Introduction
Liver cancer remains one of the leading causes of
cancer-related mortality globally (865,269 new cases and 757,948
deaths in 2022), with a high incidence in regions affected by
chronic liver diseases such as hepatitis B and C infection
(1–3). Despite advances in early detection and
treatment strategies, the prognosis for patients with liver cancer
remains poor, mainly due to the aggressive nature of the tumor,
including its ability to metastasize and develop resistance to
conventional therapies, such as surgical resection, ablation
therapy and transarterial chemoembolization (4). Therefore, there is a key need to
identify novel biomarkers and mechanistic regulators of liver
cancer that may inform future therapeutic development.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
that post-transcriptionally regulate gene expression and are
involved in several biological processes, including cell
proliferation, differentiation, apoptosis and metabolism (5,6). In
cancer, miRNAs can function as either oncogenes or tumor
suppressors, depending on their target genes and the cellular
context (7). miR-448 has been
identified as a tumor suppressor in several types of cancer,
including breast, gastric and liver cancer, where it is implicated
in the regulation of cell proliferation, migration and invasion
(8–11). However, the precise molecular
mechanisms underlying its role in liver cancer progression remain
to be elucidated.
One of the hallmarks of cancer cells is altered
metabolism, with a particular emphasis on the reprogramming of
glucose metabolism. The Warburg effect, characterized by the
upregulation of glycolysis even in the presence of oxygen, is a
common metabolic shift observed in several types of cancer,
including liver cancer, ovarian cancer, colorectal cancer and
non-small cell lung cancer (12–15).
This metabolic alteration supports the high energy demands of
rapidly proliferating cancer cells, and facilitates tumor
progression and metastasis (16).
Previous studies have suggested that miRNAs, including miR-448,
miR-126-3p and miR-423-5p, may modulate glycolysis as well as lipid
and amino acid metabolism, although the direct association between
miR-448 and glycolytic regulation in liver cancer remains to be
elucidated (17–19).
Ras-related protein Rab-7a (RAB7A), a member of the
small GTPase family, serves a key role in vesicular trafficking and
endosomal maturation. It has been reported to regulate numerous
cellular processes, including autophagy, metabolism and cell
migration (20). Furthermore, RAB7A
has been shown to be upregulated in several types of cancer,
including hepatocellular carcinoma (HCC), pancreatic adenocarcinoma
and colon adenocarcinoma, and its elevated expression is associated
with poor prognosis and aggressive tumor behavior (21–23).
Recent studies have suggested that RAB7A may influence glycolytic
metabolism in cancer cells, although its role in liver cancer
glycolysis is yet to be fully elucidated (24,25).
It was hypothesized in the present study that
exosomal miR-448 (EXO-miR-448) suppresses liver cancer progression
by directly targeting RAB7A and inhibiting glycolytic metabolism.
Therefore, the current study aimed to assess the expression levels
of RAB7A and miR-448 in liver cancer and normal liver cell lines,
to explore the relationship between EXO-miR-448 and RAB7A, and to
evaluate the effects of EXO-miR-448 on proliferation, migration,
invasion and glycolytic activity in liver cancer cells.
Materials and methods
Cell lines and culture conditions
All cell lines (HepG2, Hep3B, SK-HEP-1 and THLE-2)
were purchased from the American Type Culture Collection. The
identity of all cell lines was confirmed by the supplier using
short tandem repeat profiling. In addition, all cell lines were
routinely cultured for <10 passages and tested negative for
mycoplasma contamination using PCR-based assays prior to use. Cells
were maintained at 37°C in a humidified atmosphere containing 5%
CO2 using DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin solution. The use of commercially
obtained human cell lines was approved by the Ethics Committee of
The Second Affiliated Hospital of Hainan Medical University
(Haikou, China) and all procedures were performed in accordance
with institutional guidelines.
Exosome isolation and
identification
Exosomes were isolated from HepG2 cell culture
supernatants using ultracentrifugation. Briefly, the cell culture
supernatant was sequentially centrifuged at 300 × g for 10 min at
4°C, 2,000 × g for 10 min at 4°C and 10,000 × g for 30 min at 4°C
to remove cells and debris, followed by ultracentrifugation at
100,000 × g for 70 min at 4°C to pellet exosomes. Exosomes were
identified by transmission electron microscopy (TEM; JEOL, Ltd.),
nanoparticle tracking analysis (NTA; ZetaView®; Particle
Metrix GmbH), and western blotting of exosomal markers and a
negative control (CD63: 1:500, cat. no. ab68418, Abcam; CD9: 1:500,
cat. no. WL06408, Wanleibio Co., Ltd.; calnexin:1:500, cat. no.
WL03062, Wanleibio Co., Ltd.). For TEM analysis, isolated exosomes
were examined without fixation, embedding or sectioning. A 10 µl
exosome suspension was placed onto a carbon-coated copper grid and
allowed to adsorb for 10 min at 25°C. Excess liquid was removed
with filter paper, followed by negative staining with 2%
phosphotungstic acid for 2 min at 25°C. The grids were then air
dried and examined using TEM. For NTA, the sample chamber was first
rinsed with deionized water, and the instrument was calibrated
using 110 nm polystyrene microspheres. The chamber was then washed
with PBS, and exosome samples were diluted 1:200 in PBS before
measurement using the ZetaView instrument.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cultured cells (THLE-2,
SK-HEP-1, Hep3B and HepG2) or isolated exosomes using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. For mRNA
analysis, RNA was reverse-transcribed into cDNA using a
PrimeScript™ RT reagent kit (Takara Bio, Inc.), whereas
for miRNA analysis, cDNA was synthesized using a miRNA First-Strand
cDNA Synthesis Kit (Tailing Reaction) (cat. no. B532451; Sangon
Biotech Co., Ltd.), according to the manufacturers' protocols.
Subsequently, qPCR was performed on a 7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). miRNA
expression was analyzed using 2X Fast Taq Plus PCR Master Mix
(Biosharp Life Sciences) supplemented with SYBR Green (Beijing
Solarbio Science & Technology Co., Ltd.) under the following
conditions: 95°C for 10 min, followed by 40 cycles at 95°C for 20
sec, 60°C for 20 sec and 70°C for 10 sec, with melting curve
analysis performed at the end of amplification. mRNA expression was
analyzed using SYBR® Premix Ex Taq™ II
(Takara Bio, Inc.) according to the manufacturer's protocol, with
the following thermocycling conditions: Initial denaturation at
95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C
for 34 sec. The relative expression levels of miR-448 were
normalized to U6, whereas those of RAB7A, sphingosine-1-phosphate
phosphatase 1 (SGPP1), vasohibin 2 (VASH2), mitochondrial pyruvate
carrier 1 (MPC1) and thymosin β 4 X-linked (TMSB4X) were normalized
to β-actin using the 2−ΔΔCq method (26). Primer sequences for mRNA targets and
β-actin were synthesized by Sangon Biotech Co., Ltd. and are listed
in Table I. The forward primer for
hsa-miR-448 was synthesized by General Biotech (Anhui) Co., Ltd.
The reverse primer for miR-448 and the U6 primer were supplied as
built-in components of the miRNA First-Strand cDNA Synthesis
Kit.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence,
5′-3′ |
|---|
| RAB7A | F:
GACTGCTGCGTTCTGGT |
|
| R:
CCTCCGTTTCCTGCTTA |
| β-actin | F:
GGCACCCAGCACAATGAA |
|
| R:
TAGAAGCATTTGCGGTGG |
| hsa-miR-448 | F:
TTGCATATGTAGGATGTCCCAT |
| SGPP1 | F:
CTCCATTCATCATCATCGGGCTTC |
|
| R:
CTAGTATCTCGGCTGTGTCTCCTC |
| VASH2 | F:
AGGAACGCCGCCTTCTTGG |
|
| R:
CATGTAATTCTGGATCGCCTGGAG |
| MPC1 | F:
TGCCCTCTGTTGCTATTCTTTGAC |
|
| R:
AGCCGCCCTCCCTGGATG |
| TMSB4X | F:
CTGACAAACCCGATATGGCTGAG |
|
| R:
GCCTGCTTGCTTCTCCTGTTC |
RNase A and Triton X-100 treatment for
verification of EXO-miR-448
To determine whether miR-448 in the culture
supernatant was encapsulated within exosomes, the conditioned
medium of HepG2 cells was collected and centrifuged at 300 × g for
10 min and 2,000 × g for 10 min at 4°C to remove cells and cellular
debris. The supernatants were then treated with RNase A (100 µg/ml)
at 37°C for 60 min to degrade free extracellular miRNA, or with
RNase A (100 µg/ml) plus Triton X-100 (1%) at 37°C for 30 min to
disrupt vesicle membranes and degrade encapsulated miRNA.
Subsequently, the levels of miR-448 in the supernatants were
determined by RT-qPCR using the aforementioned procedure.
Preparation and verification of
miR-448 enriched exosomes
HepG2 cells were used as donor cells to generate
miR-448 enriched exosomes. HepG2 cells were seeded in 6-well plates
at a density of 2×105 cells/well. Cells were transfected
with miR-448 mimics or control mimics (mimics NC) (JTS Scientific
Co., Ltd.) using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.), with 75 pmol mimics/well in a final
volume of 2.5 ml, and incubated at 37°C for 48 h according to the
manufacturer's instructions. The sequences were as follows: miR-448
mimics, sense 5′-UUGCAUAUGUAGGAUGUCCCAU-3′, antisense
5′-GGGACAUCCUACAUAUGCAAUU-3′; And mimics NC, sense
5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′.
After 48 h, the culture supernatant was collected and exosomes were
isolated by differential ultracentrifugation (300 × g for 10 min,
2,000 × g for 10 min, 10,000 × g for 30 min and 100,000 × g for 70
min, all at 4°C). Exosomes derived from miR-448 mimic-transfected
donor cells and mimics NC-transfected donor cells were designated
EXO-miR-448 and EXO-NC, respectively. For functional assays, HepG2
cells were treated with EXO-miR-448 or EXO-NC (25 µg/ml) at 37°C.
Cells were analyzed at 0, 24, 48 and 72 h for the Cell Counting
Kit-8 (CCK-8) assay, whereas for EdU, migration and invasion
assays, cells were analyzed after 48 h of exosome treatment. To
confirm the successful enrichment of miR-448 in exosomes, total RNA
was extracted from the isolated exosomes, and RT-qPCR was performed
to quantify miR-448 expression levels.
Western blot analysis
Cells (THLE-2, SK-HEP-1, Hep3B and HepG2) or
isolated exosomes were lysed using RIPA buffer (Beyotime
Biotechnology) containing protease inhibitors and protein
concentrations were determined using the BCA Protein Assay Kit
(Thermo Fisher Scientific, Inc.). Equal amounts of protein samples
(40 µg/lane) were separated by SDS-PAGE on a 5% stacking gel, and 8
or 13% separating gels, and transferred onto PVDF membranes
(MilliporeSigma). Subsequently, the membranes were blocked with 5%
skim milk at 37°C for 2 h, and were incubated overnight at 4°C with
the following primary antibodies: RAB7A (1:1,000; cat. no. A12308;
ABclonal Biotech Co., Ltd.), CD63, CD9, calnexin and β-actin
(1:1,000; cat. no. WL01372; Wanleibio Co., Ltd.). The membranes
were then incubated with an HRP-conjugated secondary antibody (goat
anti-rabbit IgG-HRP; 1:5,000; cat. no. WLA023; Wanleibio Co., Ltd.)
at 37°C for 2 h. Protein bands were visualized using the Clarity™
Western ECL Substrate (Bio-Rad Laboratories, Inc.) and were
analyzed using ImageJ software (version 1.54; National Institutes
of Health).
Transfection and siRNA knockdown
assays
HepG2 cells were seeded at a density of
~2×105 cells/well in 6-well plates and transfected with
RAB7A small interfering (si)RNAs (si-RAB7A-1, si-RAB7A-2 and
si-RAB7A-3) or NC siRNA using Lipofectamine 3000. The final
concentration of siRNAs in each well was 50 nM, and transfection
was performed at 37°C for 48 h according to the manufacturer's
instructions. After 48 h, transfection efficiency was confirmed
using RT-qPCR and western blotting. The siRNA with the highest
knockdown efficiency was selected for subsequent experiments and
termed si-RAB7A. The siRNA sequences are listed in Table II.
 | Table II.Sequences of siRNAs used for RAB7A
knockdown. |
Table II.
Sequences of siRNAs used for RAB7A
knockdown.
| siRNA | Sequence,
5′-3′ |
|---|
| siRNA NC | S:
UUCUCCGAACGUGUCACGUTT |
|
| AS:
ACGUGACACGUUCGGAGAATT |
| RAB7A siRNA-1 | S:
CACAAUAGGAGCUGACUUUTT |
|
| AS:
AAAGUCAGCUCCUAUUGUGTT |
| RAB7A siRNA-2 | S:
UUAUCAUCCUGGGAGAUUCTT |
|
| AS:
GAAUCUCCCAGGAUGAUAATT |
| RAB7A siRNA-3 | S:
CAGUAUGUGAAUAAGAAAUTT |
|
| AS:
AUUUCUUAUUCACAUACUGTT |
Dual-luciferase reporter assay
The 3′-UTR fragment of RAB7A containing the putative
miR-448 binding site (wild-type; WT) and the corresponding mutant
(MUT) were cloned into the pmirGLO dual-luciferase reporter vector
(Promega Corporation). HepG2 cells were co-transfected with pmirGLO
constructs and either miR-448 mimics or mimics NC, as
aforementioned, using Lipofectamine 3000. After 48 h, luciferase
activity was measured using the Dual-Luciferase Reporter Assay
System (Promega Corporation) and normalized against Renilla
luciferase activity.
Cell viability and proliferation
assays
HepG2 cell viability and proliferation were assessed
using CCK-8 (Dojindo Laboratories, Inc.) and EdU incorporation
assays (Beyotime Biotechnology). For CCK-8, absorbance was measured
at 450 nm at the indicated time points (0, 24, 48, 72 and 96 h).
For the EdU assay, cells were incubated with EdU at a final
concentration of 10 µM at 37°C for 2 h. Cells were then fixed with
4% paraformaldehyde at 25°C for 15 min and permeabilized with PBS
containing 0.3% Triton X-100 at 25°C for 15 min. EdU staining was
performed using the Click reaction cocktail at 25°C for 30 min in
the dark, and nuclei were counterstained with DAPI at 25°C for 3
min, followed by three washes with PBS (5 min each).
EdU+ cells were observed and counted under a
fluorescence microscope (Olympus Corporation) according to the
manufacturer's instructions.
Cell migration and invasion
assays
Migration and invasion were evaluated using 24-well
Transwell chambers (pore size, 8 µm; Corning, Inc.) coated with or
without Matrigel (BD Biosciences), respectively. For the invasion
assay, Matrigel (BD Biosciences) was thawed at 4°C overnight and
diluted on ice, and 50 µl was added to the upper chamber and
allowed to polymerize at 37°C for 30 min. Cells were harvested and
resuspended in serum-free medium, and 1×105 cells in 200
µl were seeded into the upper chamber per well, whereas the lower
chamber was filled with 800 µl DMEM containing 10% FBS. The
chambers were then incubated at 37°C with 5% CO2 for 48
h, after which, the Transwell chambers were washed with PBS, fixed
with 4% paraformaldehyde for 20 min at 25°C and stained with 0.5%
crystal violet for 5 min at 25°C. Images of the cells were then
captured and the cells were counted under a fluorescence microscope
(Olympus Corporation) at ×200 magnification.
Measurement of glycolysis-related
metabolites
HepG2 cells were homogenized in PBS, followed by
centrifugation at 10,000 × g for 10 min at 4°C. The supernatants
were then collected, kept on ice and subsequently used for lactate
measurement with an L-lactate assay kit (cat. no. A019-2-1; Nanjing
Jiancheng Bioengineering Institute) according to the manufacturer's
protocol. Intracellular ATP levels were determined using an ATP
assay kit (cat. no. S0026; Beyotime Biotechnology) according to the
manufacturer's protocol. Glucose uptake was assessed using a
fluorescent glucose analog
[2-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino-2-deoxy-D-glucose
(2-NBDG); Invitrogen; Thermo Fisher Scientific, Inc.]. Cells were
incubated with 2.5 µg/ml 2-NBDG for 30 min at 37°C, washed with PBS
and analyzed using a NovoCyte flow cytometer (Agilent Technologies,
Inc.). Data were processed using NovoExpress software (version
1.5.0; Agilent Technologies, Inc.).
Measurement of extracellular
acidification rate (ECAR)
The ECAR, as an indicator of glycolytic activity,
was measured using a Seahorse XF8 extracellular flux analyzer
(Agilent Technologies, Inc.). Briefly, HepG2 cells from the
different treatment groups were seeded into XF8 cell culture
microplates at a density of 1×104 cells/well and allowed
to adhere for 12 h. The culture medium was then replaced with
Seahorse XF assay medium according to the manufacturer's
instructions, and cells were incubated at 37°C in a
non-CO2 incubator for 60 min to allow equilibration.
After baseline measurements were recorded, glucose (10 mM),
oligomycin (1 µM) and 2-deoxy-D-glucose (50 mM) were sequentially
injected at the indicated time points by the analyzer, and ECAR was
continuously monitored. ECAR data were analyzed using Wave software
(version 2.6; Agilent Technologies, Inc.) and used to evaluate
glycolytic function.
Rescue experiment
To validate that RAB7A mediates the effects of
miR-448, HepG2 cells were seeded to reach 70–80% confluence and
transfected with a RAB7A overexpression (OE) plasmid (RAB7A OE) or
the corresponding empty vector based on the pcDNA3.1 backbone
(Changsha Zeqiong Biotechnology Co., Ltd.) using Lipofectamine 3000
according to the manufacturer's instructions at 37°C. After 24 h of
transfection, cells were treated with EXO-miR-448 (25 µg/ml) at
37°C. After a further 48 h, cell proliferation, migration, invasion
and glycolytic activity were re-assessed as described in the
aforementioned methods.
Bioinformatics analysis
RAB7A expression in HCC tissues compared with that
in normal tissues was analyzed using The Cancer Genome Atlas
(TCGA)-Liver Hepatocellular Carcinoma dataset (https://portal.gdc.cancer.gov/). RNA-seq TPM
expression data from a total of 424 tumor samples and 50 adjacent
normal tissue samples were assessed. Among these samples, 50 normal
samples were paired with their corresponding tumor samples, whereas
the remaining tumor samples were unpaired. Potential downstream
target genes of miR-448, as well as the predicted miR-448 binding
site within the 3′-UTR of RAB7A, were identified using the
TargetScanHuman database (version 8.0; http://www.targetscan.org). Genes with high context
++ scores were selected for further analysis.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 9.0; Dotmatics). All data are presented as
the mean ± SD from ≥3 independent experiments. For paired data, a
paired Student's t-test was used. Unpaired Student's t-test or
one-way ANOVA followed by Tukey's post hoc test was used for
comparison among groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-448 expression is downregulated in
liver cancer cells and enriched in exosomes
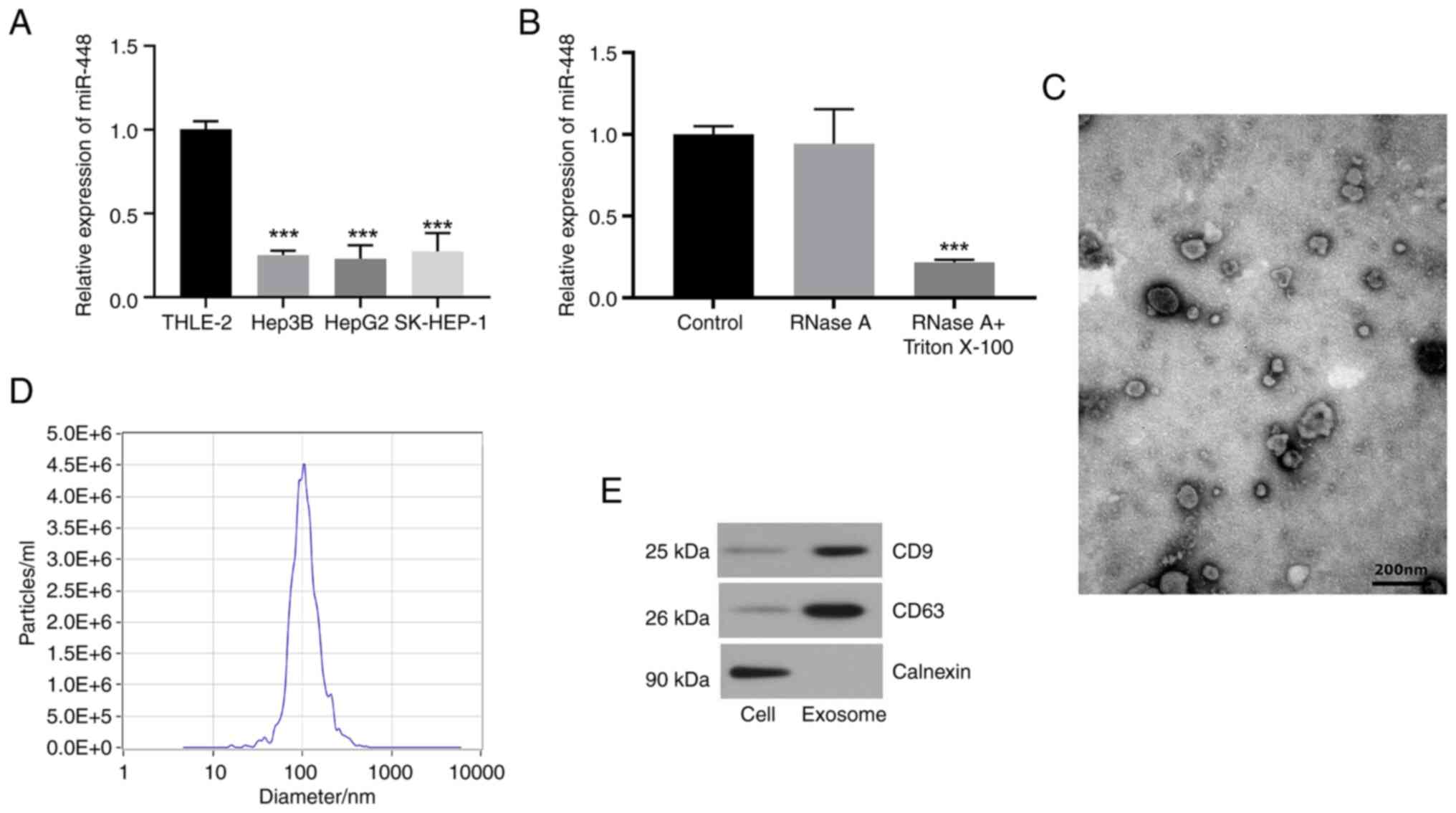
RT-qPCR analysis revealed that miR-448 expression
was significantly downregulated in liver cancer cell lines (HepG2,
Hep3B and SK-HEP-1) compared with in normal THLE-2 liver cells
(Fig. 1A). Subsequent experiments
involving RNase A treatment of the supernatant of HepG2 cells
revealed no significant alteration in miR-448 expression levels;
however, treatment with both RNase A and Triton X-100 resulted in a
significant reduction of miR-448 levels (Fig. 1B). These findings indicated that
miR-448 was predominantly contained within exosomes, rather than
being directly released into the extracellular milieu. Furthermore,
exosomes were isolated from the culture medium of HepG2 cells and
characterized by TEM and NTA, confirming their typical cup-shaped
morphology and a median particle size of ~104 nm, consistent with
the expected range for exosomes (Fig.
1C and D). Western blotting demonstrated successful exosome
isolation by detecting the exosomal markers CD63 and CD9, with
calnexin serving as a negative control (Fig. 1E). These results suggested that
miR-448 is not only downregulated in liver cancer cells but also
exists in exosomes, potentially serving a role in cell-to-cell
communication within the tumor microenvironment.
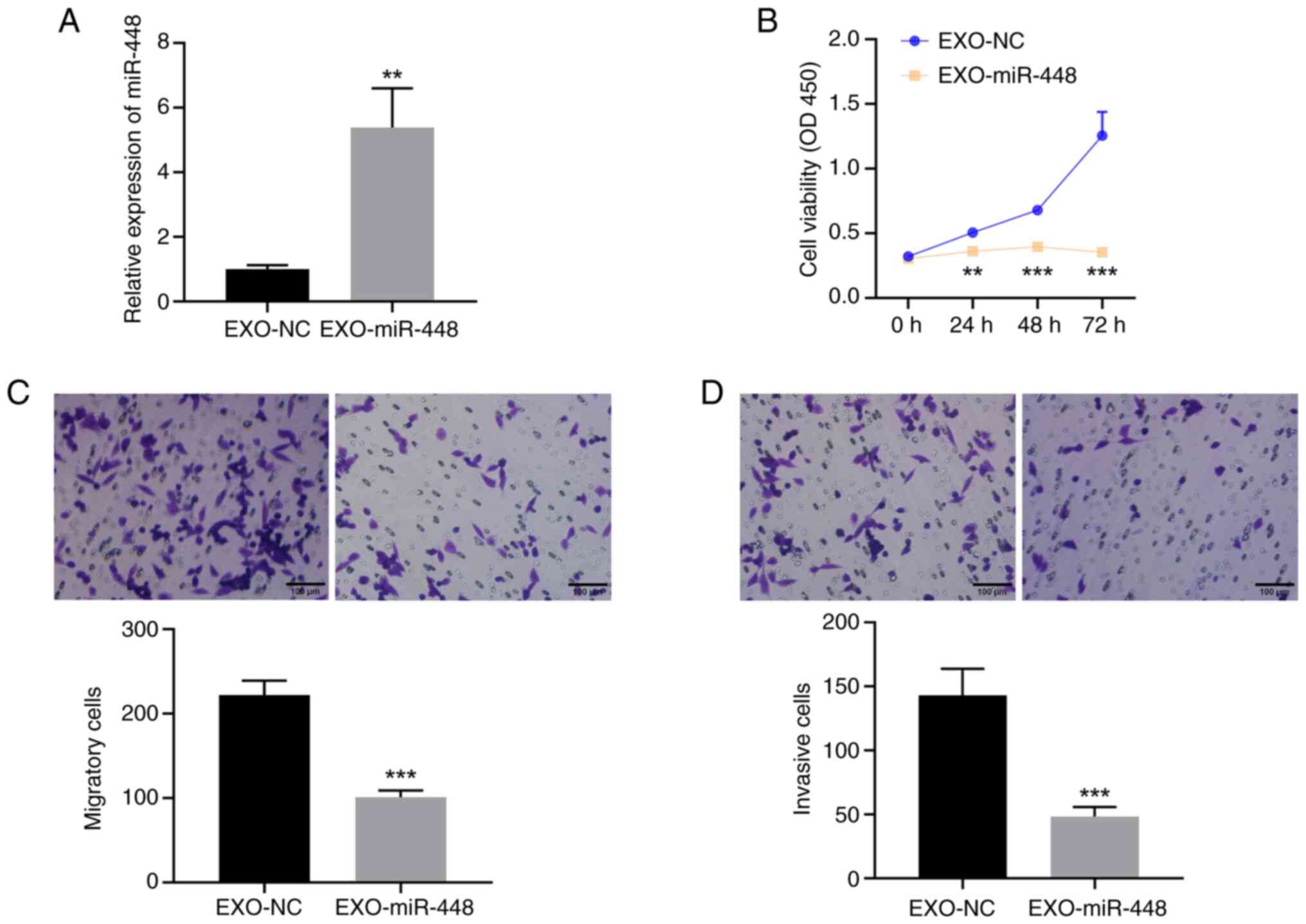
EXO-miR-448 inhibits liver cancer cell
proliferation, migration and invasion
To explore the functional role of miR-448 in liver
cancer, HepG2 cells were transfected with either miR-448 mimics or
mimics NC, and exosomes were collected and named EXO-miR-448 and
EXO-NC, respectively. The enrichment of miR-448 in exosomes was
confirmed by RT-qPCR (Fig. 2A). To
evaluate the biological effects of EXO-miR-448, cell viability,
migration and invasion were assessed in HepG2 cells treated with
EXO-miR-448 or EXO-NC. The CCK-8 assay (Fig. 2B) demonstrated that EXO-miR-448
significantly reduced cell viability compared with EXO-NC.
Furthermore, Transwell migration (Fig.
2C) and invasion assays (Fig.
2D) revealed that EXO-miR-448 treatment led to a significant
decrease in both migration and invasion of HepG2 cells compared
with those in the EXO-NC group. These results suggested that
EXO-miR-448 effectively inhibits the proliferation, migration and
invasion of liver cancer cells.
RAB7A is a direct downstream target of
EXO-miR-448 in liver cancer cells
To identify the downstream targets of miR-448,
bioinformatics analysis was performed and several potential target
genes were predicted, including RAB7A, TMS4BX, VASH2, SGPP1 and
MPC1, all of which exhibited relatively high binding scores
(Fig. 3A). To further concentrate
the candidates, RT-qPCR was performed in liver cancer cells treated
with EXO-miR-448 or EXO-NC to evaluate the mRNA expression levels
of these predicted targets. Although all five genes showed a modest
decrease following EXO-miR-448 treatment, RAB7A demonstrated the
most significant downregulation (Fig.
3B). According to previous studies, RAB7A is associated with
exosome trafficking, suggesting its potential functional relevance
(20,27,28).
Based on these findings, RAB7A was selected as the putative target
of miR-448 for further investigation. The predicted binding site of
miR-448 on the 3′-UTR of RAB7A is presented in Fig. 3C. A dual-luciferase reporter assay
demonstrated that transfection with miR-448 mimics significantly
decreased the luciferase activity of the RAB7A WT reporter
construct, indicating a direct interaction between miR-448 and
RAB7A (Fig. 3D). To confirm the
efficiency of RAB7A OE, western blot analysis was performed in
cells transfected with the RAB7A OE construct alone. The results
demonstrated a significant increase in RAB7A protein levels
compared with those in the vector group (Fig. 3E), confirming successful
transfection. Furthermore, treatment with EXO-miR-448 suppressed
RAB7A protein expression compared with that in the EXO-NC + vector
group (Fig. 3F).
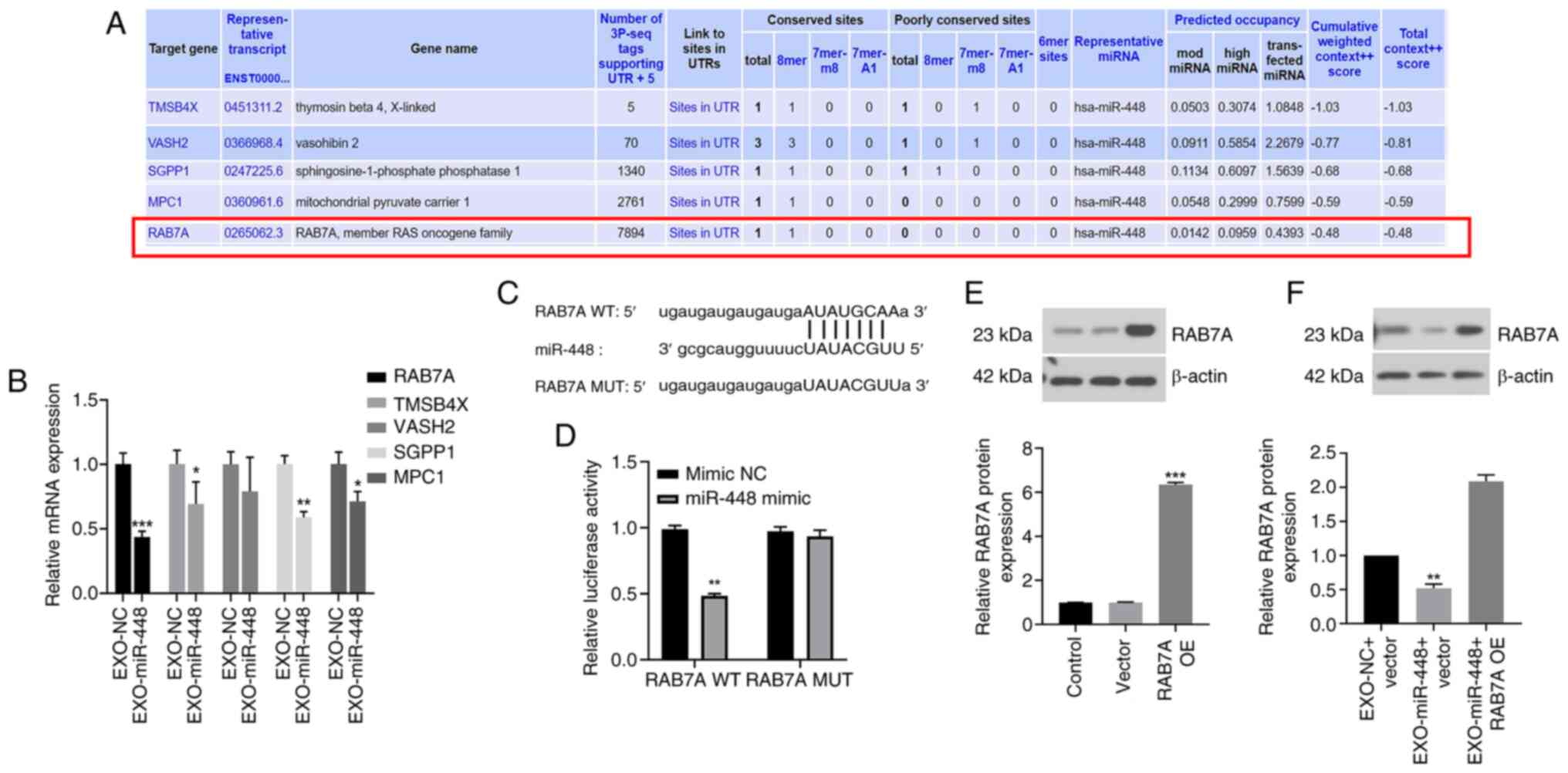 | Figure 3.RAB7A as a direct downstream target
of miR-448 in liver cancer. (A) Bioinformatics analysis predicted
potential downstream targets of miR-448, including RAB7A, TMSB4X,
VASH2, SGPP1 and MPC1. (B) Relative mRNA expression levels of
candidate targets in HepG2 cells treated with EXO-miR-448 or
EXO-NC, assessed using reverse transcription-quantitative PCR.
*P<0.05, **P<0.01 and ***P<0.001 vs. EXO-NC. (C) Predicted
binding sites of miR-448 in the 3′-UTR of RAB7A (WT and MUT). (D)
Dual-luciferase reporter assay validating the interaction between
miR-448 and RAB7A. **P<0.01 vs. mimic NC. (E) Western blotting
showing markedly increased RAB7A protein levels in HepG2 cells
transfected with the RAB7A OE construct alone. ***P<0.001 vs.
vector. (F) Western blotting showing reduced RAB7A protein levels
in HepG2 cells after EXO-miR-448 treatment. **P<0.01 vs. EXO-NC
+ vector. EXO, exosomal; miR, microRNA; MPC1, mitochondrial
pyruvate carrier 1; MUT, mutant; NC, negative control; OE,
overexpression; RAB7A, Ras-related protein Rab-7a; SGPP1,
sphingosine-1-phosphate phosphatase 1; TMSB4X, thymosin β 4
X-linked; VASH2, vasohibin 2; WT, wild-types. |
EXO-miR-448 inhibits liver cancer cell
proliferation, migration and invasion by targeting RAB7A
To further assess the functional role of EXO-miR-448
in liver cancer, its downstream target gene RAB7A was analyzed.
Data from TCGA indicated that RAB7A expression was significantly
higher in HCC tissues compared with that in normal liver tissues,
consistent with the present hypothesis that RAB7A serves a key role
in liver cancer progression (Fig. 4A
and B). Furthermore, western blotting and RT-qPCR analysis
demonstrated that RAB7A expression was significantly upregulated in
different liver cancer cells compared with in THLE-2 normal liver
cells (Fig. 4C and D). To assess
this, RAB7A expression was silenced in liver cancer cells using
siRNA targeting RAB7A, and successful knockdown was confirmed using
RT-qPCR and western blotting (Fig. 4E
and F). Subsequently, the biological effects of RAB7A silencing
on liver cancer cells were evaluated. The results revealed that
RAB7A knockdown significantly reduced cell proliferation, migration
and invasion, as assessed using CCK-8, EdU and Transwell migration
and invasion assays, respectively (Fig.
4G-J).
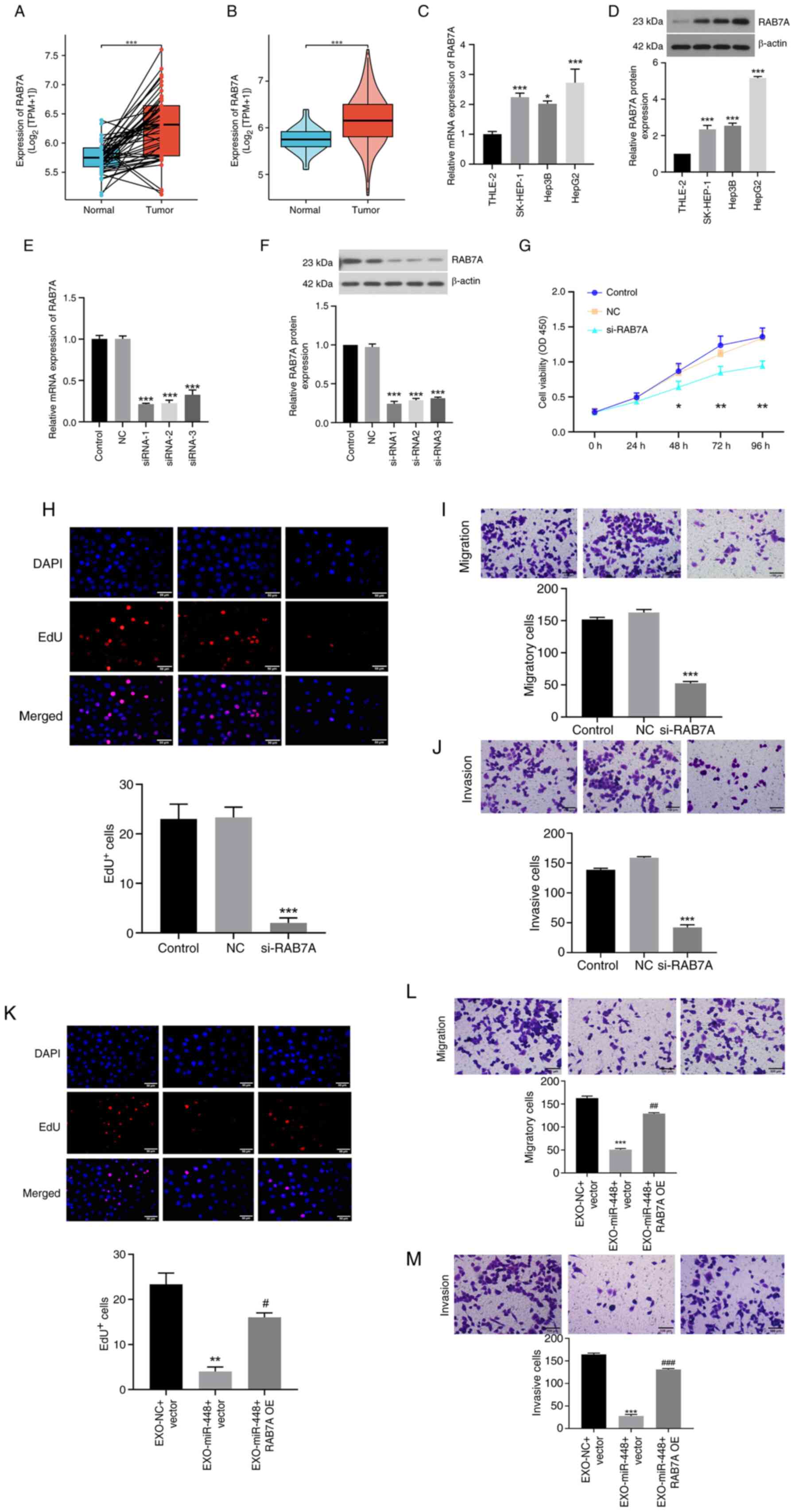 | Figure 4.EXO-miR-448 inhibits liver cancer
cell proliferation, migration and invasion by targeting RAB7A. (A)
Paired comparison of RAB7A expression between HCC tumor tissues and
their matched adjacent normal tissues (n=50 pairs). ***P<0.001.
(B) Unpaired comparison of all HCC tumor samples (n=424) vs. normal
liver tissues (n=50). ***P<0.001. (C) RAB7A mRNA expression in
normal liver cells (THLE-2) and liver cancer cell lines (HepG2,
Hep3B and SK-HEP-1). *P<0.05 and ***P<0.001 vs. THLE-2. (D)
RAB7A protein expression in normal liver cells (THLE-2) and liver
cancer cell lines (HepG2, Hep3B and SK-HEP-1). ***P<0.001 vs.
THLE-2. (E) Validation of RAB7A knockdown efficiency in HepG2 cells
using reverse transcription-quantitative PCR. ***P<0.001 vs. NC.
(F) Validation of RAB7A knockdown efficiency in HepG2 cells using
western blotting. ***P<0.001 vs. NC. (G) Cell Counting Kit-8
assay showing reduced proliferation of HepG2 cells after RAB7A
knockdown. *P<0.05 and **P<0.01 vs. NC. (H) EdU assay showing
reduced proliferation of HepG2 cells after RAB7A knockdown (scale
bar, 50 µm). ***P<0.001 vs. NC. (I) Transwell migration assay
showing suppressed migratory ability of HepG2 cells following RAB7A
knockdown (scale bar, 100 µm). ***P<0.001 vs. NC. (J) Transwell
invasion assay showing suppressed invasive ability of HepG2 cells
following RAB7A knockdown (scale bar, 100 µm). ***P<0.001 vs.
NC. (K) Rescue experiment showing that RAB7A OE reverses the
inhibitory effect of EXO-miR-448 on HepG2 cell proliferation. scale
bar, 50 µm. **P<0.01 vs. EXO-NC + vector; #P<0.05
vs. EXO-miR-448 + vector. (L) Rescue experiment showing that RAB7A
OE reverses the inhibitory effect of EXO-miR-448 on HepG2 cell
migration. Scale bar, 100 µm. ***P<0.001 vs. EXO-NC + vector;
##P<0.01 vs. EXO-miR-448 + vector. (M) Rescue
experiment showing that RAB7A OE reverses the inhibitory effect of
EXO-miR-448 on HepG2 cell invasion. Scale bar, 100 µm.
***P<0.001 vs. EXO-NC + vector; ###P<0.001 vs.
EXO-miR-448 + vector. EXO, exosomal; HCC, hepatocellular carcinoma;
miR, microRNA; NC, negative control; OE, overexpression; RAB7A,
Ras-related protein Rab-7a; siRNA, small interfering RNA; TPM,
transcripts per million. |
To further explore the impact of EXO-miR-448 on
RAB7A-regulated cell behavior, RAB7A was overexpressed in liver
cancer cells and the results demonstrated that RAB7A OE rescued the
inhibitory effects of EXO-miR-448 on cell proliferation, migration
and invasion (Fig. 4K-M). These
findings suggested that EXO-miR-448 may suppress liver cancer cell
proliferation, migration and invasion by directly targeting
RAB7A.
EXO-miR-448 inhibits glycolysis in
liver cancer cells by targeting RAB7A
To further investigate the mechanism through which
EXO-miR-448 affects liver cancer progression, the potential role of
RAB7A in regulating glycolysis was explored in liver cancer cells.
Glycolysis is a key metabolic pathway that is often upregulated in
cancer cells, including liver cancer, to support rapid cell
proliferation and metastasis (29,30).
To evaluate the effect of EXO-miR-448 on glycolysis in liver cancer
cells, HepG2 cells were treated with EXO-miR-448.
Glycolysis-associated indicators, including lactate production,
glucose uptake and ATP levels, were significantly decreased
following EXO-miR-448 treatment (Fig.
5A-C). Furthermore, ECAR, an indicator of glycolytic activity
and capacity, was markedly reduced upon EXO-miR-448 exposure
(Fig. 5D), further supporting the
inhibitory effect of EXO-miR-448 on glycolysis in liver cancer
cells. Furthermore, silencing RAB7A expression using siRNA resulted
in a similar decrease in glycolytic activity, including
significantly reduced lactate production, ATP levels, glucose
uptake and ECAR, suggesting that RAB7A is involved in the
regulation of glycolysis (Fig.
5E-H). Notably, RAB7A OE reversed the inhibitory effects of
EXO-miR-448 on glycolysis, as demonstrated by the significant
restoration of lactate production, glucose uptake, ATP levels and
ECAR (Fig. 5I-L). Overall, the
findings indicated that EXO-miR-448 may regulate glycolysis in
liver cancer cells through the direct targeting of RAB7A and this
mechanism may contribute to the antitumor effects of miR-448 in
liver cancer.
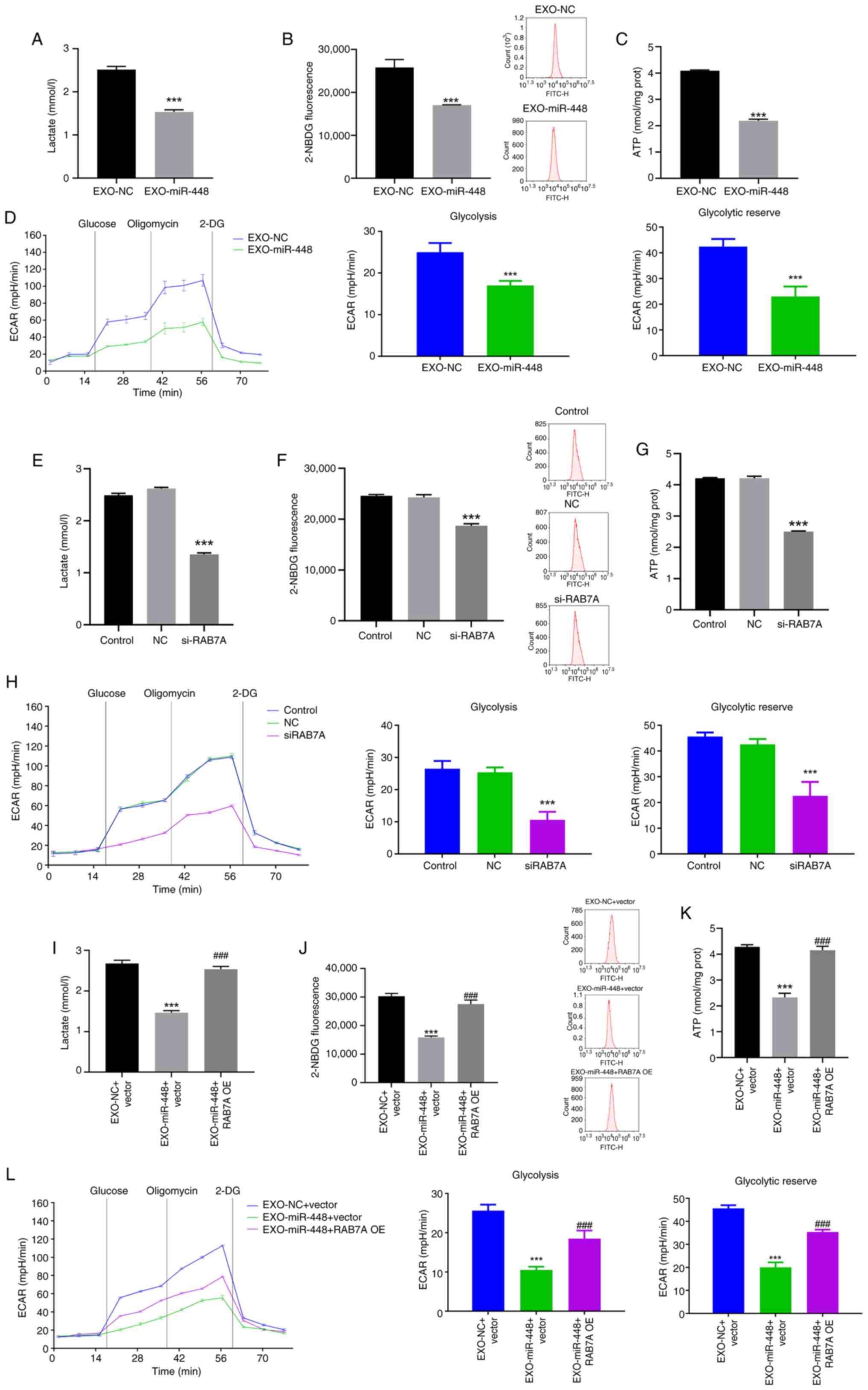 | Figure 5.EXO-miR-448 suppresses liver cancer
progression by targeting RAB7A to inhibit glycolysis. (A) Lactate
production in HepG2 cells treated with EXO-miR-448 or EXO-NC,
determined by a colorimetric assay. ***P<0.001 vs. EXO-NC. (B)
Glucose uptake in HepG2 cells treated with EXO-miR-448 or EXO-NC,
assessed using the 2-NBDG assay. ***P<0.001 vs. EXO-NC. (C)
Intracellular ATP levels in HepG2 cells treated with EXO-miR-448 or
EXO-NC, determined using an ATP assay kit. ***P<0.001 vs.
EXO-NC. (D) ECAR in HepG2 cells treated with EXO-miR-448 or EXO-NC,
measured using a Seahorse XF Analyzer. ***P<0.001 vs. EXO-NC.
(E) Lactate production in HepG2 cells transfected with si-RAB7A or
control siRNA. ***P<0.001 vs. NC. (F) Glucose uptake in HepG2
cells transfected with si-RAB7A or control siRNA. ***P<0.001 vs.
NC. (G) Intracellular ATP levels in HepG2 cells transfected with
si-RAB7A or control siRNA. ***P<0.001 vs. NC. (H) ECAR in HepG2
cells transfected with si-RAB7A or control siRNA. ***P<0.001 vs.
NC. (I) Lactate production in HepG2 cells following EXO-miR-448
treatment with or without RAB7A OE. ***P<0.001 vs. EXO-NC +
vector; ###P<0.001 vs. EXO-miR-448 + vector. (J)
Glucose uptake in HepG2 cells following EXO-miR-448 treatment with
or without RAB7A OE. ***P<0.001 vs. EXO-NC + vector;
###P<0.001 vs. EXO-miR-448 + vector. (K) ATP levels
in HepG2 cells following EXO-miR-448 treatment with or without
RAB7A OE. ***P<0.001 vs. EXO-NC + vector;
###P<0.001 vs. EXO-miR-448 + vector. (L) ECAR
analysis showing glycolytic activity in HepG2 cells following
EXO-miR-448 treatment with or without RAB7A OE. ***P<0.001 vs.
EXO-NC + vector; ###P<0.001 vs. EXO-miR-448 + vector.
miR, microRNA; NC, negative control; ECAR, extracellular
acidification rate; si, small interfering RNA; RAB7A, Ras-related
protein Rab-7a; NBDG,
N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino-2-deoxy-D-glucose; EXO,
exosomal; OE, overexpression; 2-DG, 2-deoxy-D-glucose. |
Discussion
Liver cancer remains one of the most aggressive
malignancies. As HCC accounts for 75–85% of primary liver cancer
cases, its clinical characteristics largely define the behavior of
liver cancer. Notably, the 5-year recurrence rate of HCC after
curative resection reaches 40–70%, and long-term survival remains
poor, underscoring its high recurrence, metastatic potential and
resistance to therapy (31–33). Despite advancements in targeted
therapies, prognosis remains poor due to the complexity of its
pathogenesis and the lack of effective early detection methods
(34,35). Altered metabolism, particularly the
upregulation of glycolysis (the Warburg effect), is a hallmark of
cancer cells and enables them to meet the bioenergetic and
biosynthetic demands of rapid proliferation and metastasis, a
feature particularly prominent in liver cancer (36). The aim of the present study was to
assess the role of EXO-miR-448 in liver cancer progression,
focusing on its potential to target RAB7A and regulate
glycolysis.
The results of the present study demonstrated that
miR-448 was significantly downregulated in liver cancer cell lines
compared with in normal liver cells, consistent with previous
studies demonstrating reduced miR-448 expression in HCC tissues and
its association with hepatocarcinogenesis (9,37).
miRNAs can be actively exported via extracellular vesicles, with
exosomes serving as predominant carriers for intercellular transfer
of bioactive molecules (38).
Aberrant expression levels of exosomal miRNAs have been implicated
in multiple malignancies, including lung, breast, colorectal,
liver, gastric and pancreatic cancer, where they regulate key
tumor-associated processes such as proliferation, angiogenesis,
metastasis and chemoresistance (39). The present study revealed that
miR-448 was markedly packaged into exosomes derived from HepG2
cells. Functionally, miR-448 has been recognized as a tumor
suppressor in several cancer types (40,41),
including non-small-cell lung cancer, where it targets sirtuin 1 to
inhibit proliferation, migration and epithelial-mesenchymal
transition (42). Consistently, the
results of the present study indicated that EXO-miR-448 may
suppress the proliferation, migration and invasion of liver cancer
cells.
A key finding of the present study was that miR-448
targets RAB7A, a small GTPase involved in endosomal maturation,
autophagy and intracellular trafficking. Beyond its role in
vesicular trafficking and autophagy, RAB7A has been reported to
promote tumor metastasis through multiple mechanisms in different
malignancies. In HCC, RAB7A OE activates the PI3K/AKT signaling
pathway and induces epithelial-mesenchymal transition, thereby
enhancing migration and invasion (43). In breast cancer, RAB7A knockdown
significantly suppresses proliferation, migration and xenograft
tumor growth, confirming its essential contribution to metastatic
behavior (44). In melanoma, RAB7A
interacts with the endolysosomal channel two pore segment channel 2
to modulate the GSK-3β/β-catenin/microphthalmia-associated
transcription factor axis, which drives invasive growth and distant
metastasis (45). In colon
adenocarcinoma, elevated RAB7A expression has been reported to be
associated with increased tumor size, invasion depth, lymph node
metastasis and poor overall survival (22). Furthermore, RAB7A upregulation by
oncogenic miR-3200 has been reported to promote telomere remodeling
and activate mTOR and autophagy-associated pathways, further
supporting its role as a facilitator of malignant progression and
dissemination in liver cancer (46). Using dual-luciferase reporter
assays, the results of the present study demonstrated that miR-448
can bind to the 3′-UTR of RAB7A, and EXO-miR-448 treatment was
further shown to significantly reduce RAB7A mRNA and protein
expression. Functional assays further revealed that RAB7A knockdown
significantly suppressed cell proliferation, migration and
invasion, whereas RAB7A OE rescued the inhibitory effects of
EXO-miR-448, indicating that suppression of RAB7A is a key
mechanism by which EXO-miR-448 exerts its tumor-suppressive effects
in liver cancer.
Notably, the present study revealed that EXO-miR-448
may inhibit glycolysis in liver cancer cells, as demonstrated by
significantly reduced lactate production, glucose uptake, ATP
levels and ECAR. Consistently, a previous study reported that
miR-448 suppresses HCC cell viability and glycolytic activity by
directly targeting insulin-like growth factor-1 receptor, leading
to decreased glucose uptake, lactate production and ATP generation
(47). In the present study,
silencing RAB7A produced similar effects, supporting its role as a
mediator of EXO-miR-448 driven metabolic suppression. Furthermore,
RAB7A OE reversed the inhibitory effects of EXO-miR-448 on
glycolysis, restoring lactate production, glucose uptake, ATP
levels and ECAR, thereby further demonstrating the functional
association between miR-448 and RAB7A in regulating cancer cell
metabolism. These findings are further supported by previous
studies that demonstrated RAB7A enhances aerobic glycolysis and
yes-associated protein 1-driven transcriptional activity to promote
HCC growth and metastasis (25,48,49).
Furthermore, mechanistic evidence from lipopolysaccharide-activated
macrophages indicates that phosphorylation of RAB7A at Ser72
disrupts EGFR endocytosis and sustains glycolytic flux via the
pyruvate kinase M2/hypoxia-inducible factor-1α pathway, while
disruptions in vesicular trafficking have similarly been reported
to alter cellular energy metabolism (50). Notably, RAB7A dysregulation among
autophagy-associated genes affected by vacuolar protein sorting 51
deficiency can lead to enhanced glycolysis and impaired oxidative
phosphorylation (51). Overall, the
ability of RAB7A to coordinate vesicular trafficking, autophagy,
nutrient sensing and metabolic stress adaptation may underlie its
dual regulatory roles in tumor metabolism and metastatic
progression (23,52,53).
Recent studies have further highlighted the
intersection of glycolysis, exosome biology and RAB7A in cancer.
Lactate accumulation has been reported to drive HCC metastasis by
enhancing exosome biogenesis via RAB7A lactylation, thereby
associating glycolytic metabolism with RAB7A-mediated vesicular
trafficking (54). Furthermore,
bone marrow stromal cell-derived exosomal miR-196a-5p has been
reported to suppress glycolysis and leukemia progression by
targeting 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3,
underscoring the role of exosome-delivered miRNAs in metabolic
regulation (55). The findings of
the present study fit within this framework, identifying miR-448 as
a novel regulator that inhibits RAB7A-mediated glycolysis via
exosomal delivery, thereby attenuating liver cancer.
Nevertheless, the present study had certain
limitations. For example, it is limited by its exclusive use of
in vitro assays. While RAB7A OE rescue experiments
strengthen the functional interpretation of the miR-448/RAB7A axis,
the lack of in vivo validation precludes direct conclusions
regarding therapeutic efficacy. Future research should incorporate
animal models or patient-derived xenografts to confirm in
vivo relevance and explore the translational potential of
miR-448 mimics or RAB7A inhibitors in liver cancer.
In conclusion, the results of the present study
revealed that EXO-miR-448 may inhibit glycolysis and suppress
malignant phenotypes in liver cancer cells by directly targeting
RAB7A. These findings provide novel mechanistic insights into
miRNA-mediated metabolic regulation in liver cancer and suggest
that modulation of the miR-448/RAB7A axis could be a potential
therapeutic approach, pending validation in in vivo models
in the future.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science
Foundation of China (grant no. 82260125), the Zhongyuan Thousand
Talent Program-Youth Outstanding Talent (grant no. 2018), the
Research Program of the Education Department of Henan Province
(grant no. 21A320001), the Research Program of the Health
Commission of Henan Province (grant no. YXKC2020043) and the Hainan
Provincial Natural Science Foundation of China (grant nos.
822QN473, 822MS181 and 823RC591).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YC, ZW, FC and WL contributed to the present study
conception and design, prepared materials, collected and analyzed
the data. YC and QW wrote the first draft of the manuscript, and QW
also participated in data organization and statistical analysis.
HW, TZ and MN substantially contributed to the acquisition,
analysis and interpretation of experimental data. ZW and WL confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The use of commercially obtained human cell lines
was approved by the Ethics Committee of The Second Affiliated
Hospital of Hainan Medical University (Haikou, China) and all
procedures were performed in accordance with institutional
guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Marengo A, Rosso C and Bugianesi E: Liver
cancer: Connections with obesity, fatty liver, and cirrhosis. Annu
Rev Med. 67:103–117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoshida Y, Fuchs BC, Bardeesy N, Baumert
TF and Chung RT: Pathogenesis and prevention of hepatitis C
virus-induced hepatocellular carcinoma. J Hepatol. 61 (Suppl
1):S79–S90. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alawyia B and Constantinou C:
Hepatocellular carcinoma: A narrative review on current knowledge
and future prospects. Curr Treat Options Oncol. 24:711–724. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khameneh SC, Razi S, Lashanizadegan R,
Akbari S, Sayaf M, Haghani K and Bakhtiyari S: MicroRNA-mediated
metabolic regulation of immune cells in cancer: An updated review.
Front Immunol. 15:14249092024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu X, Li Y, Zhang R, Chen X, Shen J, Yuan
M, Chen Y, Chen M, Liu S, Wu J and Sun Q: Jianpi Yangzheng
decoction suppresses gastric cancer progression via modulating the
miR-448/CLDN18.2 mediated YAP/TAZ signaling. J Ethnopharmacol.
311:1164502023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu H, Zhou X, Ma C, Chang H, Li H, Liu F
and Lu J: Low expression of miR-448 induces EMT and promotes
invasion by regulating ROCK2 in hepatocellular carcinoma. Cell
Physiol Biochem. 36:487–498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin Z, Zhu Y and Liu Y: The role of
miR-448 in cancer progression and its potential therapeutic
applications. J Cancer Res Clin Oncol. 143:679–687. 2017.
|
|
11
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Y, Lin F, Wan T, Chen A, Wang H,
Jiang B, Zhao W, Liao S, Wang S, Li G, et al: ZEB1 enhances Warburg
effect to facilitate tumorigenesis and metastasis of HCC by
transcriptionally activating PFKM. Theranostics. 11:5926–5938.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Ouyang F, Gao A, Zeng T, Li M, Li
H, Zhou W, Gao Q, Tang X, Zhang Q, et al: ESM1 enhances fatty acid
synthesis and vascular mimicry in ovarian cancer by utilizing the
PKM2-dependent Warburg effect within the hypoxic tumor
microenvironment. Mol Cancer. 23:942024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jing Z, Liu Q, He X, Jia Z, Xu Z, Yang B
and Liu P: NCAPD3 enhances Warburg effect through c-myc and E2F1
and promotes the occurrence and progression of colorectal cancer. J
Exp Clin Cancer Res. 41:1982022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng L, Zhao Y, Tan J, Hou J, Jin X, Liu
DX, Huang B and Lu J: PRMT1 promotes Warburg effect by regulating
the PKM2/PKM1 ratio in non-small cell lung cancer. Cell Death Dis.
15:5042024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Paul S, Ghosh S and Kumar S: Tumor
glycolysis, an essential sweet tooth of tumor cells. Semin Cancer
Biol. 86:1216–1230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lan F, Qin Q, Yu H and Yue X: Effect of
glycolysis inhibition by miR-448 on glioma radiosensitivity. J
Neurosurg. 132:1456–1464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luce A, Lombardi A, Ferri C, Zappavigna S,
Tathode MS, Miles AK, Boocock DJ, Vadakekolathu J, Bocchetti M,
Alfano R, et al: A proteomic approach reveals that miR-423-5p
modulates glucidic and amino acid metabolism in prostate cancer
cells. Int J Mol Sci. 24:6172022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chu M, Zhao Y, Feng Y, Zhang H, Liu J,
Cheng M, Li L, Shen W, Cao H, Li Q and Min L: MicroRNA-126
participates in lipid metabolism in mammary epithelial cells. Mol
Cell Endocrinol. 454:77–86. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guerra F and Bucci C: Role of the RAB7
protein in tumor progression and cisplatin chemoresistance. Cancers
(Basel). 11:10962019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ju L, Luo Y, Cui X, Zhang H, Chen L and
Yao M: CircGPC3 promotes hepatocellular carcinoma progression and
metastasis by sponging miR-578 and regulating RAB7A/PSME3
expression. Sci Rep. 14:76322024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shan Z, Chen X, Chen H and Zhou X:
Investigating the impact of Ras-related protein RAB7A on colon
adenocarcinoma behavior and its clinical significance. Biofactors.
51:e700062025. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Q, Bai Y, Shi X, Guo D, Wang Y, Wang
Y, Guo WZ and Zhang S: High RAS-related protein Rab-7a (RAB7A)
expression is a poor prognostic factor in pancreatic
adenocarcinoma. Sci Rep. 12:174922022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Güleç Taşkıran AE, Hüsnügil HH, Soltani
ZE, Oral G, Menemenli NS, Hampel C, Huebner K, Erlenbach-Wuensch K,
Sheraj I, Schneider-Stock R, et al: Post-transcriptional regulation
of Rab7a in lysosomal positioning and drug resistance in
nutrient-limited cancer cells. Traffic. 25:e129562024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang JY, Zhu X, Liu Y and Wu X: The
prognostic biomarker RAB7A promotes growth and metastasis of liver
cancer cells by regulating glycolysis and YAP1 activation. J Cell
Biochem. 125:e306212024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ostrowski M, Carmo NB, Krumeich S, Fanget
I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et
al: Rab27a and Rab27b control different steps of the exosome
secretion pathway. Nat Cell Biol. 12:19–30. 1–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mulligan RJ and Winckler B: Regulation of
endosomal trafficking by Rab7 and its effectors in neurons: Clues
from charcot-marie-tooth 2B disease. Biomolecules. 13:13992023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, Dai
W and Guo C: Emerging roles and the regulation of aerobic
glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res.
39:1262020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wen T, Jin C, Facciorusso A, Donadon M,
Han HS, Mao Y, Dai C, Cheng S, Zhang B, Peng B, et al:
Multidisciplinary management of recurrent and metastatic
hepatocellular carcinoma after resection: An international expert
consensus. Hepatobiliary Surg Nutr. 7:353–371. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abdelhamed W and El-Kassas M:
Hepatocellular carcinoma recurrence: Predictors and management.
Liver Res. 7:321–332. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI
|
|
34
|
Shin H and Yu SJ: A concise review of
updated global guidelines for the management of hepatocellular
carcinoma: 2017–2024. J Liver Cancer. 25:19–30. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang F, Hilakivi-Clarke L, Shaha A, Wang
Y, Wang X, Deng Y, Lai J and Kang N: Metabolic reprogramming and
its clinical implication for liver cancer. Hepatology.
78:1602–1624. 2023.PubMed/NCBI
|
|
37
|
Katayama Y, Maeda M, Miyaguchi K, Nemoto
S, Yasen M, Tanaka S, Mizushima H, Fukuoka Y, Arii S and Tanaka H:
Identification of pathogenesis-related microRNAs in hepatocellular
carcinoma by expression profiling. Oncol Lett. 4:817–823. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li B, Cao Y, Sun M and Feng H: Expression,
regulation, and function of exosome-derived miRNAs in cancer
progression and therapy. FASEB J. 35:e219162021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu X, Tang H, Liu G, Wang H, Shu J and Sun
F: miR-448 suppressed gastric cancer proliferation and invasion by
regulating ADAM10. Tumour Biol. 37:10545–10551. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bamodu OA, Huang WC, Lee WH, Wu A, Wang
LS, Hsiao M, Yeh CT and Chao TY: Aberrant KDM5B expression promotes
aggressive breast cancer through MALAT1 overexpression and
downregulation of hsa-miR-448. BMC Cancer. 16:1602016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qi H, Wang H and Pang D: miR-448 promotes
progression of non-small-cell lung cancer via targeting SIRT1. Exp
Ther Med. 18:1907–1913. 2019.PubMed/NCBI
|
|
43
|
Liu Y, Ma J, Wang X, Liu P, Cai C, Han Y,
Zeng S, Feng Z and Shen H: Lipophagy-related gene RAB7A is involved
in immune regulation and malignant progression in hepatocellular
carcinoma. Comput Biol Med. 158:1068622023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xie J, Yan Y, Liu F, Kang H, Xu F, Xiao W,
Wang H and Wang Y: Knockdown of Rab7a suppresses the proliferation,
migration, and xenograft tumor growth of breast cancer cells.
Biosci Rep. 39:BSR201804802019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Abrahamian C, Tang R, Deutsch R, Ouologuem
L, Weiden EM, Kudrina V, Blenninger J, Rilling J, Feldmann C, Kuss
S, et al: Rab7a is an enhancer of TPC2 activity regulating melanoma
progression through modulation of the GSK3β/β-Catenin/MITF-axis.
Nat Commun. 15:100082024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Song S, Xie S, Liu X, Li S, Wang L, Jiang
X and Lu D: miR-3200 accelerates the growth of liver cancer cells
by enhancing Rab7A. Noncoding RNA Res. 8:675–685. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y, Chen X, Yao N, Gong J, Cao Y, Su
X, Feng X and Tao M: MiR-448 suppresses cell proliferation and
glycolysis of hepatocellular carcinoma through inhibiting IGF-1R
expression. J Gastrointest Oncol. 13:355–367. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Qi C, Zou L, Wang S, Mao X, Hu Y, Shi J,
Zhang Z and Wu H: Vps34 inhibits hepatocellular carcinoma invasion
by regulating endosome-lysosome trafficking via Rab7-RILP and
Rab11. Cancer Res Treat. 54:182–198. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang CC, Meng GX, Dong ZR and Li T: Role
of Rab GTPases in hepatocellular carcinoma. J Hepatocell Carcinoma.
8:1389–1397. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang X, Chen C, Ling C, Luo S, Xiong Z,
Liu X, Liao C, Xie P, Liu Y, Zhang L, et al: EGFR tyrosine kinase
activity and Rab GTPases coordinate EGFR trafficking to regulate
macrophage activation in sepsis. Cell Death Dis. 13:9342022.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Aygun D and Yücel Yılmaz D: From gene to
pathways: Understanding novel Vps51 variant and its cellular
consequences. Int J Mol Sci. 26:57092025. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Edinger AL, Cinalli RM and Thompson CB:
Rab7 prevents growth factor-independent survival by inhibiting
cell-autonomous nutrient transporter expression. Dev Cell.
5:571–582. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Deffieu MS, Cesonyte I, Delalande F,
Boncompain G, Dorobantu C, Song E, Lucansky V, Hirschler A,
Cianferani S, Perez F, et al: Rab7-harboring vesicles are carriers
of the transferrin receptor through the biosynthetic secretory
pathway. Sci Adv. 7:eaba78032021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jiang C, He X, Chen X, Huang J, Liu Y,
Zhang J, Chen H, Sui X, Lv X, Zhao X, et al: Lactate accumulation
drives hepatocellular carcinoma metastasis through facilitating
tumor-derived exosome biogenesis by Rab7A lactylation. Cancer Lett.
627:2176362025. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fan B, Wang L, Hu T, Zheng L and Wang J:
Exosomal miR-196a-5p secreted by bone marrow mesenchymal stem cells
inhibits ferroptosis and promotes drug resistance of acute myeloid
leukemia. Antioxid Redox Signal. 42:933–953. 2025. View Article : Google Scholar : PubMed/NCBI
|