Introduction
Prostate cancer (PCa) presents a major global health
concern for men (1). The widely
used biomarker, prostate-specific antigen (PSA), however, lacks the
specificity to reliably distinguish cancer from benign conditions,
such as benign prostatic hyperplasia and prostatitis. Consequently,
elevated PSA levels frequently lead to unnecessary prostate biopsy,
which are invasive procedures associated with potential
complications including infection, bleeding, and significant
patient anxiety (2). This
difficulty highlights a pressing need for identifying biomarkers
that better reflect the underlying tumor pathophysiology.
Circular (circ)RNAs, a class of non-coding RNAs that
have been demonstrated to fulfil several key regulatory roles
(3,4), are promising candidates as biomarkers
due to their unique stability and presence in biological samples
(5–7). Specifically, their covalently closed
loop structure renders them resistant to exonuclease-mediated
degradation, making them ideal candidates for liquid biopsy.
Compared with traditional tissue biopsy, liquid biopsies based on
stable circulating biomarkers offer a non-invasive, repeatable, and
real-time method to monitor tumor dynamics, which is crucial for
early detection and disease management.
Hsa_circ_0006168 has previously been reported as an
oncogene in esophageal squamous cell carcinoma (ESCC) and
glioblastoma (8,9). Therefore, it was hypothesized that it
might also be dysregulated in PCa. The combination of data-driven
evidence from PCa and functional reports in other cancers
highlighted a critical research gap. Despite the growing interest
in circRNAs (3–7), the specific expression profile and
diagnostic potential of hsa_circ_0006168 in the context of
prostatic malignancies remain largely unexplored. Therefore, the
present study aimed to investigate the role and clinical
significance of hsa_circ_0006168 in the pathophysiology of PCa. The
present study is the first, to the best of our knowledge, to assess
the expression level of hsa_circ_0006168 in serum samples from
patients with PCa to explore its clinical significance as a
biomarker for early diagnosis and prognosis evaluation.
Materials and methods
Patients and serum samples
The present study was approved by the Ethics
Committee of Nantong First People's Hospital (Nantong, China;
approval no. 2023-KY007-1). All patients provided written informed
consent to participate specifically in the present research. A
total of 90 patients with PCa from Nantong First People's Hospital,
who were treated between October 2022 and May 2023, were recruited
into the PCa group. The inclusion criteria were as follows: i)
Confirmed diagnosis of PCa via biopsy; ii) first-time treatment of
the patient, undergoing radical prostatectomy or other treatments,
such as radiotherapy or chemotherapy; and iii) complete clinical
data available. The exclusion criteria included the following: i)
Diagnosis of other concurrent malignancies; ii) diagnosis of
immune, infectious or acute diseases; iii) inadequate organ
function; and iv) incomplete clinical data available. Additionally,
30 patients with benign prostatic hyperplasia (BPH) and 60
age-matched healthy individuals undergoing routine physical
examination were enrolled as the BPH group and control group,
respectively, during the same period. For the BPH group, inclusion
criteria were: i) Clinically confirmed diagnosis of BPH. Exclusion
criteria were: i) Evidence or suspicion of PCa (confirmed via
histopathology or clinical assessment); ii) History of any
malignancy; and iii) incomplete clinical data. For the control
group, Inclusion criteria were: i) Age-matched individuals
undergoing routine physical examination. Exclusion criteria were:
i) History of malignancy; ii) known urological diseases; and iii)
abnormal PSA levels or Digital Rectal Exam (DRE) results.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) assay
Patient blood samples were collected in the morning
after fasting, and 4 ml venous blood was drawn. The samples were
subsequently centrifuged at 2,000 × g at 4°C for 10 min, and the
supernatant was collected and placed in RNase-free microcentrifuge
tubes. These tubes were stored at −80°C for later use. Total RNA
was extracted using a plasma RNA extraction kit (BioTeke
Corporation) and diluted with 30 µl nuclease-free water per sample.
Total RNA from the RWPE-1, LNCaP, DU145 and PC-3 cell lines was
extracted using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. cDNA was synthesized from
the total RNA using a BL699A Reverse Transcription kit (with DNase;
cat. no. BL699A; Biosharp Life Sciences) with random hexamer
primers under the following conditions: 25°C for 10 min, 42°C for
15 min and 85°C for 5 sec. All RNA and cDNA samples were stored at
−80°C prior to subsequent analysis.
The relative expression of hsa_circ_0006168 was
determined using a LightCycler® 480 Instrument II (Roche
Diagnostics) with 2X Universal SYBR Green Fast qPCR Mix (cat. no.
RK21203; Abclonal Biotech Co., Ltd.). The following thermocycling
conditions were used: Initial denaturation at 95°C for 30 sec,
followed by 45 cycles of denaturation at 95°C for 5 sec and
annealing/extension at 60°C for 30 sec. The RT-qPCR primer
sequences for hsa_circ_0006168 were specifically designed to span
the back-splice junction to ensure amplification of the circular
transcript. The sequences were as follows: Forward,
5′-TGCCAAGCTTCATAATCTGGT-3′ and reverse,
5′-CCTTTGGCATCCCTATTAGTCTT-3′, with an expected amplified product
length of 60 bp. The identity of the amplicon spanning the
back-splice junction was confirmed using Sanger sequencing
(Fig. S1). For GAPDH, the RT-qPCR
primers were as follows: Forward, 5′-CCTTCATTGACCTCAACTA-3′ and
reverse, 5′-TGGAAGATGGTGATGGGATT-3′. The relative expression levels
were calculated using the 2−ΔΔCq method (10), with GAPDH as the internal
control.
Cell culture
The human normal prostatic epithelial cell line
RWPE-1, and the PCa cell lines PC-3, DU-145 and LNCaP, were
cultured in RPMI-1640 medium (Invitrogen™; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere
containing 5% CO2. To validate hsa_circ_0006168
expression in vitro, total RNA was extracted from these
cells and analyzed via the RT-qPCR method described above.
Bioinformatics analysis
CircRNA expression data and the clinical information
of PCa tissue samples were retrieved from the Gene Expression
Omnibus (GEO) database (ncbi.nlm.nih.gov/geo/) datasets GSE113153
(11) and GSE155792. The GSE113153
dataset comprised data from five pairs of PCa tissues, with samples
categorized based on the Gleason score (12): >8, poorly differentiated cancer;
and <6, well-differentiated cancer. The GSE155792 dataset
included the data from one PCa tissue and its adjacent normal
tissue. Differential expression analysis was performed using R
software (version 4.3.2; The R Foundation), with the criteria set
as log2 fold change ≥1 and P≤0.05 for identifying
differentially expressed circRNAs.
Database analysis
The target genes of hsa_circ_0006168 were predicted
using the Cancer-Specific CircRNA Database (http://gb.whu.edu.cn/CSCD/). To determine the
potential biological functions of differentially expressed mRNAs in
the competing endogenous (ce)RNA network in PCa, Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional
enrichment analyses were performed using the ClusterProfiler
package in R software (version 4.3.2; R Foundation for Statistical
Computing). To identify differentially expressed microRNAs
(miRNAs/miRs), miRNA expression data from two public GEO datasets
were also analyzed: GSE119338 (13)
and GSE14857 (14).
Statistical analysis
Statistical analysis was performed using SPSS 20.0
(IBM Corp.) and R software (version 4.3.2; The R Foundation).
Measurement data with a normal distribution are expressed as the
mean ± SD. Unpaired t-test was utilized to compare differences
between two groups. For comparisons among three or more groups,
one-way analysis of variance was performed, followed by Tukey's
post hoc test. Correlation analysis was performed using Pearson's
correlation test. The χ2 test was used to analyze the
association between hsa_circ_0006168 levels and the
clinicopathological features of patients with PCa. The survival
package in R software (version 4.3.2) was used to perform
univariate and multivariate Cox regression analysis of
clinicopathological factors and hsa_circ_0006168 expression levels.
Additionally, nomograms and survival curves were plotted to
determine whether clinicopathological factors and hsa_circ_0006168
expression levels could serve as independent prognostic indicators.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Upregulation of hsa_circ_0006168 in
the serum of patients with PCa
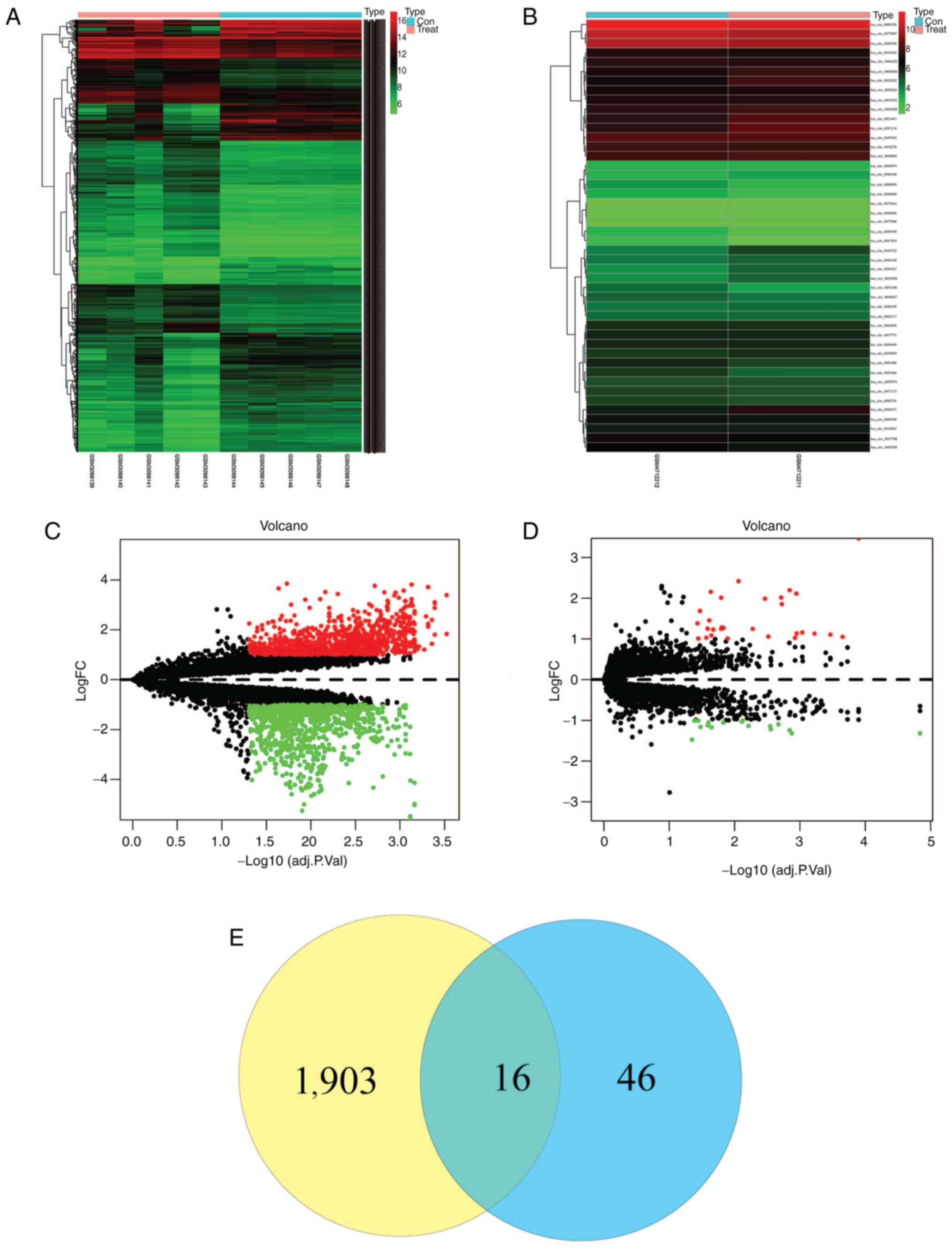
Bioinformatics analysis of the public GEO datasets
GSE155792 and GSE113153 identified hsa_circ_0006168 as a
significantly upregulated circRNA in PCa tissues (Fig. 1). To validate this finding, the
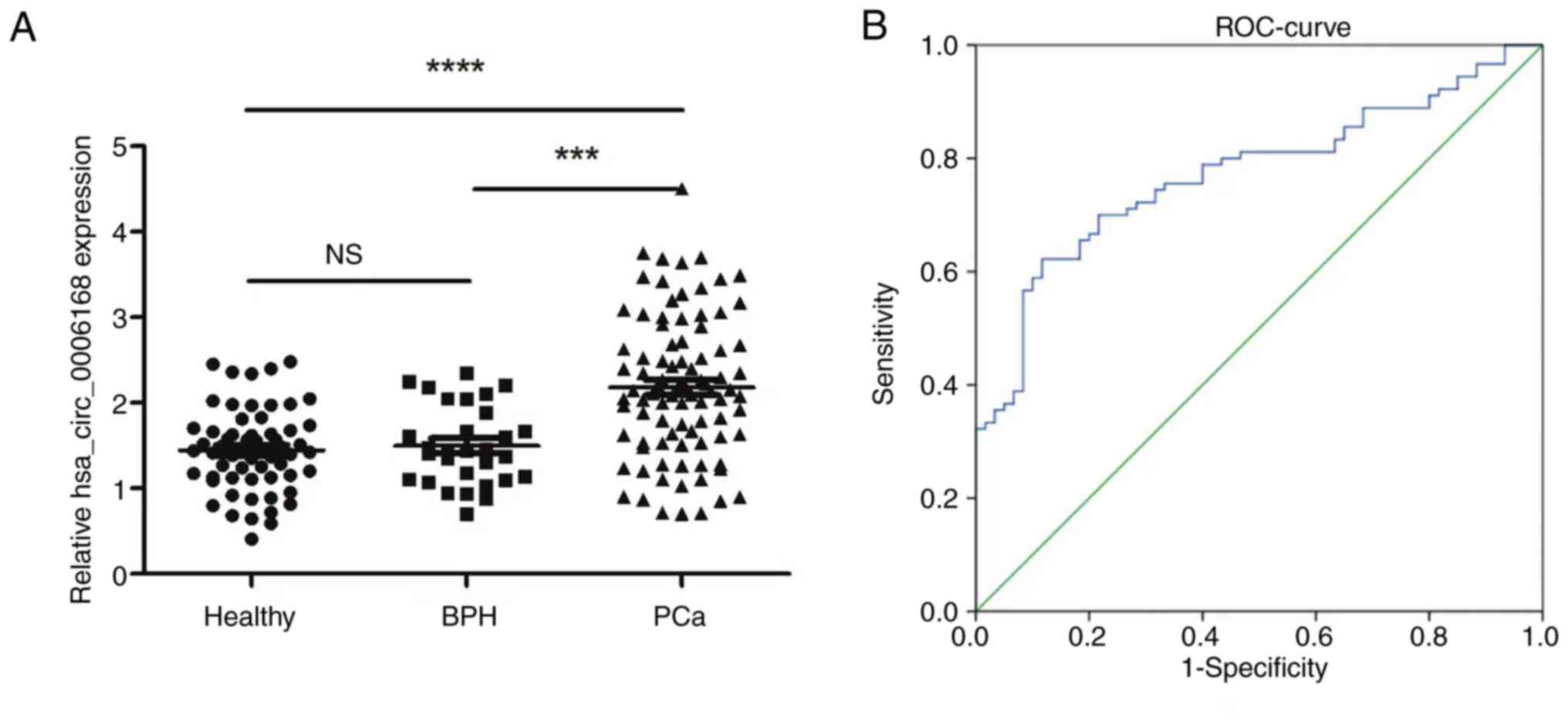
expression level of serum hsa_circ_0006168 was assessed in patients
with PCa, which demonstrated that the serum levels of
hsa_circ_0006168 was significantly elevated compared with in
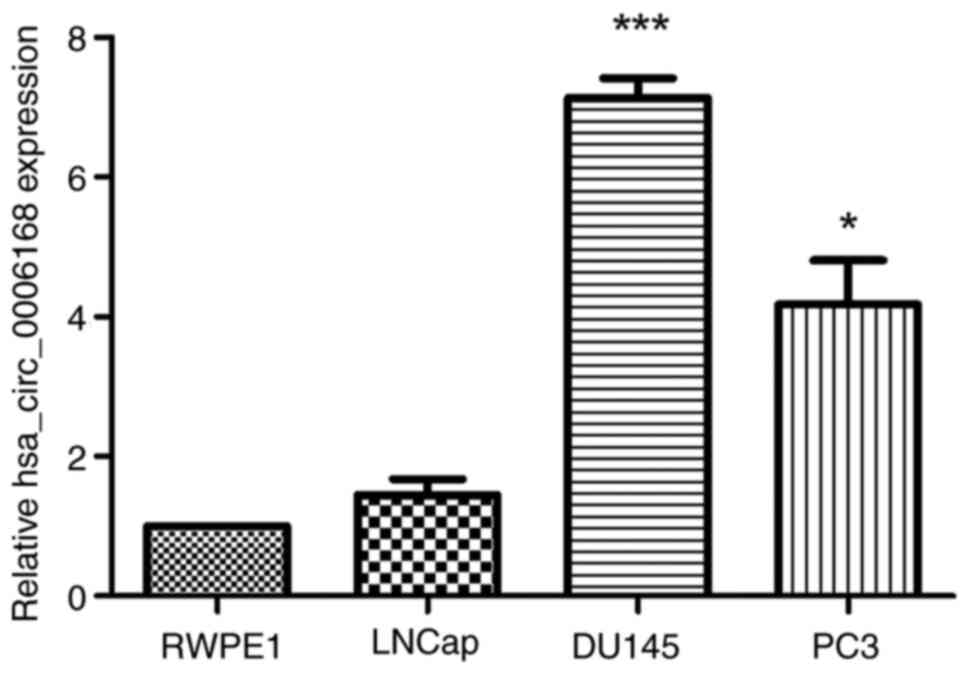
patients with BPH and healthy controls (Fig. 2A). Furthermore, hsa_circ_0006168
expression levels were evaluated in the normal prostatic epithelial
cell line RWPE-1, and the PCa cell lines LNCaP, DU145 and PC-3. The
results demonstrated that the expression of hsa_circ_0006168 was
significantly elevated in the DU145 and PC-3 cell lines compared
with the normal RWPE-1 cell line (Fig.
3).
Diagnostic value of serum
hsa_circ_0006168
To evaluate the diagnostic potential of serum
hsa_circ_0006168, receiver operating characteristic curve analysis
was performed. Serum hsa_circ_0006168 was revealed to demonstrate
good diagnostic performance in distinguishing patients with PCa
from controls, with an area under the curve of 0.773 (95%
confidence interval, 0.698–0.847), a sensitivity of 74.4% and a
specificity of 88.3% at the optimal cut-off value (Fig. 2B).
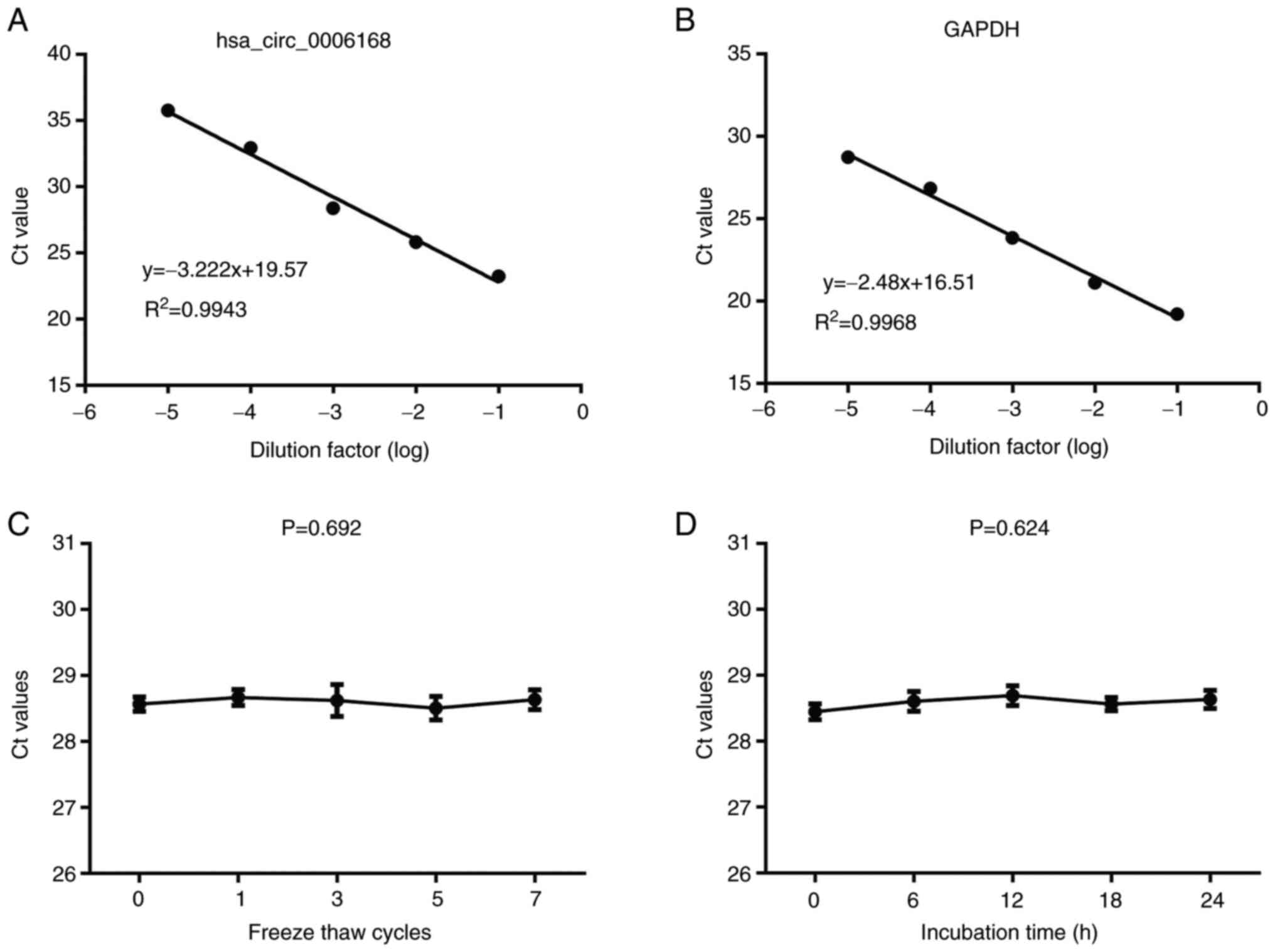
Evaluation of hsa_circ_0006168
detection in the serum
RT-qPCR for detecting serum hsa_circ_0006168 was
performed, and the analysis was demonstrated to be highly reliable:
The assay demonstrated marked linearity (R2=0.9943) and
high reproducibility (Fig. 4A and
B; Table I). Notably, in terms
of clinical application, hsa_circ_0006168 levels were revealed to
remain stable in serum samples exposed to prolonged ambient
temperatures and multiple freeze-thaw cycles (Fig. 4C and D), confirming its suitability
as a liquid biopsy biomarker.
 | Table I.Intra- and inter assay coefficients of
variation of hsa_circ_0006168 and GAPDH. |
Table I.
Intra- and inter assay coefficients of
variation of hsa_circ_0006168 and GAPDH.
| Parameter | hsa_circ_0006168 | GAPDH |
|---|
| Intra-assay |
|
|
| Mean ±
SD | 28.62±0.37 | 18.35±0.32 |
| CV,
% | 1.46 | 1.85 |
| Inter-assay |
|
|
| Mean ±
SD | 28.90±0.75 | 18.81±0.53 |
| CV,
% | 2.37 | 2.24 |
Association with clinicopathological
factors
Analysis of clinicopathological parameters revealed
that a high expression of hsa_circ_0006168 was significantly
associated with higher Gleason scores, suggesting an association
with worse tumor differentiation, and therefore, indicating its
potential role in PCa progression. However, no significant
associations were demonstrated between a high expression of the
circRNA and age, tumor-node-metastasis (TNM) stage (15), bone metastasis or PSA level
(Table II).
 | Table II.Association between hsa_circ_0006168
expression and clinicopathological features of patients with
prostate cancer. |
Table II.
Association between hsa_circ_0006168
expression and clinicopathological features of patients with
prostate cancer.
|
|
| hsa_circ_0006168
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Total (n=90)
(%) | Low (n=32) (%) | High (n=58)
(%) | χ2
value | P-value |
|---|
| Age |
|
|
| 0.148 | 0.700 |
| >60
years | 51 (56.7) | 19 (59.4) | 32 (55.2) |
|
|
| ≤60
years | 39 (43.3) | 13 (40.6) | 26 (44.8) |
|
|
| PSA, ng/ml |
|
|
| 0.170 | 0.680 |
|
>4 | 48 (53.3) | 18 (56.2) | 30 (51.7) |
|
|
|
<4 | 42 (46.7) | 14 (43.8) | 28 (48.3) |
|
|
| Gleason score |
|
|
| 4.098 | 0.043a |
| ≥7 | 60 (66.7) | 17 (53.1) | 43 (74.1) |
|
|
|
<7 | 30 (33.7) | 15 (46.9) | 15 (25.9) |
|
|
| Bone
metastasis |
|
|
| 0.030 | 0.862 |
|
Yes | 32 (35.6) | 11 (34.4) | 21 (36.2) |
|
|
| No | 58 (64.4) | 21 (65.6) | 37 (63.8) |
|
|
| TNM stage |
|
|
| 0.357 | 0.550 |
| I and
II | 46 (51.1) | 15 (46.9) | 31 (53.4) |
|
|
| III and
IV | 44 (48.9) | 17 (53.1) | 27 (46.6) |
|
|
Cox regression analysis of prognostic
factors
In the univariate Cox regression analysis, the TNM
stage, Gleason score, presence of bone metastasis and high
expression of hsa_circ_0006168 were all revealed to be significant
predictors of poor prognosis. However, in the multivariate
analysis, only the TNM stage remained as an independent prognostic
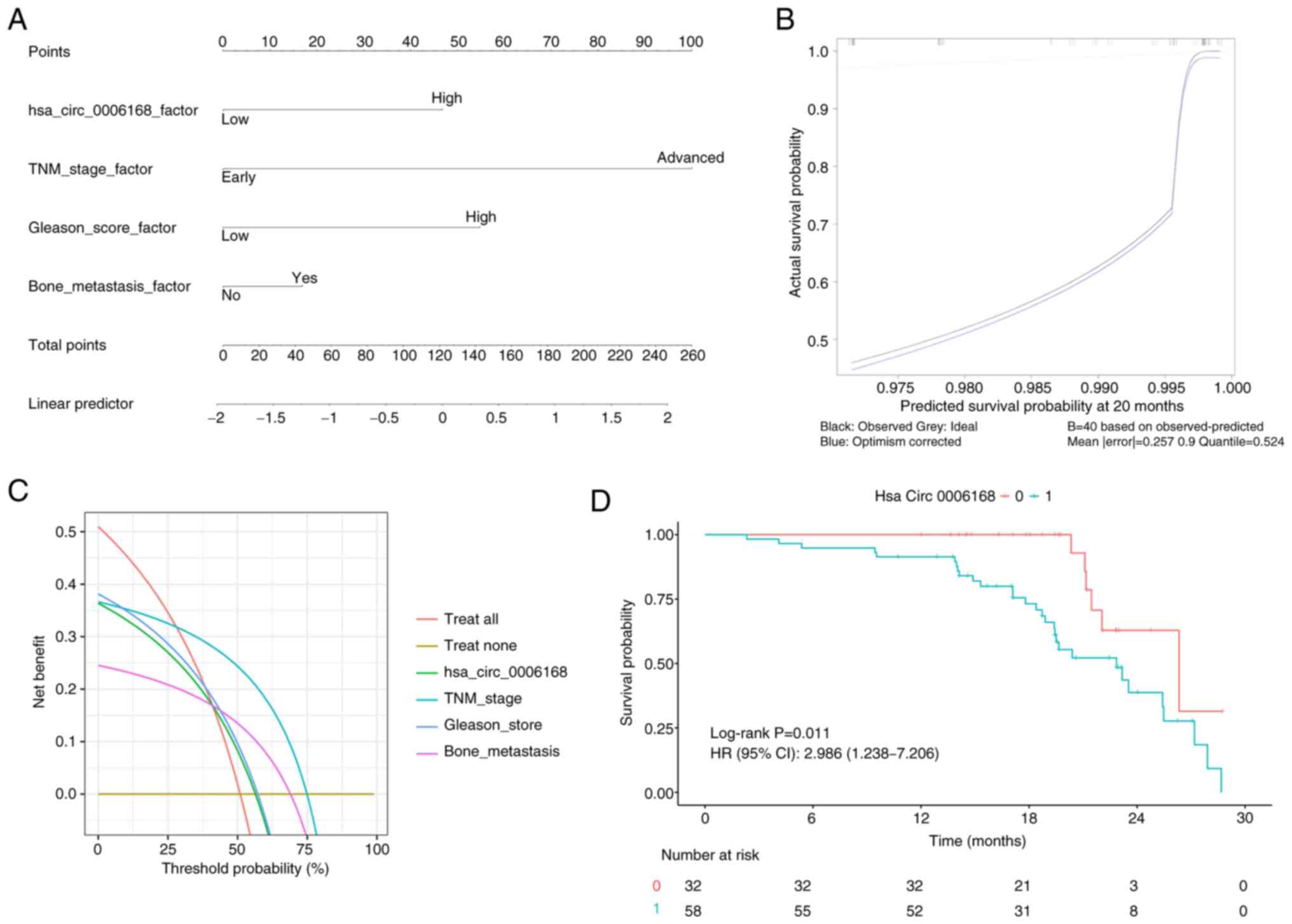
factor (Table III). A nomogram
was then constructed based on these factors, which demonstrated a
strong discriminative ability for predicting patient survival
(C-index, 0.7886), with good calibration and clinical utility
(Fig. 5A-C). Kaplan-Meier survival
analysis demonstrated that patients with high hsa_circ_0006168
expression had significantly poorer overall survival compared with
the low expression group (Fig.
5D).
 | Table III.Univariate and multivariate Cox
regression analysis of hsa_circ_0006168 expression levels and
clinicopathological factors. |
Table III.
Univariate and multivariate Cox
regression analysis of hsa_circ_0006168 expression levels and
clinicopathological factors.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| hsa_circ_0006168
expression | 2.989
(1.235–7.227) | 0.015 | 2.114
(0.858–5.212) | 0.104 |
| Age | 0.632
(0.332–1.203) | 0.191 | – | – |
| TNM stage | 7.017
(3.028–16.233) | <0.001 | 4.958
(1.811–13.577) | 0.002 |
| Gleason score | 2.697
(1.033–7.047) | 0.041 | 2.404
(0.866–6.669) | 0.092 |
| Bone
metastasis | 2.687
(1.355–5.327) | 0.005 | 1.311
(0.634–2.710) | 0.465 |
| PSA | 0.918
(0.465–1.812) | 0.805 | – | – |
Exploration of the potential
underlying regulatory mechanism
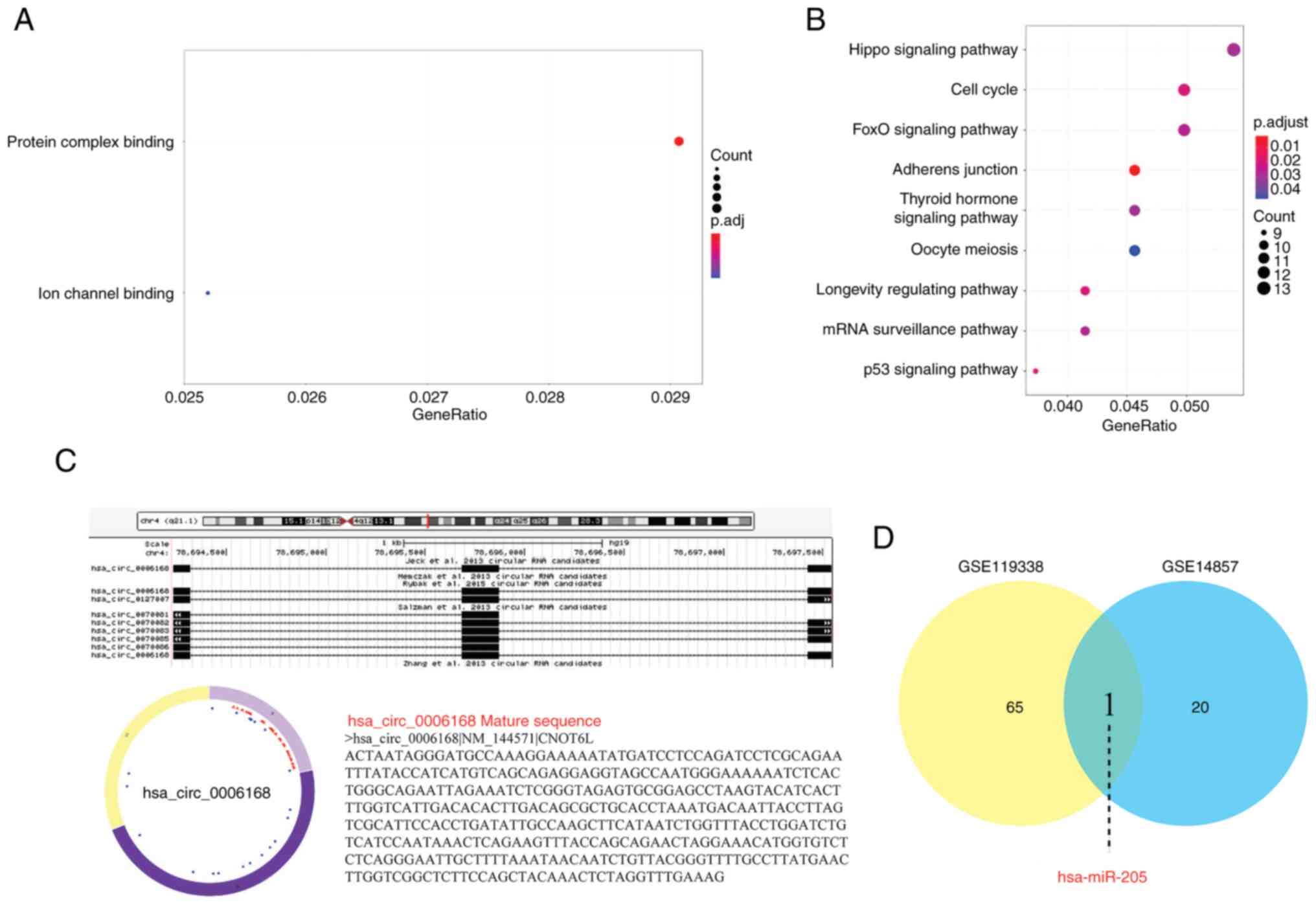
To explore potential mechanisms, a preliminary
bioinformatics analysis was performed. GO and KEGG enrichment
analyses of predicted target genes for hsa_circ_0006168 indicated a
primary association with the p53 signaling pathway (Fig. 6A and B; Table IV). Moreover, the sequence and
predicted structure of hsa_circ_0006168 were identified (Fig. 6C). The analysis of two public miRNA
datasets then identified several differentially expressed miRNAs
(Tables V and VI), with miR-205 noted in both (Fig. 6D), suggesting it as a potential
interaction partner. Collectively, these exploratory in
silico findings form a working hypothesis that hsa_circ_0006168
may function via the p53 pathway and/or by sponging miR-205, a
hypothesis that requires future experimental validation.
 | Table IV.Gene Ontology and Kyoto Encyclopedia
of Genes and Genomes analysis of downstream target genes associated
with has_circ_0006168. |
Table IV.
Gene Ontology and Kyoto Encyclopedia
of Genes and Genomes analysis of downstream target genes associated
with has_circ_0006168.
| ID | Description | Gene ID | P-value |
|---|
| GO:0003712 | Transcription
coregulator activity |
HMGB2/ZBTB18/HCFC2/YAF2/MIER1/PHF12/HMGA1/HNRNPU/RYBP/ATXN7L3/LIMD1/KMT2D/SETD3/RCOR1/DDIT3/BTG1/SOX12/RBM14/TMF1/NPAT/TCERG1/BCL9L/CTNNB1 | 0.000264 |
| GO:0017016 | Ras GTPase
binding |
CDC42EP1/CDC42SE1/PREX2/RNF41/RCC2/TMEM127/PARD6B/FGD4/RASGRP1/ARHGEF28/TIAM1/TNPO1/AP1G1/ABI2/PLEKHG5/BICD2/TBC1D16/HACE1/RAB11FIP4/KCTD10/NET1/EXOC2 |
5.01×10−5 |
| GO:0031267 | Small GTPase
binding |
CDC42EP1/CDC42SE1/PREX2/RNF41/RCC2/TMEM127/PARD6B/FGD4/RASGRP1/ARHGEF28/TIAM1/TNPO1/AP1G1/ABI2/PLEKHG5/BICD2/TBC1D16/HACE1/RAB11FIP4/KCTD10/NET1/EXOC2 |
7.89×10−5 |
| GO:0140297 | DNA-binding
transcription factor binding |
HIF1A/RARB/ACTB/FOS/MIER1/SP1/HMGA1/GTF2I/DHX33/CRTC3/ETS2/RB1/SETD3/DDIT3/ESR1/TMF1/TCF12/TCERG1/FOXP1/CTNNB1 |
3.46×10−5 |
| GO:0003713 | Transcription
coactivator activity |
HMGB2/ZBTB18/HCFC2/YAF2/HMGA1/ATXN7L3/KMT2D/SETD3/DDIT3/SOX12/RBM14/TMF1/NPAT/TCERG1/BCL9L/CTNNB1 | 0.000133 |
| GO:0061629 | RNA polymerase
II-specific DNA-binding transcription factor binding |
HIF1A/RARB/ACTB/FOS/MIER1/SP1/HMGA1/GTF2I/ETS2/RB1/SETD3/ESR1/TMF1/TCERG1/FOXP1/CTNNB1 | 0.000133 |
| GO:0017048 | Rho GTPase
binding |
CDC42EP1/CDC42SE1/PREX2/RCC2/PARD6B/FGD4/ARHGEF28/TIAM1/ABI2/PLEKHG5/HACE1/KCTD10/NET1 |
3.02×10−5 |
| GO:0008022 | Protein C-terminus
binding |
PRKAA1/YWHAQ/CD2AP/PRRC2C/DAB2/FOXN3/SP1/DLG4/FIGN/RBFOX1/NPAT/CTNNB1 | 0.000542 |
| GO:0003730 | mRNA 3′-UTR
binding |
HNRNPR/ELAVL4/NOVA1/SERBP1/HNRNPU/ZFP36L2/CPEB2/RNPS1/ZFP36L1 | 0.000683 |
| GO:0010485 | H4 histone
acetyltransferase activity |
KANSL1L/NAA50/KANSL1/NAA40 | 0.000334 |
| hsa04010 | MAPK signaling
pathway |
HSPA1B/RPS6KA6/CSF1R/CRKL/ERBB3/MAX/FOS/EPHA2/TRADD/MKNK2/RAP1B/PPP3R1/RASGRP1/MAP3K4/DDIT3/MAPK1/GADD45A/RAC3/TAOK1/MAP3K7 |
1.22×10−5 |
| hsa04218 | Cellular
senescence |
CDKN1A/E2F3/CALM2/PTEN/CCND2/CHEK1/CCNE1/PPP3R1/RB1/ZFP36L2/HIPK3/ZFP36L1/MAPK1/GADD45A/PPP1CB |
2.59×10−6 |
| hsa05166 | Human T cell
leukemia virus 1 infection |
CDKN1A/CREB5/E2F3/FOS/PTEN/CCND2/CHEK1/CCNE1/PPP3R1/CRTC3/ETS2/RB1/MAPK1/NRP1 | 0.000564 |
| hsa05224 | Breast cancer |
FZD6/CDKN1A/E2F3/FOS/PTEN/SP1/RB1/ESR1/MAPK1/GADD45A/JAG1/FRAT2/CTNNB1 |
3.04×10−5 |
| hsa05203 | Viral
carcinogenesis |
YWHAQ/YWHAZ/CDKN1A/CREB5/CCND2/TRADD/CHEK1/CCNE1/TRAF1/RB1/YWHAE/MAPK1/SYK | 0.000804 |
| hsa04110 | Cell cycle |
ORC4/YWHAQ/YWHAZ/CDKN1A/E2F3/CCND2/CHEK1/CCNE1/RB1/YWHAE/GADD45A | 0.000141 |
| hsa04934 | Cushing
syndrome |
FZD6/CDKN1A/CREB5/E2F3/RAP1B/CCNE1/SP1/KMT2D/RB1/MAPK1/CTNNB1 | 0.000844 |
| hsa04390 | Hippo signaling
pathway |
FZD6/YWHAQ/YWHAZ/ACTB/CCND2/PARD6B/DLG4/LIMD1/YWHAE/PPP1CB/CTNNB1 | 0.000938 |
| hsa05160 | Hepatitis C |
YWHAQ/YWHAZ/CDKN1A/E2F3/TRADD/OCLN/RB1/YWHAE/MAPK1/CLDN16/CTNNB1 | 0.000938 |
| hsa04114 | Oocyte meiosis |
YWHAQ/YWHAZ/RPS6KA6/CALM2/CCNE1/PPP3R1/CPEB2/YWHAE/MAPK1/PPP1CB | 0.000827 |
| hsa05222 | Small cell lung
cancer |
CDKN1A/RARB/E2F3/MAX/PTEN/CCNE1/TRAF1/RB1/GADD45A | 0.000244 |
| hsa04115 | p53 signaling
pathway |
CDKN1A/RRM2/PTEN/CCND2/SESN3/CHEK1/CCNE1/GADD45A | 0.000248 |
 | Table V.Differentially expressed miRs in the
GSE119338 dataset. |
Table V.
Differentially expressed miRs in the
GSE119338 dataset.
| Gene | logFC | P-value | Gene
regulation |
|---|
| hsa-miR-034c | 1.88339 | 0.002460 | Down |
| hsa-miR-376b | 1.81105 | 0.003005 | Down |
| hsa-miR-376a | 1.77345 | 0.003255 | Down |
| hsa-miR-368 | 1.71005 | 0.003275 | Down |
| hsa-miR-030a
3p | 1.67436 | 0.003350 | Down |
| hsa-miR-137 | 1.63115 | 0.003378 | Down |
| hsa-miR-133a | 1.72425 | 0.003670 | Down |
| hsa-miR-210 | 1.83033 | 0.003706 | Down |
| hsa-miR-034b | 1.65755 | 0.003752 | Down |
| hsa-miR-193b | 1.61622 | 0.003861 | Down |
| hsa-miR-133b | 1.63867 | 0.004235 | Down |
| hsa-miR-022 | 1.91686 | 0.004770 | Down |
| hsa-miR-205 | 2.08786 | 0.004826 | Down |
| hsa-miR-203 | 1.99239 | 0.004882 | Down |
| hsa-miR-031 | 1.98816 | 0.005144 | Down |
| hsa-miR-030e
3p | 1.49580 | 0.005833 | Down |
| hsa-miR-010b | 1.53879 | 0.005835 | Down |
| hsa-miR-030d | 1.53824 | 0.006043 | Down |
| hsa-miR-030e
5p | 1.83719 | 0.006113 | Down |
| hsa-miR-030c | 1.59348 | 0.006288 | Down |
| hsa-miR-024 | 1.74432 | 0.006604 | Down |
| hsa-miR-001 | 1.50516 | 0.006745 | Down |
| hsa-miR-152 | 1.70493 | 0.006880 | Down |
| hsa-miR-030a
5p | 1.49181 | 0.007102 | Down |
| hsa-miR-224 | 1.92611 | 0.007991 | Down |
| hsa-miR-423 | 1.64733 | 0.009312 | Down |
| hsa-miR-365-1 | 1.58153 | 0.009464 | Down |
| hsa-miR-030b | 1.34008 | 0.010272 | Down |
| hsa-miR-424 | 1.45159 | 0.010713 | Down |
| hsa-miR-029c | 1.56767 | 0.010900 | Down |
| hsa-miR-449 | 1.27469 | 0.013276 | Down |
| hsa-miR-221 | 1.38374 | 0.013456 | Down |
| hsa-miR-503 | 1.33977 | 0.013816 | Down |
| hsa-miR-027b | 1.53227 | 0.014072 | Down |
| hsa-miR-023a | 1.47580 | 0.014885 | Down |
| hsa-miR-135b | 1.72096 | 0.015631 | Down |
| hsa-miR-452 | 1.41561 | 0.019488 | Down |
| hsa-miR-021 | 1.57238 | 0.019811 | Down |
| hsa-miR-023b | 1.40925 | 0.020878 | Down |
| hsa-miR-027a | 1.34136 | 0.022885 | Down |
| hsa-miR-029a | 1.27442 | 0.026287 | Down |
| hsa-miR-095 | 1.16648 | 0.028329 | Down |
| hsa-miR-149 | 1.19165 | 0.028613 | Down |
| hsa-miR-034a | 1.08216 | 0.029093 | Down |
| hsa-miR-193 | 1.43661 | 0.030047 | Down |
| hsa-miR-222 | 1.15869 | 0.033629 | Down |
| hsa-miR-029b | 1.40342 | 0.034209 | Down |
| hsa-miR-107 | 1.02251 | 0.037370 | Down |
| hsa-miR-009 | 1.21235 | 0.040226 | Down |
| hsa-miR-130a | 1.39363 | 0.041765 | Down |
| hsa-miR-189 | 1.20643 | 0.045449 | Down |
| hsa-miR-130b | 1.25535 | 0.049841 | Down |
|
hsa-miR-103-1-pre | 1.65357 | 0.022734 | Up |
|
hsa-miR-452-pre | 1.65577 | 0.023460 | Up |
|
hsa-miR-491-pre | 1.49639 | 0.030418 | Up |
|
hsa-miR-485-pre | 1.58863 | 0.036311 | Up |
|
hsa-miR-492-pre | 1.24900 | 0.037825 | Up |
|
hsa-miR-512-1-pre | 1.33137 | 0.037889 | Up |
|
hsa-miR-506-pre | 1.54105 | 0.043464 | Up |
|
hsa-miR-495-pre | 1.33179 | 0.043764 | Up |
| hsa-miR-485-3p | 1.11936 | 0.044769 | Up |
| hsa-miR-512-5p | 1.07008 | 0.045630 | Up |
|
hsa-miR-513-2-pre | 1.10115 | 0.047218 | Up |
| hsa-miR-515-5p | 1.16960 | 0.047827 | Up |
|
hsa-miR-513-1-pre | 1.20450 | 0.049643 | Up |
 | Table VI.Differentially expressed microRNAs in
the GSE14857 dataset. |
Table VI.
Differentially expressed microRNAs in
the GSE14857 dataset.
| Gene | logFC | P-value | Gene
regulation |
|---|
| hsa-miR-205 | 4.02160 | 0.000121 | Down |
| hsa-miR-591 | 2.81675 | 0.000442 | Down |
| hsa-miR-668 | 2.00971 | 0.003108 | Down |
| hsa-miR-31 | 2.31899 | 0.003232 | Down |
| hsa-miR-578 | 3.86374 | 0.011947 | Down |
| hsa-miR-490 | 1.30685 | 0.010993 | Down |
| hsa-miR-589 | 2.49788 | 0.016980 | Down |
| hsa-miR-499 | 1.24839 | 0.032533 | Down |
| hsa-miR-217 | 3.04896 | 0.031024 | Down |
| hsa-miR-588 | 3.08085 | 0.031731 | Down |
| hsa-miR-570 | 9.13298 | 0.018255 | Down |
|
hsa-miR-526ba | 1.74544 | 0.017158 | Up |
| hsa-miR-767-3p | 1.32825 | 0.017129 | Up |
| hsa-miR-183 | 1.04655 | 0.000855 | Up |
| hsa-miR-511 | 1.54791 | 0.039622 | Up |
| hsa-miR-600 | 5.08612 | 0.024913 | Up |
| hsa-miR-663 | 1.04562 | 0.028394 | Up |
|
hsa-miR-182a | 1.35082 | 0.033078 | Up |
| hsa-miR-216 | 2.58817 | 0.045927 | Up |
| hsa-miR-563 | 1.01419 | 0.034812 | Up |
Discussion
PCa is a leading cause of morbidity and mortality
among men worldwide, with an estimated 1.5 million new cases and
397,000 deaths reported in 2022 (1). Although several treatment options have
been reported to notably improve outcomes in early-stage disease,
advanced PCa with distant metastasis remains challenging to manage
(2). Therefore, there is a pressing
need to explore novel diagnostic and therapeutic approaches to
enable both early detection of the disease and the design of
personalized treatment strategies.
Hsa_circ_0006168 (alternatively known as circCNOT6L)
is located at chr4:78694234-78697546 and has a length of 3,312
nucleotides. It was first identified by Shi et al (8), who revealed that hsa_circ_0006168
expression is markedly elevated in ESCC tissues and cell lines
compared with that in normal controls, and its high expression was
associated with lymph node metastasis and advanced TNM staging in
patients with ESCC. The present study screened for differentially
expressed circRNAs using publicly available circRNA expression
data, and the findings were validated in serum samples. The results
obtained demonstrated that the expression level of hsa_circ_0006168
was significantly higher in the serum of patients with PCa compared
with that in BPH and healthy controls, suggesting the potential of
hsa_circ_0006168 as a non-invasive diagnostic biomarker for
PCa.
Moreover, a positive association was observed
between high hsa_circ_0006168 expression and the Gleason score,
demonstrating that elevated hsa_circ_0006168 levels are associated
with lower PCa differentiation and a worse prognosis. Furthermore,
hsa_circ_0006168 expression levels were evaluated in the normal
prostatic epithelial cell line RWPE-1, and the PCa cell lines
LNCaP, DU145 and PC-3. Notably, the expression of hsa_circ_0006168
was demonstrated to be significantly elevated in the DU145 and PC-3
cell lines, which are androgen-independent, poorly differentiated
and exhibit high metastatic potential (16). This increased expression may be
associated with androgen receptor (AR) activity as AR positivity is
strongly associated with castration-resistant prostate cancer
(CRPC). In this cancer type, traditional endocrine therapy remains
effective for 18–24 months (17);
however, disease progression, driven by AR mutations and even low
androgen levels, can promote PCa malignancy (18). The bioinformatics analysis performed
in the present study predicted that the target genes of
hsa_circ_0006168 are enriched in the p53 signaling pathway. In PCa,
inactivation of p53 is a key event that drives tumor progression,
metastasis and therapeutic resistance, making it a critical
biomarker throughout the natural history of the disease (19–21).
Future studies should use larger sample sizes and diverse ethnic
and geographical representation, and employ knockdown or
overexpression of hsa_circ_0006168 experiments. Moreover, its
interaction with ARs should be investigated both in vitro
and in vivo to elucidate its regulatory role in PCa
progression.
CircRNAs are widely distributed in eukaryotic cells
and contain multiple miRNA response elements, enabling them to act
as ceRNAs through competitively binding to miRNAs (22). Following processing by nucleases,
mature miRNAs are assembled into RNA-induced silencing complexes,
where they target mRNAs via base-pairing complementarity, leading
to mRNA degradation or translational repression (23–25).
As ceRNAs, circRNAs can mitigate the inhibitory effects of miRNAs
on target mRNAs, thereby regulating the expression and function of
associated genes. For example, a bioinformatics analysis performed
previously predicted that circGNG4 can enhance EYA transcriptional
coactivator and phosphatase 3/c-Myc expression through sponging
miR-223, thereby promoting the malignant progression of PCa
(26). Similarly, Chao et al
(27) performed a fluorescence
in situ hybridization analysis to localize a novel circRNA,
circSOBP, primarily in the cytoplasm of PCa cells, where its
overexpression was reported to inhibit cell invasion and migration.
Dual-luciferase assays reported that circSOBP could also bind to
miR-141-3p, thereby modulating the myosin phosphatase target
subunit 1/phosphorylated myosin regulatory light chain 2 axis to
suppress PCa progression. The finding in the present study that
hsa_circ_0006168 is upregulated and associated with more aggressive
features of PCa is consistent with the oncogenic roles reported for
other circRNAs within this specific cancer type. For example,
circHIPK3 has been reported to be overexpressed in PCa, where it
promotes cell proliferation and invasion by acting as a ceRNA to
sponge miR-193a-3p (11).
Similarly, circ0005276 has also been reported to be upregulated in
PCa tissues and facilitates cell proliferation and migration.
However, it functions through a different mechanism by interacting
with the Fused in Sarcoma protein to transcriptionally activate
X-linked Inhibitor of Apoptosis (28). This suggests that a class of
circRNAs may typically be involved in the malignant progression of
PCa. However, in comparison with the aforementioned studies, the
present study is the first to have characterized hsa_circ_0006168
as a non-invasive, serum-detectable marker, and also to have
associated it with the AR/miR-205 axis, to the best of our
knowledge, thereby providing a unique perspective on its role in
PCa.
In the present study, differentially expressed
miRNAs in PCa tissues and cells were analyzed using miRNA
expression data from the datasets GSE119338 and GSE14857,
identifying a downregulation in miR-205 expression. This finding
aligns with an previous study by Gandellini et al (29), who reported a decreased expression
of miR-205 in PCa cells and tissues, demonstrating its
tumor-suppressive role through inhibiting epithelial-mesenchymal
transition. Hagman et al (30) further reported that miR-205 binds to
the 3′-untranslated region of the AR, reducing both the
transcription and protein levels of the AR, suggesting the
therapeutic potential of miR-205 in CRPC. Additionally, Kalogirou
et al (31) reported that
squalene epoxidase (SQLE) and several cholesterol biosynthesis
enzymes are overexpressed in advanced PCa, which was associated
with poor survival. They also reported that miR-205 regulates SQLE
expression, with high miR-205 levels found to inhibit SQLE and to
suppress cholesterol synthesis. These findings highlighted a strong
association between miR-205 expression, AR regulation and PCa
progression. Given the mechanism associated with ceRNAs, we
hypothesize that an elevated expression of hsa_circ_0006168 may
lead to its competitively binding with miR-205, thereby reducing
its levels and promoting PCa progression. However, it must be
emphasized that this remains merely an inference based on
bioinformatics analysis, and the putative direct interaction needs
to be rigorously confirmed by future functional experiments,
including performing dual-luciferase reporter assays.
In the present study, multivariate Cox regression
analysis confirmed TNM stage as an independent predictor of
survival. Although high hsa_circ_0006168 expression, Gleason score
and the presence of bone metastasis were reported to be significant
risk factors according to the univariate analysis, they lost
prognostic independence when adjusted for TNM stage. This is likely
due to collinearity (32), a
statistical phenomenon where predictor variables are highly
associated. In this context, the TNM staging system is a
comprehensive index that already incorporates information on tumor
grade and metastatic status, such as Gleason score and bone
metastasis. Consequently, its prognostic value is encompassed by
the staging system. Far from undermining its biological importance,
however, this result suggests that hsa_circ_0006168 upregulation
may represent a key molecular event in the tumor progression and
invasion pathways that determine a higher TNM stage. Future studies
are warranted to dissect the precise association between
hsa_circ_0006168 expression and pathological stage.
Moreover, the present study had several limitations.
First, the small, single-center sample size limits the
generalizability of the conclusions. Therefore, the findings
require validation in a larger, independent cohort to confirm their
robustness and rule out overfitting before considering clinical
application. Secondly, the proposed molecular mechanism is based
solely on bioinformatics. Fully elucidating the biological role of
hsa_circ_0006168 requires extensive experimental validation,
including in vitro functional studies in cell lines and
in vivo animal models, to investigate its impact on tumor
progression.
In conclusion, the results of the present study
indicate that hsa_circ_0006168 is highly expressed in PCa, and that
this is associated with tumor malignancy. This suggests that it may
be a promising biomarker for the adjuvant diagnosis and prognostic
monitoring of PCa.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YD analyzed and interpreted data and wrote the
manuscript. MTL designed the study and performed the statistical
analysis. NY and LG performed the experiments. LPC conceived the
study and revised the manuscript. All authors have read and
approved the final manuscript. LPC and YD confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Nantong First People's Hospital (approval no.
2023-KY007-1). All patients provided written informed consent. The
study was performed in accordance with the ethical guidelines of
the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Sung H,
Ferlay J, Siegel RL, Soerjomataram I and Jemal A: Global cancer
statistics 2022: GLOBOCAN estimates of incidence and mortality
worldwide for 36 cancers in 185 countries. CA Cancer J Clin.
74:229–263. 2024.PubMed/NCBI
|
|
2
|
US Preventive Services Task Force, ;
Grossman DC, Curry SJ, Owens DK, Bibbins-Domingo K, Caughey AB,
Davidson KW, Doubeni CA, Ebell M, Epling JW Jr, et al: Screening
for prostate cancer: US preventive services task force
recommendation statement. JAMA. 319:1901–1913. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li S, Hu W, Deng F, Chen S, Zhu P, Wang M,
Chen X, Wang Y, Hu X, Zhao B, et al: Identification of circular RNA
hsa_circ_0001599 as a novel biomarker for large-artery
atherosclerotic stroke. DNA Cell Boil. 40:457–468. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lv J, Ren L, Han S, Zhang J, Zhao X, Zhang
Y, Fang H, Zhang L, Yang H, Wang S, et al: Peripheral blood
hsa-circRNA5333-4: A novel biomarker for myasthenia gravis. Clin
Immunol. 224:1086762021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi J, Liu C, Chen C, Guo K, Tang Z, Luo
Y, Chen L, Su Y and Xu K: Circular RNA circMBOAT2 promotes prostate
cancer progression via a miR-1271-5p/mTOR axis. Aging (Albany NY).
12:13255–13280. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi Y, Guo Z, Fang N, Jiang W, Fan Y, He
Y, He Y, Ma Z and Chen Y: hsa_circ_0006168 sponges miR-100 and
regulates mTOR to promote the proliferation, migration and invasion
of esophageal squamous cell carcinoma. Biomed Pharmacother.
117:1091512019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang T, Mao P, Feng Y, Cui B, Zhang B,
Chen C, Xu M and Gao K: Blocking hsa_circ_0006168 suppresses cell
proliferation and motility of human glioblastoma cells by
regulating hsa_circ_0006168/miR-628-5p/IGF1R ceRNA axis. Cell
Cycle. 20:1181–1194. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen D, Lu X, Yang F and Xing N: Circular
RNA circHIPK3 promotes cell proliferation and invasion of prostate
cancer by sponging miR-193a-3p and regulating MCL1 expression.
Cancer Manag Res. 11:1415–1423. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gleason DF: Classification of prostatic
carcinomas. Cancer Chemother Rep. 50:125–128. 1966.PubMed/NCBI
|
|
13
|
Verma S, Pandey M, Shukla GC, Singh V and
Gupta S: Integrated analysis of miRNA landscape and cellular
networking pathways in stage-specific prostate cancer. PLoS One.
14:e02240712019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schaefer A, Jung M, Mollenkopf HJ, Wagner
I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G and Jung
K: Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th edition.
Springer; New York, NY: 2017
|
|
16
|
Sobel RE and Sadar MD: Cell lines used in
prostate cancer research: A compendium of old and new lines-part 1.
J Urol. 173:342–359. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Le TK, Duong QH, Baylot V, Fargette C,
Baboudjian M, Colleaux L, Taïeb D and Rocchi P:
Castration-resistant prostate cancer: From uncovered resistance
mechanisms to current treatments. Cancers (Basel). 15:50472023.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Watson PA, Arora VK and Sawyers CL:
Emerging mechanisms of resistance to androgen receptor inhibitors
in prostate cancer. Nat Rev Cancer. 15:701–711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD,
Gao CC, Ma L, Chen JS, Liu KL, Zheng B, et al: Dysregulation of
p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate
cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha
pathway. Oncogene. 38:2516–2532. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ofner H, Kramer G, Shariat SF and Hassler
MR: TP53 deficiency in the natural history of prostate cancer.
Cancers (Basel). 17:6452025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Teroerde M, Nientiedt C, Duensing A,
Hohenfellner M, Stenzinger A and Duensing S: Chapter 8: Revisiting
the role of p53 in prostate cancer. Prostate Cancer. Bott SRJ and
Ng KL: Exon Publications; Brisbane, Australia: pp. 113–123.
2021
|
|
22
|
To KKW, Zhang H and Cho WC: Competing
endogenous RNAs (ceRNAs) and drug resistance to cancer therapy.
Cancer Drug Resist. 7:372024.PubMed/NCBI
|
|
23
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu S, Lian Z, Zhang S, Xu Y and Zhang H:
CircGNG4 promotes the progression of prostate cancer by sponging
miR-223 to enhance EYA3/c-myc expression. Front Cell Dev Biol.
9:6841252021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chao F, Song Z, Wang S, Ma Z, Zhuo Z, Meng
T, Xu G and Chen G: Novel circular RNA circSOBP governs amoeboid
migration through the regulation of the miR-141-3p/MYPT1/p-MLC2
axis in prostate cancer. Clin Transl Med. 11:e3602021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng Y, Yang Y, Zhao X, Fan Y, Zhou L,
Rong J and Yu Y: Circular RNA circ0005276 promotes the
proliferation and migration of prostate cancer cells by interacting
with FUS to transcriptionally activate XIAP. Cell Death Dis.
10:7922019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gandellini P, Folini M, Longoni N, Pennati
M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta
P, et al: miR-205 exerts tumor-suppressive functions in human
prostate through down-regulation of protein kinase cepsilon. Cancer
Res. 69:2287–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hagman Z, Haflidadottir BS, Ceder JA,
Larne O, Bjartell A, Lilja H, Edsjö A and Ceder Y: miR-205
negatively regulates the androgen receptor and is associated with
adverse outcome of prostate cancer patients. Br J Cancer.
108:1668–1676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kalogirou C, Linxweiler J, Schmucker P,
Snaebjornsson MT, Schmitz W, Wach S, Krebs M, Hartmann E, Puhr M,
Müller A, et al: MiR-205-driven downregulation of cholesterol
biosynthesis through SQLE-inhibition identifies therapeutic
vulnerability in aggressive prostate cancer. Nat Commun.
12:50662021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gregorich M, Strohmaier S, Dunkler D and
Heinze G: Regression with highly correlated predictors: Variable
omission is not the solution. Int J Environ Res Public Health.
18:42592021. View Article : Google Scholar : PubMed/NCBI
|