Introduction
Endometrial cancer (EC) is one of the most common
malignancies in women, ranking fourth in high-income countries and
sixth globally, with ~417,000 new cases diagnosed worldwide in
2020. Its incidence has increased by >130% over the past three
decades, with ~97,370 mortalities reported worldwide in 2020
(1). Risk factors for EC include
obesity, a high glycaemic index diet, early menarche, late
menopause, advanced age, diabetes, the use of oestrogen,
nulliparity, polycystic ovary syndrome and a sedentary lifestyle
(2–4). Prolonged exposure to unopposed
oestrogen, particularly through hormone replacement therapy without
concomitant progestin, increases the risk of developing EC by
4.5-8.0-fold (2). The pathogenesis
of EC is categorised into two primary subtypes in a study by
Bokhman (5). Type I is associated
with oestrogen and offers a more favourable prognosis, whereas type
II represents a high-grade variant with a poorer prognosis,
typically observed in older individuals (aged 55–60 years)
(6).
According to the Human Genome Project, >90% of
the genome is considered to be transcribed (7). Research indicates that only 2% of
these synthesised transcripts can encode proteins, while >75%
consists of non-protein-coding RNAs (8). Non-coding RNAs (ncRNAs) are classified
into two categories based on their size, namely long ncRNAs
(lncRNAs) and small ncRNAs. These categories can be further divided
based on size, cellular localisation or function (9). In particular, lncRNAs and microRNAs
(miRs) serve crucial roles in epigenetic regulation, control of
gene expression levels and the regulation of cellular processes
including proliferation, apoptosis, migration, invasion and the
epithelial-mesenchymal transition (9–13).
Previous studies demonstrate an association between the expression
of ncRNA and various pathological conditions, including
neurological, cardiological and cancer-related diseases (12–14).
lncRNAs are generally longer than 200 nucleotides.
They are considered to contribute to carcinogenesis by binding to
transcription factors or repressors, acting as signalling molecules
to activate or inhibit the expression of genes and engaging with
protein complexes and cofactors (13). Moreover, lncRNAs also serve a role
in post-transcriptional regulation by participating in mRNA
processing. Additionally, lncRNAs function as sponges, preventing
miR from binding to their target mRNA (15).
The study by Rinn et al (14) was the first to report and
characterize Hox antisense intergenic RNA (HOTAIR) as a lncRNA.
Studies demonstrate that HOTAIR helps regulate gene activity, which
has led to investigations on its potential connection to various
types of cancer, including lung, gastric, glioma, pancreatic,
cervical (16–20) and EC (21,22).
The expression of HOTAIR increases in patients with EC, and its
elevation is associated with advanced disease, metastasis and drug
resistance (16,17,22).
However, the molecular effects of HOTAIR in EC and the potential
clinical application of its serum levels have not been fully
elucidated.
miRs are small ncRNA sequences, ~22 nucleotides in
length, involved in regulating processes such as development, cell
differentiation, apoptosis and proliferation. Their primary
function is gene silencing (23).
miRs can act as either tumour suppressors or oncogenes under
specific conditions (24). While
oncogenic miRs enhance the tumour cell phenotype, tumour suppressor
miRs diminish it (25). The
abundant presence of miRs in faeces, sputum, pleural effusion and
urine suggests that alterations in miR expression levels are
readily detectable.
Previous studies indicate that miR-646 is linked to
various types of cancer, including lung (26), laryngeal (27), clear cell carcinoma (28), retinoblastoma (29) and osteosarcoma (30), with a notable role in
gastrointestinal (31) and
gynaecological tumours (32). It is
hypothesized to function as a tumour suppressor in these
malignancies, and in EC, it primarily reduces tumorigenic potential
through the downregulation of nucleophosmin 1 (NPM1) (32,33).
In vitro studies by Zhou et al (33) demonstrates that HOTAIR increases
NPM1 levels by suppressing the expression of miR-646, which
promotes cancer cell proliferation, invasion and migration.
At present, there is not a reliable non-invasive
biomarker that is routinely used for the diagnosis or monitoring of
EC. HOTAIR is a potential biomarker candidate due to its
overexpression and association with advanced stages of the disease
(33). Additionally, miR-646 may
also serve as a detectable marker in body fluids (34). Experimental evidence from Zhou et
al (33) suggests a possible
regulatory interaction between HOTAIR and miR-646; however, this
association is yet to be confirmed in clinical serum samples.
Therefore, the present study aimed to investigate the serum
expression levels of HOTAIR and miR-646 in patients with EC and
investigated their potential as non-invasive diagnostic or
prognostic biomarkers.
Materials and methods
Subjects
The study population consisted of women aged ≥18
years with histologically confirmed EC, diagnosed by endometrial
biopsy and managed surgically. All participants provided written
informed consent prior to inclusion. Patients with biopsy-proven EC
of any histopathological subtype who underwent hysterectomy with
surgical staging and subsequent histopathological evaluation of the
surgical specimens were included. The control group included women
who attended the general gynaecology outpatient clinic for
non-malignant gynaecological conditions and who had no history of
malignancy. Patients were excluded if they declined to provide
informed consent, were medically inoperable, refused surgical
treatment, had a final pathology not confirming EC, had synchronous
or other primary malignancies, had recurrent disease, or had
received prior oncologic treatments such as chemotherapy,
radiotherapy or immunotherapy. The present study examined the
association between HOTAIR and miR-646 gene expression levels and
the development of EC. Additionally, the association between
prognostic factors of EC, such as grade and clinical T stage, and
the expression of HOTAIR and miR-646 was investigated. In total, 71
patients that were admitted to the Department of Obstetrics,
Gynaecology-Oncology Division, Istanbul Medical Faculty, Istanbul
University (Istanbul, Turkey) and diagnosed with EC based on biopsy
results, were included in the present study. Additionally, 62
healthy women who sought services at the Department of Obstetrics
and Gynaecology, Istanbul Medical Faculty, Istanbul University, for
routine gynaecological examination or benign conditions were
included in the control group. Patients and healthy control
individuals were enrolled in the present study between August 2021
and March 2023. The study protocol was reviewed and approved by the
Institutional Review Board and Ethics Committee of Istanbul
University (Istanbul, Turkey; approval no. 272576; approval date,
01.07.2021). Sample collection and experimental analyses were
completed by June 2023. Blood samples were collected after both the
patients and control individuals were informed verbally and in
writing.
Patients and controls were matched based on age,
body mass index (BMI) and smoking status. An individual was classed
as a smoker if they had a minimum of 10 packs/year. An individual
was classed as a non-smoker if they had either quit smoking more
than a year before the present study or had never smoked. All
participants in both groups were female. The median ages were 57
years (range, 29–81 years) in the patient group and 54 years
(range, 34–77 years) in the control group. The mean age ± standard
deviation (SD) was 57.7±11.0 years for the patient group and
54.8±10.1 years for the control group. Tumour stage and grade were
determined according to surgical staging. In the present study, EC
was classified as low grade [International Federation of Gynecology
and Obstetrics (FIGO) grades 1–2 endometrioid)] and high grade
(FIGO grade 3 endometrioid and non-endometrioid types), which was
consistent with accepted dichotomy in the literature (35). Previous studies define high-grade EC
to include grade 3 endometrioid tumours, serous, clear cell,
undifferentiated and carcinosarcoma subtypes (6,36,37).
These tumours are characterized by more aggressive biological
behaviour, a poorer prognosis compared with low-grade (FIGO grade
1–2 endometrioid) endometrial carcinomas and as high risk by major
clinical guidelines, including the European Society of
Gynaecological Oncology, the European Society for Radiotherapy and
Oncology, and the European Society of Pathology guidelines for EC
(2021) (38) and the National
Comprehensive Cancer Network Clinical Practice Guidelines in
Oncology: Uterine Neoplasms (version 3.2025) (39). Stages IA and IB were regarded as
early stages, while stages II–IV were considered advanced stages.
Additionally, cancer antigen 125 (CA125) levels, menopausal status,
hypertension, diabetes mellitus, recurrence, lymph node
involvement, disease-free survival, overall survival and
cancer-related mortality were evaluated.
Reverse transcription-quantitative PCR
(RT-qPCR)
For RNA extraction, venous blood samples were
allowed to clot for 30 min, and were then centrifuged at 1,500 × g
for 10 min at 4°C. The separated serum samples were stored at −80°C
until further analysis. RNA isolation was carried out using the
miRNeasy Serum/Plasma Advanced kit (cat. no. 217204; Qiagen GmbH).
The concentration and purity of the extracted RNA samples were
evaluated using an Eon Biotek instrument. Following measurement,
RNA samples were stored at −80°C. For miRNA expression level
analysis, reverse transcription was carried out using the miRCURY
LNA RT kit (cat. no. 339340; Qiagen GmbH) according to the
manufacturer's protocol. The reaction was carried out at 42°C for 1
h, followed by enzyme inactivation at 95°C for 5 min. For lncRNA
expression level analysis, cDNA synthesis was conducted using the
RT2 PreAMP cDNA Synthesis kit (cat. no. 330451; Qiagen
GmbH) following the manufacturer's protocol. The reverse
transcription step was carried out at 37°C for 1 h, followed by
enzyme inactivation at 95°C for 5 min. RT-qPCR was carried out on a
Rotor-Gene Q Real-Time PCR System (Qiagen GmbH) using the miRCURY
LNA SYBR Green PCR kit (cat. no. 339345; Qiagen GmbH) for the
detection of hsa-miR-646, and the RT2 SYBR Green ROX
FAST Mastermix kit (cat. no. 330620; Qiagen GmbH) for the analysis
of lncRNA HOTAIR expression levels. PCR amplification was carried
out according to the manufacturer's protocols. For lncRNA, initial
denaturation was carried out at 95°C for 10 min, followed by 40
cycles of 95°C for 10 sec and 60°C for 1 min. For miR, initial
denaturation was carried out at 95°C for 2 min, followed by 40
cycles of 95°C for 10 sec and 56°C for 1 min.
The primers used in the present study were obtained
from Qiagen GmbH. They included the following: RT2 qPCR
Assay for GAPDH (cat. no. PPH00150F), RT2 lncRNA qPCR
Assay for HOTAIR (cat. no. LPH07360A), miRCURY LNA miRNA PCR Assay
for hsa-miR-646 (cat. no. YP00204546) and miRCURY LNA™ miRNA PCR
Assay for hsa-U6 (cat. no. YP02119464). The exact sequences of
these primers are proprietary and not publicly available from the
manufacturer. Gene expression levels were quantified using the
2−ΔΔCq method (40).
Relative expression levels were normalized to GAPDH for lncRNA and
to U6 for miR, in accordance with previously published studies
(22,27,41–43).
Statistical analysis
SPSS version 21 (IBM Corp.) was used for data
analysis. The distribution of data in the present study was
assessed using the Shapiro-Wilk test. Variables with normal
distribution were analysed using one-way ANOVA followed by the
Bonferroni post hoc test, while non-normally distributed variables
were analysed using the Mann-Whitney U test. In the present data,
‘age’ had a normal distribution, whereas ‘BMI’ did not follow a
normal distribution. The Chi-square test was used to analyse the
association between qualitative data. Spearman's correlation test
was used to investigate the association between two datasets.
Receiver operating characteristic (ROC) curve analysis was carried
out to determine whether an independent variable could distinguish
between cases of EC and healthy controls. The ROC curve was used to
identify the optimal predictive value and to calculate the
sensitivity and specificity of the predictive value. The
Kaplan-Meier method was used to compare survival between groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
There were no significant differences between the
control group and patients with EC in terms of age, BMI, smoking,
hypertension or diabetes mellitus status. Among the patients, 46
cases (64.8%) had low-grade tumours (grades 1 and 2), while 25
cases (35.2%) had high-grade tumours (grade 3), of which 2 cases
(8%) were of non-endometrioid histology (1 case had serous
carcinoma and another case had clear cell carcinoma). Early-stage
disease (stages IA and B) was present in 84.5% of the patients,
whereas 15.5% had advanced-stage disease. Tumour recurrence was
observed in 14.1% of cases, and lymph node involvement was
identified in 87.3% of patients. Mean disease-free survival and
overall survival were 30.06±10.42 and 32.18±7.98 months,
respectively, while cancer-related mortality was 10.2% (Table I).
 | Table I.Comparison of demographic and
clinical characteristics between the control and patient
groups. |
Table I.
Comparison of demographic and
clinical characteristics between the control and patient
groups.
| Parameter | Healthy control
individuals (n=62) | Patients with
endometrial cancer (n=71) | P-value |
|---|
| Age, years (mean ±
SD) | 54.77±10.09 | 57.64±11.03 | 0.118 |
| BMI,
kg/m2 (mean ± SD) | 31.20±6.37 | 32.24±8.36 | 0.261 |
| CA125, median
(min-max) | - | 13.0
(5.0-745.0) |
|
| Smoking status, N
(%) |
|
| 0.249 |
|
Non-smoker | 39 (63.9) | 52 (73.2) |
|
|
Smoker | 22 (36.1) | 19 (26.8) |
|
| Menopause status, N
(%) |
|
| - |
|
Premenopausal | - | 24 (33.8) |
|
|
Postmenopausal | - | 47 (66.2) |
|
| Tumour grade, N
(%) |
|
| - |
| Low
grade (grades 1 and 2) | - | 46 (64.8) |
|
| High
grade (grade 3 endometrioid and non-endometrioid histology
types) | - | 25 (35.2) |
|
| Stage, N (%) |
|
| - |
| Early
stage (IA and B) | - | 60 (84.5) |
|
|
Advanced stage (II–IV) | - | 11 (15.5) |
|
| Recurrence, N
(%) |
|
| - |
| No | - | 61 (85.9) |
|
|
Yes | - | 10 (14.1) |
|
| Lymph node
involvement, N (%) |
|
| - |
|
Yes | - | 62 (87.3) |
|
| No | - | 9 (12.7) |
|
| Hypertension, N
(%) |
|
| 0.070 |
|
Yes | 13 (21.0) | 25 (35.2) |
|
| No | 49 (79.0) | 46 (64.8) |
|
| Diabetes mellitus,
N (%) |
|
| 0.150 |
|
Yes | 8 (12.9) | 16 (22.5) |
|
| No | 54 (87.1) | 55 (77.5) |
|
| Disease-free
survival, months (mean ± SD) | - | 30.06±10.42 | - |
| Overall survival,
months (mean ± SD) | - | 32.18±7.98 | - |
| Cancer-related
mortality, N (%) |
|
| - |
| No | - | 53 (89.9) |
|
|
Yes | - | 6 (10.2) |
|
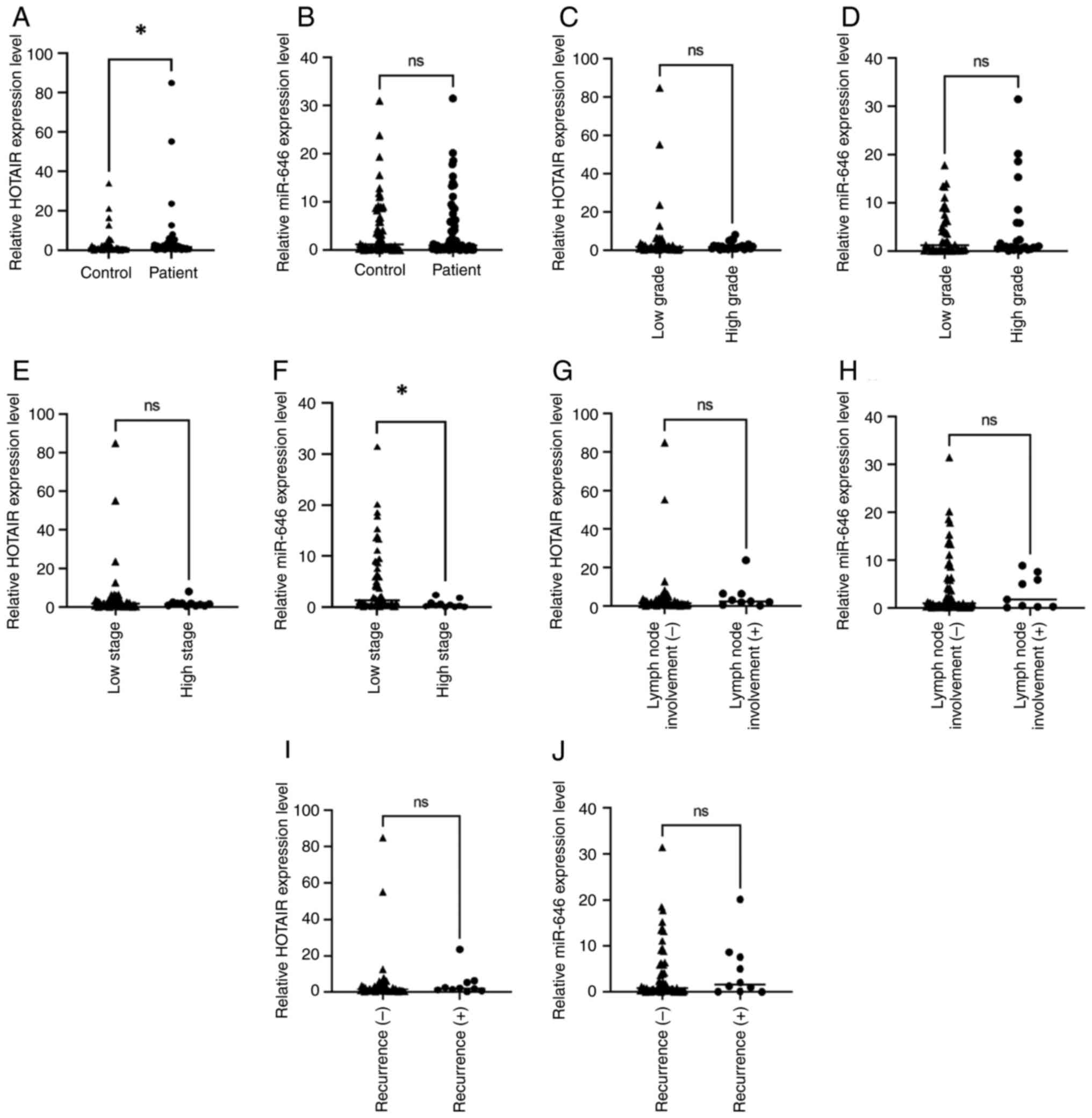
The expression of HOTAIR was significantly increased
in patients with EC compared with the control group (P=0.020).
However, the expression of miR-646 did not differ significantly
between the patient and the control groups (P=0.579). Additionally,
no significant differences in HOTAIR or miR-646 expression levels
were revealed between low- and high-grade tumours (P=0.230 and
P=0.377, respectively). The HOTAIR expression levels did not vary
significantly between low and high clinical tumour stages
(P=0.466). However, the miR-646 expression level was significantly
reduced in the high tumour stage group compared with the low tumour
stage group (P=0.045). Recurrence status and lymph node involvement
showed no significant association with the expression levels of
HOTAIR (P=0.321 and P=0.192, respectively) or miR-646 (P=0.741 and
P=0.942, respectively) (Fig. 1).
Spearman's correlation analysis did not demonstrate a significant
association between serum miR-646 and HOTAIR expression levels
(Table II). Furthermore, there
were no significant correlations between the miR-646 or HOTAIR
expression levels and CA125 concentrations (Table III).
 | Table II.Spearman correlation between HOTAIR
and miR-646. |
Table II.
Spearman correlation between HOTAIR
and miR-646.
| Parameter |
| r-value | P-value |
|---|
| HOTAIR | miR-646 | 0.105 | 0.212 |
 | Table III.Spearman correlation of HOTAIR and
miR-646 with CA125. |
Table III.
Spearman correlation of HOTAIR and
miR-646 with CA125.
|
| Correlation with
CA125 |
|---|
|
|
|
|---|
| Parameter | r-value | P-value |
|---|
| HOTAIR | 0.091 | 0.288 |
| miR-646 | 0.053 | 0.385 |
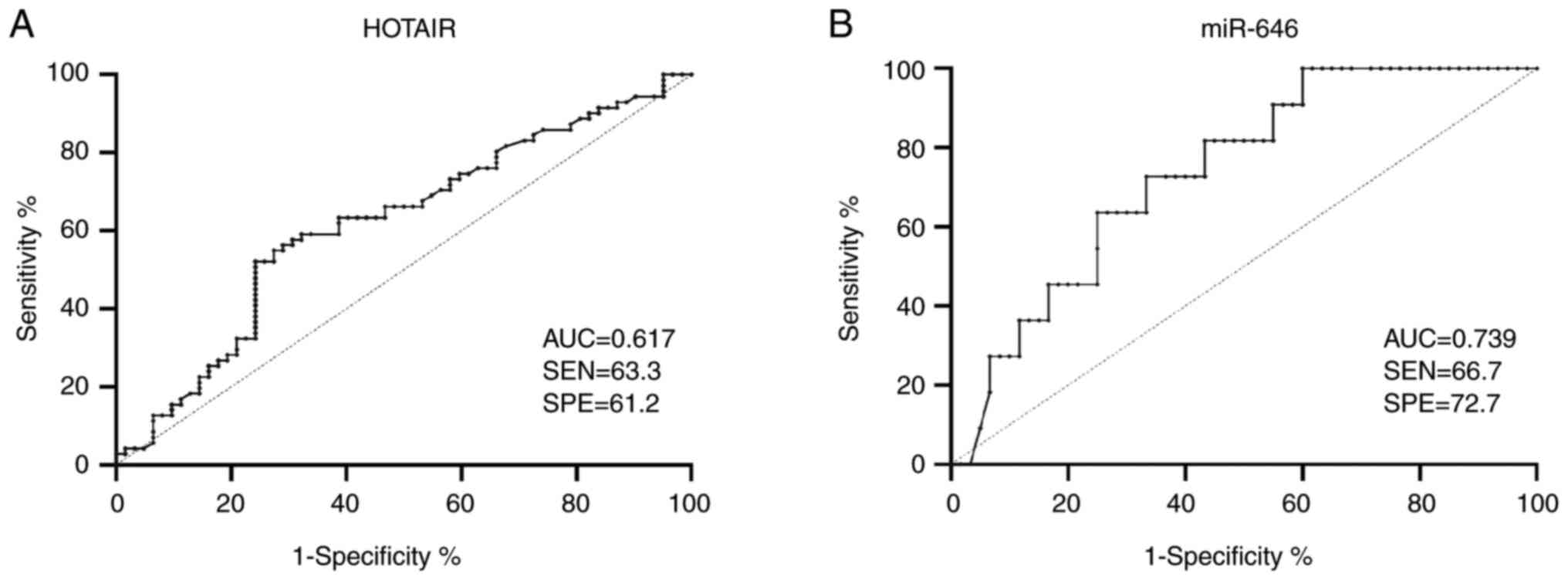
ROC analysis was carried out to investigate the
optimal cut-off values for serum HOTAIR and miR-646 expression
levels to distinguish between patients with EC and the healthy
controls. For HOTAIR, an expression level threshold of 1.08 was
revealed to differentiate between patients with EC and healthy
controls, with a sensitivity of 63.3% and a specificity of 61.2%.
Therefore, patients were classified as being HOTAIR-positive (with
a value of ≥1.08) or HOTAIR-negative (with a value of <1.08).
For miR-646, an expression level cut-off value of 0.655 was
demonstrated to distinguish between patients with advanced-stage
and early-stage EC, with a sensitivity of 66.7% and a specificity
of 72.7%. Therefore, patients were categorized as being
miR-646-positive (≥0.655) or miR-646-negative (<0.655). These
classification criteria are presented in Fig. 2.
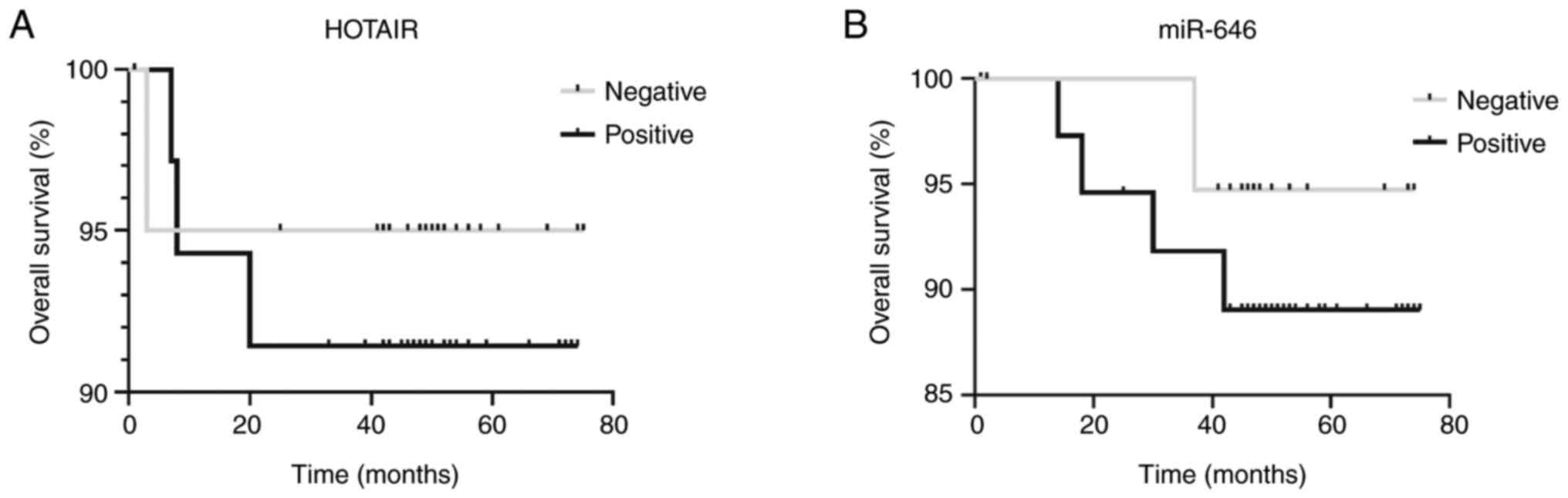
No statistically significant differences in overall
survival (in months) were observed between the HOTAIR-positive and
-negative groups or between the miR-646-positive and -negative
groups (Fig. 3).
Discussion
Although the incidence rate of EC has increased by
>130% over the past three decades, with >417,000 new cases
and ~97,370 mortalities reported worldwide in 2020 (1), reliable non-invasive diagnostic
biomarkers are still unavailable. Advancing diagnostic methods and
therapeutic strategies depends on elucidating the molecular
pathways involved in this disease. ncRNAs, including lncRNAs and
miRs, have garnered attention as potential biomarkers across
various types of cancer such as breast, gastric and gynaecological
cancer (29–32). While numerous studies have
investigated the expression of HOTAIR and miR-646 in EC tissue
samples, information regarding their circulating levels in serum is
still insufficient (33,34).
ncRNA species, such as miRs, lncRNAs and circular
RNAs (circRNAs), serve crucial roles in the positive and negative
regulation of the expression of genes. Over the past two decades,
numerous studies highlight the involvement of ncRNAs in various
aspects of cell biology, particularly in cancer development,
progression and drug resistance (43–48).
These molecules participate in virtually all cellular processes and
have fundamental roles at every stage of tumorigenesis. Altered
expression levels of mRNAs, lncRNAs, circRNAs and miRs are reported
in numerous types of cancer, such as triple-negative and oestrogen
receptor-positive breast cancer, serving as potential biomarkers
for diagnosis and therapeutic targets (43,47–50).
Furthermore, ncRNAs have been studied as promising biomarkers for
early detection and prognostic evaluation in various malignancies,
including lung (26), gastric
(31), breast (43), prostate (51), liver (52), colorectal (53), bladder (54), pancreatic (55) and haematological disorders (56). This evidence suggests that
circulating lncRNAs in peripheral blood may potentially serve as
non-invasive biomarkers for cancer diagnosis and monitoring.
The expression of miR is tissue-specific and serves
a key role in maintaining cellular homeostasis. Their stability in
circulation and cancer-specific expression patterns have led to
their investigation as promising non-invasive biomarkers in various
malignancies including breast, lung, colorectal and prostate cancer
(57–61). Although the majority of studies
focus on other types of cancer, such as ovarian, breast and
colorectal cancer (59–61), evidence suggests that specific miRs
and lncRNAs exhibit altered expression profiles in EC tissues
compared with normal endometrium, indicating potential roles in
diagnosis and prognosis (62).
HOTAIR is an important prognostic biomarker that is
overexpressed in various types of cancer including breast, ovarian,
colorectal and lung cancer (16–20).
Its increased expression level is linked to higher levels of
suppressor of zeste 12 (SUZ12), a gene involved in epigenetic
regulation (63). This suggests
that HOTAIR may serve a role in modifying chromatin structure in
tumours (64,65). HOTAIR may also mediate genome-wide
epigenetic changes by recruiting chromatin-modifying complexes such
as the polycomb repressive complex 2 (containing enhancer of zeste
homolog 2, SUZ12 and embryonic ectoderm development), which
catalyses histone 3 lysine 27 trimethylation, and the lysine
specific demethylase 1/corepressor for element-1-silencing
transcription factor/RE1-silencing transcription factor complex,
which demethylates the active histone hallmark histone H3 lysine 4
trimethylation (63,66). This mechanism may explain its
overexpression in breast, colorectal, hepatocellular,
gastrointestinal and pancreatic tumours (66). Located within the HOXC gene cluster,
HOTAIR enhances tumour invasiveness and metastasis (63). Furthermore, Bhan et al
(67) reports that the expression
of HOTAIR is transcriptionally regulated by oestradiol. HOTAIR is
similarly upregulated in EC in tumour tissues compared with normal
endometrium. The expression of HOTAIR is associated with higher
tumour grades, lymph node metastasis and poor clinical prognosis
(21,22,68).
Functional studies demonstrate that downregulation of HOTAIR can
inhibit cell proliferation and invasion in vitro and in
vivo (68,69). Furthermore, HOTAIR contributes to
chemoresistance by promoting cisplatin resistance by regulating
autophagy pathways in EC cells (70).
In the present study, serum HOTAIR expression levels
were significantly increased in patients with EC compared with the
control group. Although this difference was statistically
significant, its diagnostic accuracy (sensitivity and specificity)
was modest, limiting its immediate clinical applicability. In
contrast to previous studies on tumour tissue (17,22,33,68–70)
the results of the present study demonstrated no correlation
between serum HOTAIR expression levels and disease stage, tumour
grade or lymph node metastasis. Additionally, survival analysis
revealed no significant association with patient outcomes. These
discrepancies may be attributed to differences in the patient
populations, or they may indicate that tissue-level associations
may not be fully captured in serum-based assessments.
miR-646 is associated with numerous types of cancer
including non-small cell lung, clear cell renal, retinoblastoma,
laryngeal squamous cell, colorectal, gastric and breast cancer and
is generally downregulated in malignancies, where it contributes to
tumour growth, metastasis and invasion (25–31).
By contrast, its upregulation suppresses these processes. Previous
studies by Liu et al (32)
and Zhou et al (33)
demonstrate that miR-646 inhibits the expression of NPM1 and exerts
tumour-suppressive effects in EC cell lines including HEC-1A and
Ishikawa. The study by Zhou et al (33) demonstrates that miR-646 expression
levels are reduced in both EC tissues and cancer cell lines
compared with normal endometrial tissues and cells, respectively.
Furthermore, HOTAIR reduces miR-646 expression levels in
vitro, and a positive association between HOTAIR and NPM1,
alongside a negative association between NPM1 and miR-646, is
observed in EC tissues (32,33).
The results of the present study found no
statistically significant difference in serum miR-646 expression
levels between patients with EC and healthy controls. Additionally,
miR-646 expression levels were not demonstrated to be associated
with tumour grade, recurrence, lymph node metastasis or overall
survival. However, patients with advanced-stage EC exhibited
significantly lower miR-646 levels compared with those with
early-stage EC. This suggested a possible association between
miR-646 downregulation and disease progression, which may be
potentially mediated through the NPM1 pathway. However, the
expression of NPM1 was not assessed in the present
investigation.
The associations between serum levels of HOTAIR and
miR-646 with CA-125 were further investigated; however, no
significant associations were revealed. ROC curve analysis of
HOTAIR revealed an area under the curve of 0.617, with an optimal
threshold value of 1.08, sensitivity of 63.3% and specificity of
61.2%. This suggested a modest diagnostic accuracy. Although these
values indicated a modest diagnostic accuracy, they fall below the
commonly accepted criteria for clinical applicability (AUC
>0.80; sensitivity and specificity >80%) (71), which suggested that serum HOTAIR
alone was insufficient for diagnostic use; however, it may serve as
a potential adjunct biomarker. Future research focusing on exosomal
HOTAIR expression levels may enhance diagnostic performance.
Regarding miR-646, ROC analysis differentiating
early from advanced stages of EC demonstrated an optimal cut-off
value of 0.655, with sensitivity and specificity values of 66.7 and
72.7%, respectively. Although these metrics were below the levels
required for a clinically useful diagnostic test, they suggested
that miR-646 may have a potential utility in disease staging,
particularly if exosomal miR-646 expression levels are investigated
in future studies. Although there was not a significant association
between HOTAIR or miR-646 levels and tumour grade, lymph node
metastasis or recurrence status, these findings should be
interpreted cautiously. It is possible that serum-based assessments
do not accurately reflect the tumour microenvironment, or that the
small sample size of the present study limited the ability of the
present study to detect these associations. The novelty of the
present study was the evaluation of serum-based expression levels
of HOTAIR and miR-646 as potential non-invasive biomarkers in EC,
which, to the best of our knowledge, has not been previously
reported in clinical patient samples.
The present study had several limitations. Firstly,
in the present study, the expression of HOTAIR or miR-646 in
exosomes was not assessed, which may provide a more accurate
reflection of tumour-derived ncRNAs. Secondly, the small number of
patients with lymph node metastasis and recurrence limited the
possibility of conducting subgroup analyses. Thirdly, the absence
of associations between miR-646 and certain clinicopathological
features may be due to the characteristics of the cohort used in
the present study or the lack of exosomal data. Finally, the use of
GAPDH and U6 as endogenous controls for normalization may represent
another limitation of the present study. Although these reference
genes are used in serum-based analyses of lncRNA and miR to ensure
comparability with previous studies, their stability in circulation
is not absolute and may introduce technical variability. Future
studies should incorporate spike-in controls, such as synthetic
cel-miR-39 or Universal spike-in oligonucleotides, for both HOTAIR
and miR-646 to monitor RNA extraction and reverse transcription
efficiency (72), and use stable
endogenous RNAs (such as U6 for miR-646 (73) and GAPDH or 18S ribosomal RNA for
HOTAIR (74) to improve
normalisation and enhance the reproducibility of results. In
conclusion, the results of the present study reinforced the
relevance of HOTAIR and miR-646 in EC. Serum HOTAIR may contribute
to diagnosis, while serum miR-646 may reflect disease stage.
However, both biomarkers require validation in larger, more diverse
patient cohorts and studies incorporating exosomal analysis in
order to clarify their clinical applicability in the diagnosis,
monitoring and prognosis of EC. In addition to their potential use
as biomarkers, the findings of the present study suggested that
both HOTAIR and miR-646 may serve as targets for new treatment
strategies in EC. The increased expression of HOTAIR could possibly
be reduced by drugs that block its function, and the decreased
expression of miR-646 in advanced stages may possibly be restored
using miR-646 mimics. The results of the present study may be
useful not only for diagnosis and prognosis but also for the
development of novel therapies.
Acknowledgements
Not applicable.
Funding
The present study was supported by Istanbul University
Scientific Research Projects Coordination Unit (BAP) (grant no.
TYL-2021-38002).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
RH, HK, YM, HS, ST, CK and AFA conceived and
designed the experiments. RH, HK, YM, HS, ST, CK and AFA confirm
the authenticity of all the raw data. RH, HK, YM, HS, ST, CK and
AFA performed the experiments, including literature database
searching where applicable, conducted data analysis, prepared
figures and tables, and contributed to the drafting of the
manuscript and its critical revision for important intellectual
content. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Istanbul Faculty of Medicine, Istanbul University
(approval date, 01.07.2021; approval no. 272576; Istanbul, Turkey).
Written informed consent was obtained from all participants, and
the study was conducted in accordance with the Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
ORCID IDs: Rızki Harismunandar, https://orcid.org/0009-0004-9966-9595;
Hande Karpuzoğlu, https://orcid.org/0000-0001-9603-5838; Yağmur
Minareci, https://orcid.org/0000-0003-1420-9318; Hamdullah
Sözen, https://orcid.org/0000-0003-1894-1688; Samet Topuz,
https://orcid.org/0000-0002-9069-0185; Canan
Küçükgergin, https://orcid.org/0000-0002-1797-5889; A. Fatih Aydın,
https://orcid.org/0000-0002-3336-4332.
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence tools were used to improve the readability and
language of the manuscript or to generate images, and subsequently,
the authors revised and edited the content produced by the
artificial intelligence tools as necessary, taking full
responsibility for the ultimate content of the present
manuscript.
References
|
1
|
Baker-Rand H and Kitson SJ: Recent
advances in endometrial cancer prevention, early diagnosis and
treatment. Cancers (Basel). 16:10282024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zaino RJ: Introduction to endometrial
cancer. Molecular Pathology of Gynecologic Cancer. Giordano A,
Bovicelli A and Kurman RJ: Humana Press; Totowa, NJ: pp. 51–72.
2007, View Article : Google Scholar
|
|
3
|
Patni R: Current concepts in endometrial
cancer. Endometrial Carcinoma: Evolution and Overview. Patni R:
Springer; Singapore: pp. 1–10. 2017
|
|
4
|
Pokharna S: Epidemiology and prevention of
endometrial carcinoma. Current Concepts in Endometrial Cancer.
Patni R: Springer; Singapore: pp. 11–18. 2017, View Article : Google Scholar
|
|
5
|
Bokhman JV: Two pathogenetic types of
endometrial carcinoma. Gynecol Oncol. 15:10–17. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilczyński M, Danielska J and Wilczyński
JR: An update of the classical Bokhman's dualistic model of
endometrial cancer. Menopause Rev. 15:63–68. 2016. View Article : Google Scholar
|
|
7
|
Collins FS and Mansoura MK: The human
genome project. Cancer. 91:221–225. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu M, Fu P, Qu L, Liu J and Lin A: Long
noncoding RNAs: New critical regulators in cancer immunity. Front
Oncol. 10:5509872020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15(Spec No 1): R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhattacharjee R, Prabhakar N, Kumar L,
Bhattacharjee A, Kar S, Malik S, Kumar D, Ruokolainen J, Negi A,
Jha NK and Kesari KK: Crosstalk between long noncoding RNA and
microRNA in cancer. Cell Oncol (Dordr). 46:885–908. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slack FJ and Chinnaiyan AM: The role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Wang W, Zhu W, Dong J, Cheng Y,
Yin Z and Shen F: Mechanisms and functions of long non-coding RNAs
at multiple regulatory levels. Int J Mol Sci. 20:55732019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hajjari M and Salavaty A: HOTAIR: An
oncogenic long non-coding RNA in different cancers. Cancer Biol
Med. 12:1–9. 2015.PubMed/NCBI
|
|
17
|
Rajagopal T, Talluri S, Akshaya RL and
Dunna NR: HOTAIR lncRNA: A novel oncogenic propellant in human
cancer. Clin Chim Acta. 503:1–18. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu X, Alsager S, Zhuo Y and Shan B: HOX
transcript antisense RNA (HOTAIR) in cancer. Cancer Lett.
454:90–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F,
Yu W, Wang X, Zhang L, Yu J and Hao X: Long noncoding RNA HOTAIR
involvement in cancer. Tumour Biol. 35:9531–9538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mozdarani H, Ezzatizadeh V and Rahbar
Parvaneh R: The emerging role of the long non-coding RNA HOTAIR in
breast cancer development and treatment. J Transl Med. 18:1522020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vallone C, Rigon G, Gulia C, Baffa A,
Votino R, Morosetti G, Zaami S, Briganti V, Catania F, Gaffi M, et
al: Coding RNAs and endometrial cancer. Genes (Basel). 9:1872018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He X, Bao W, Li X, Chen Z, Che Q, Wang H
and Wan XP: The long non-coding RNA HOTAIR is upregulated in
endometrial carcinoma and correlates with poor prognosis. Int J Mol
Med. 33:325–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Romano G, Veneziano D, Acunzo M and Croce
C: Small non-coding RNA and cancer. Carcinogenesis. 38:485–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Fu LL, Wen X, Liu B, Huang J, Wang
JH and Wei YQ: Oncogenic and tumor suppressive roles of microRNAs
in apoptosis and autophagy. Apoptosis. 19:1177–1189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng Y and Croce CM: The role of microRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan Y, Chen Y, Ma D, Ji Z, Cao F, Chen Z,
Ning Y and Bai C: miR-646 is a key negative regulator of EGFR
pathway in lung cancer. Exp Lung Res. 42:286–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zang Y, Li J, Wan B and Tai Y: circRNA
circ-CCND1 promotes the proliferation of laryngeal squamous cell
carcinoma via elevating CCND1 expression by interacting with HuR
and miR-646. J Cell Mol Med. 24:2423–2433. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li W, Liu M, Feng Y, Xu YF, Huang YF, Che
JP, Wang GC, Yao XD and Zheng JH: Downregulated miR-646 in clear
cell renal carcinoma correlated with tumour metastasis by targeting
the nin one binding protein (NOB1). Br J Cancer. 111:1188–1200.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Wu J, Li Y, Jiang Y, Wang L, Chen
Y, Lv Y, Zou Y and Ding X: Circ_0000527 promotes the progression of
retinoblastoma by regulating miR-646/LRP6 axis. Cancer Cell Int.
20:3012020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang L, Liu G, Xiao S, Wang L, Liu X, Tan
Q and Li Z: Long noncoding MT1JP enhanced the inhibitory effects of
miR-646 on FGF2 in osteosarcoma. Cancer Biother Radiopharm.
35:371–376. 2020.PubMed/NCBI
|
|
31
|
Zhang P, Tang WM, Zhang H, Li YQ, Peng Y,
Wang J, Liu GN, Huang XT, Zhao JJ, Li G, et al: miR-646 inhibits
cell proliferation and EMT-induced metastasis by targeting FOXK1 in
gastric cancer. Br J Cancer. 117:525–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Chen S, Zong ZH, Guan X and Zhao Y:
CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the
development of endometrial cancer. J Cell Mol Med. 24:6898–6907.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou YX, Wang C, Mao LW, Wang YL, Xia LQ,
Zhao W, Shen J and Chen J: Long noncoding RNA HOTAIR mediates the
estrogen-induced metastasis of endometrial cancer cells via the
miR-646/NPM1 axis. Am J Physiol Cell Physiol. 314:C690–C701. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang WT, Zhao YN, Yan JX, Weng MY, Wang Y,
Chen YQ and Hong SJ: Differentially expressed microRNAs in the
serum of cervical squamous cell carcinoma patients before and after
surgery. J Hematol Oncol. 7:62014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Murali R, Davidson B, Fadare O, Carlson
JA, Crum CP, Gilks CB, Irving JA, Malpica A, Matias-Guiu X,
McCluggage WG, et al: High-grade endometrial carcinomas:
Morphologic and immunohistochemical features, diagnostic challenges
and recommendations. Int J Gynecol Pathol. 38:40–63. 2019.
View Article : Google Scholar
|
|
36
|
Zhang C and Zheng W: High-grade
endometrial carcinomas: Morphologic spectrum and molecular
classification. Semin Diagn Pathol. 39:176–186. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oliva E and Soslow RA: High-grade
endometrial carcinomas. Surg Pathol Clin. 4:199–241. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Concin N, Matias-Guiu X, Vergote I, Cibula
D, Mirza MR, Marnitz S, Lederman S, Bosse T, Chargari C, Fagotti A,
et al: ESGO/ESTRO/ESP guidelines for the management of patients
with endometrial carcinoma. Int J Gynecol Cancer. 31:12–39. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
NCCN Clinical Practice Guidelines in
Oncology (NCCN Guidelines®), . Uterine Neoplasms. Version 3.2025.
National Comprehensive Cancer Network; Plymouth Meeting, PA:
2025
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mohamad N, Khedr AMB, Shaker OG and Hassan
M: Expression of long noncoding RNA, HOTAIR, and MicroRNA-205 and
their relation to transforming growth factor β1 in patients with
alopecia Areata. Skin Appendage Disord. 9:111–120. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang B, Sun Q, Ye W, Li L and Jin P: Long
non-coding RNA CDKN2B-AS1 enhances LPS-induced apoptotic and
inflammatory damages in human lung epithelial cells via regulating
the miR-140-5p/TGFBR2/Smad3 signal network. BMC Pulm Med.
21:2002021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
İlhan B, Kuraş S, Kılıç B, Tilgen
Yasasever C, Oğuz Soydinç H, Alsaadoni H, Öztan G, Adamnejad
Ghafour A, Ucuncu M, Kunduz E and Bademler S: Exploratory analysis
of circulating serum miR-197-3p, miR-1236, and miR-1271 expression
in early breast cancer. Int J Mol Sci. 26:89442025. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Melton C, Reuter JA, Spacek DV and Snyder
M: Recurrent somatic mutations in regulatory regions of human
cancer genomes. Nat Genet. 47:710–716. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Egranova SD, Hua Q, Lina C and Yanga L:
lncRNAs as tumor cell intrinsic factors that affect cancer
immunotherapy. RNA Biol. 17:1625–1627. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kansara S, Pandey V, Lobie PE, Sethi G,
Garg M and Pandey AK: Mechanistic involvement of long noncoding
RNAs in oncotherapeutics resistance in triple-negative breast
cancer. Cells. 9:15112020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Basak P, Chatterjee S, Bhat V, Su A, Jin
H, Lee-Wing V, Liu Q, Hu P, Murphy LC and Raouf A: Long non-coding
RNA H19 acts as an estrogen receptor modulator that is required for
endocrine therapy resistance in ER+ breast cancer cells. Cell
Physiol Biochem. 51:1518–1532. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Whitfield ML, George LK, Grant GD and
Perou CM: Common markers of proliferation. Nat Rev Cancer.
6:99–106. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ding L, Wang R, Shen D, Cheng S, Wang H,
Lu Z, Zheng Q, Wang L, Xia L and Li G: Role of noncoding RNA in
drug resistance of prostate cancer. Cell Death Dis. 12:5902021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen N, Li Y and Li X: Dynamic role of
long noncoding RNA in liver diseases: Pathogenesis and diagnostic
aspects. Clin Exp Med. 25:1602025. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lin Y, Zhao W, Pu R, Lv Z, Xie H, Li Y and
Zhang Z: Long non coding RNAs as diagnostic and prognostic
biomarkers for colorectal cancer (Review). Oncol Lett. 28:4862024.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li S, Zhao Y and Chen X: Microarray
expression profile analysis of circular RNAs and their potential
regulatory role in bladder carcinoma. Oncol Rep. 45:239–253. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kawai M, Fukuda A, Otomo R, Obata S,
Minaga K, Asada M, Umemura A, Uenoyama Y, Hieda N, Morita T, et al:
Early detection of pancreatic cancer by comprehensive serum miRNA
sequencing with automated machine learning. Br J Cancer.
131:1158–1168. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wong NK, Huang CL, Islam R and Yip SP:
Long non-coding RNAs in hematological malignancies: Translating
basic techniques into diagnostic and therapeutic strategies. J
Hematol Oncol. 11:1312018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ludwig N, Leidinger P, Becker K, Backes C,
Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E
and Keller A: Distribution of miRNA expression across human
tissues. Nucleic Acids Res. 44:3865–3877. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Valihrach L, Androvic P and Kubista M:
Circulating miRNA analysis for cancer diagnostics and therapy. Mol
Aspects Med. 72:1008262020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chung YW, Bae HS, Song JY, Lee JK, Lee NW,
Kim T and Lee KW: Detection of microRNA as novel biomarkers of
epithelial ovarian cancer from the serum of ovarian cancer
patients. Int J Gynecol Cancer. 23:673–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Iorio MV and Croce CM: microRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Donkers H, Bekkers R and Galaal K:
Diagnostic value of microRNA panel in endometrial cancer: A
systematic review. Oncotarget. 11:2010–2023. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ayers D: Long non-coding RNAs: Novel
emergent biomarkers for cancer diagnostics. J Cancer Res Treat.
1:31–35. 2013.
|
|
65
|
Akman HB and Bensan AE: Noncoding RNAs and
cancer. Turk J Biol. 38:817–828. 2014. View Article : Google Scholar
|
|
66
|
Beckedorff FC, Amaral MS,
Deocesano-Pereira C and Verjovski-Almeida S: Long noncoding RNAs
and their implications in cancer epigenetics. Biosci Rep.
33:701–713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bhan A, Hussain I, Ansari KI, Kasiri S,
Bashyal A and Mandal SS: Antisense transcript long noncoding RNA
(lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol
Biol. 425:3707–3722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang J, Ke P, Guo L, Wang W, Tan H, Liang
Y and Yao S: Lentivirus-mediated RNA interference targeting the
long noncoding RNA HOTAIR inhibits proliferation and invasion of
endometrial carcinoma cells in vitro and in vivo. Int J Gynecol
Cancer. 24:635–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhou Y, Zhang Y, Shao Y, Yue X, Chu Y,
Yang C and Chen D: LncRNA HOTAIR down-expression inhibits the
invasion and tumorigenicity of epithelial ovarian cancer cells by
suppressing TGF-β1 and ZEB1. Discov Oncol. 14:2282023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sun MY, Zhu JY, Zhang CY, Zhang M, Song
YN, Rahman K, Zhang LJ and Zhang H: Autophagy regulated by lncRNA
HOTAIR contributes to the cisplatin-induced resistance in
endometrial cancer cells. Biotechnol Lett. 39:1477–1484. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Çorbacıoğlu ŞK and Aksel G: Receiver
operating characteristic curve analysis in diagnostic accuracy
studies: A guide to interpreting the area under the curve value.
Turk J Emerg Med. 23:195–198. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Roest HP, IJzermans JNM and van der Laan
LJW: Evaluation of RNA isolation methods for microRNA
quantification in a range of clinical biofluids. BMC Biotechnol.
21:482021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Aerts JL, Gonzales MI and Topalian SL:
Selection of appropriate control genes to assess expression of
tumor antigens using real-time RT-PCR. BioTechniques. 36:84–86.
8890–101. 2004. View Article : Google Scholar : PubMed/NCBI
|