635–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhou Y, Zhang Y, Shao Y, Yue X, Chu Y,
Yang C and Chen D: LncRNA HOTAIR down-expression inhibits the
invasion and tumorigenicity of epithelial ovarian cancer cells by
suppressing TGF-β1 and ZEB1. Discov Oncol. 14:2282023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sun MY, Zhu JY, Zhang CY, Zhang M, Song
YN, Rahman K, Zhang LJ and Zhang H: Autophagy regulated by lncRNA
HOTAIR contributes to the cisplatin-induced resistance in
endometrial cancer cells. Biotechnol Lett. 39:1477–1484. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Çorbacıoğlu ŞK and Aksel G: Receiver
operating characteristic curve analysis in diagnostic accuracy
studies: A guide to interpreting the area under the curve value.
Turk J Emerg Med. 23:195–198. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Roest HP, IJzermans JNM and van der Laan
LJW: Evaluation of RNA isolation methods for microRNA
quantification in a range of clinical biofluids. BMC Biotechnol.
21:482021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Aerts JL, Gonzales MI and Topalian SL:
Selection of appropriate control genes to assess expression of
tumor antigens using real-time RT-PCR. BioTechniques. 36:84–86.
8890–101. 2004. View Article : Google Scholar : PubMed/NCBI
|