Introduction
Ovarian cancer is the most lethal of gynecological
malignancies, with >70% of cases diagnosed at advanced stages.
Due to delayed detection and both intrinsic and therapy-induced
chemoresistance, ovarian cancer has a 5-year survival rate of
<50% (1). Despite incremental
advancements in cytoreductive surgery and platinum-based treatment
protocols, recurrence and refractory disease continue to pose
notable challenges, highlighting the urgent need for innovative
therapeutic targets and strategies (2).
Previously, long non-coding RNAs (lncRNAs) have
emerged as key regulators of oncogenesis, coordinating key cancer
hallmarks including sustained proliferation, migration, invasion
and drug resistance, through epigenetic, transcriptional and
post-translational mechanisms (3–5). Among
lncRNAs, LINC00473, a conserved oncogenic lncRNA, has been
implicated in enhancing tumor aggressiveness and chemoresistance in
lung, breast and hepatocellular carcinoma by disrupting key
signaling pathways, such as the Hippo/Yes-associated protein (YAP)
axis and Sox2-mediated stemness program (6–8). These
pathways are essential for maintaining the plasticity of cancer
stem cells (CSC) and DNA damage repair capacity, which are key
factors determining platinum resistance in ovarian cancer (9–11).
In ovarian cancer, the role of LINC00473 remains
largely unexplored. Research has indicated that lncRNAs serve a
pivotal role in the progression and chemoresistance of ovarian
cancer, highlighting their potential as therapeutic targets
(12–14). For example, lncRNAs such as
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and
HOX transcript antisense RNA (HOTAIR) have been demonstrated to
facilitate ovarian cancer cell proliferation and to confer
cisplatin (CDDP) resistance through diverse mechanisms (6–8).
Nonetheless, the precise function of LINC00473 in ovarian cancer
progression and its viability as a therapeutic target have yet to
be comprehensively studied.
While the mechanistic roles of certain lncRNAs such
as MALAT1 and HOTAIR have been mechanistically associated with
ovarian cancer progression and CDDP resistance, the functional
landscape of LINC00473 in this malignancy remains largely
unexplored. Based on its established roles in CSC maintenance and
therapy resistance across other types of cancer such as non-small
cell lung cancer (11), it may be
hypothesized that LINC00473 drives ovarian cancer progression by
enhancing proliferative capacity, metastatic potential and CDDP
tolerance through Sox2/YAP1-dependent signaling. To address this
knowledge gap, the present study aimed to investigate the direct
effects of LINC00473 on key malignant phenotypes in two ovarian
cancer cell lines, including proliferation, invasion, apoptosis and
CDDP resistance. Furthermore, the underlying molecular mechanisms
were preliminarily explored by analyzing Sox2/YAP1 protein
expression levels using western blotting, providing initial
evidence for the proposed signaling involvement. This integrated
approach sought to characterize both the phenotypic impact of
LINC00473 and potentially establish a foundation for further
mechanistic dissection in future research.
Materials and methods
Cell culture and transfection
SK-OV-3 and A2780 ovarian cancer cells, obtained
from Procell Life Science & Technology Co., Ltd. and iCell
Bioscience Inc. companies respectively, were routinely cultured in
McCoy's 5A medium (cat. no. PM150710; Procell Life Science &
Technology Co., Ltd.) supplemented with 10% FBS (cat. no. FCS500;
Shanghai ExCell Biology, Inc.) and Neomycin Solution Stabilized
(cat. no. P477894; 1:100; Shanghai Aladdin Biochemical Technology
Co., Ltd.). The cells were maintained under conditions of 37°C, 5%
CO2 and 98% relative humidity, growing as a monolayer
adherent to the culture surface. Once the cells reached 80–90%
confluence, they were digested with 0.25% trypsin (cat. no.
PB180226; Procell Life Science & Technology Co., Ltd.) and
collected during the logarithmic growth phase.
To enhance the inhibitory effect on LINC00473, the
knockdown of LINC00473 by using three small interfering RNA (siRNA,
GuangZhou Ribobio Co., Ltd.) sequences simultaneously targeting
distinct regions of its gene sequence (sequences listed in Table I). During transfection, the
transfection reagent Lipofectamine® 2000 (cat. no.
11668019; Invitrogen; Thermo Fisher Scientific, Inc.) was used to
deliver chemically synthesized siRNAs into ovarian cancer cells at
a final siRNA concentration of 50 nM. The transfection mixture was
incubated with cells at 37°C in a humidified atmosphere containing
5% CO2 for 6 h, after which the medium was replaced with
fresh complete medium. The transfection reagent facilitated the
formation of RNA-lipid complexes, which mediated the delivery of
siRNA into cells. The siRNA particularly bound to complementary
sequences in LINC00473, leading to its cleavage and degradation via
the RNA-induced silencing complex pathway, thus reducing its
expression levels.
 | Table I.siRNA sequences and qPCR primers
information. |
Table I.
siRNA sequences and qPCR primers
information.
| Gene | Primer | Sequence
(5′-3′) | PCR Products |
|---|
| siRNA targeting
LINC00473 | S1 |
GCACUUCCAGGAACAUCAU | – |
|
| S2 |
GCUUCCAUUGCUGGAGCUU | – |
|
| S3 |
GCCUGGUUGUUGGCAAGUA | – |
| Homo sapiens
LINC00473 | Forward |
TGCAAAGCGGACACCTAGTA | 215 bp |
|
| Reverse |
TTCTCCAGTTACCACCCACC |
|
| Homo sapiens
GAPDH | Forward |
TCAAGAAGGTGGTGAAGCAGG | 115 bp |
|
| Reverse |
TCAAAGGTGGAGGAGTGGGT |
|
The experiment was divided into three groups: i)
si-negative control (siNC) group (transfected with non-targeting
scrambled siRNA); ii) siLINC00473 group (transfected with
LINC00473-specific siRNA); and iii) untreated control group
(untransfected cells). At 6 h post-transfection, the medium was
replaced with fresh complete medium and the cells were cultured for
a further 48 h. Each group included three replicate (wells) and the
entire experiment performed as three independent biological
replicates. Cells in the logarithmic growth phase (70–80%
confluence) were collected for subsequent analyses.
LINC00473 knockdown efficiency
analysis by reverse transcription-quantitative (q) PCR
Cells in the logarithmic growth phase were collected
48 h post-transfection from the following three groups: i) siNC
group; ii) siLINC00473 group; and iii) untreated control group.
Cells were washed twice with ice-cold PBS, then lysed in 1 ml/well
TRIzol® reagent (cat. no. 15596026; Invitrogen; Thermo
Fisher Scientific, Inc.) in a 6-well plate. Total RNA was extracted
using chloroform-isopropanol precipitation and resuspended in
RNase-free water. RNA concentration and purity (A260/A280 ratio,
1.8–2.1) were verified using a NanoDrop spectrophotometer (Thermo
Fisher Scientific, Inc.).
Total RNA (1 µg) was then reverse-transcribed using
the PrimeScript™ RT Master Mix kit (Takara Biotechnology Co., Ltd.)
in a 20-µl reaction, which included a genomic DNA removal step
(37°C for 2 min) followed by cDNA synthesis (37°C for 15 min and
85°C for 5 sec). qPCR was performed using SYBR Premix Ex Taq™
(Takara Biotechnology Co., Ltd.) in a 10 µl reaction containing 0.4
µM each primer (sequences listed in Table I) and 2 µl cDNA (1:10 dilution). The
cycling conditions were as follows: 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec and 60°C for 30 sec. Melt curve analysis
(65–95°C, increment 0.5°C/sec) confirmed single-peak amplification
(15). The relative expression
level of LINC00473 was normalized to GAPDH using the
2−ΔΔCq method.
Cell Counting Kit 8 (CCK-8) assay
Logarithmic-phase cells from the siNC group,
siLINC00473 group and untreated control group were trypsinized and
resuspended in complete medium. Cells were then seeded in a 96-well
plate at a density of 2.0×104cells/well, with five
technical replicates per group. An additional cell-free blank
control (100 µl complete medium only) was included to measure
background absorbance. The outer wells of the plate were filled
with 200 µl PBS to minimize edge effects.
Enhanced CCK-8 assays were performed at 24, 48, 72
and 96 h post-seeding. At each time point, the culture medium was
aspirated and 100 µl fresh complete medium containing 10 µl CCK-8
reagent (cat. no. E-CK-A362; Wuhan Elabscience Biotechnology Co.,
Ltd.) was added to each well (final volume, 110 µl/well). The plate
was incubated at 37°C in the dark for 2 h. Absorbance (OD value) at
450 nm was measured using a microplate reader (BioTek Synergy HTX;
BioTek; Agilent Technologies, Inc.). Cell viability (%) was
calculated using the following formula:
[(ODexperimental-ODblank)/(ODcontrol-ODblank)]x100%.
A proliferation curve was generated by plotting cell viability (%)
on the y-axis against time points on the x-axis.
Cell migration and invasion
assays
Logarithmic-phase cells were washed twice with PBS,
digested with 0.25% trypsin at 37°C for 2–3 min and resuspended in
culture medium to a density of 1.0×106 cells/ml. For the
migration assay, 200 µl cell suspension in serum-free medium (0%
FBS) was added to the upper chamber of the Transwell insert, while
600 µl of medium containing 10% FBS (cat. no. 10099-141C; Gibco;
Thermo Fisher Scientific, Inc.) was added to the lower chamber.
For the invasion assay, Matrigel (cat. no. 356234;
Corning, Inc.) was thawed overnight at 4°C and diluted with
serum-free medium at a 1:8 ratio (v/v) on ice. A total of 40 µl of
diluted Matrigel was evenly applied to the upper surface of the
Transwell membrane (pore size, 8.0 µm; Corning, Inc.) using a
pre-chilled pipette tip. The chamber was then incubated at 37°C for
5 h for gel polymerization (confirmed by inverted microscopy).
Unpolymerized Matrigel was aspirated gently before cell
seeding.
Non-migrated/invaded cells on the upper surface were
removed with a cotton swab after the cells were incubated at 37°C
in a humidified atmosphere with 5% CO2 for 24 h.
Migrated/invaded cells on the lower surface were fixed with 70%
ice-cold ethanol at 4°C for 15 min, washed twice with PBS and
stained with 0.5% crystal violet in 25% methanol at room
temperature for 20 min. Excess stain was removed by rinsing three
times with distilled water and chambers were air-dried. Migrated or
invaded cells were counted in five random fields per chamber at
×200 magnification using a light microscope. Images were analyzed
with ImageJ (version 1.8.0; National Institutes of Health) by
thresholding and particle analysis.
Flow cytometric analysis of
apoptosis
Adherent cells were detached using 0.25% trypsin
(37°C; 2 min) and digestion was terminated by adding complete
medium containing 10% FBS. The cell suspension was washed twice
with ice-cold PBS (300 × g, 5 min) and lastly resuspended in 1X
Annexin V Binding Buffer (cat. no. GK10037; Shanghai HongYe Biotech
Co., Ltd.) to a density of 1×106 cells/ml. This
concentration was experimentally determined to prevent cell
aggregation during staining while maintaining sufficient event
counts for statistical analysis. Aliquots of 100 µl
(~1×105 cells) were transferred to 1.5-ml
microcentrifuge tubes, to which 5 µl Annexin V-FITC conjugate and 5
µl PI stock solution (50 µg/ml in PBS) were added sequentially. The
tubes were gently vortexed for 3 sec and incubated in the dark at
room temperature for precisely 15 min, with periodic inversion
every 5 min to ensure uniform dye distribution. Following
incubation, Binding Buffer was added to each tube to terminate
staining.
To ensure accurate gating and compensation, three
control samples were processed in parallel: Negative control
(unstained cells), single-stained controls (Annexin V or PI only)
and positive control (apoptosis-induced cells). Flow cytometric
detection was performed using a BriCyte E6 flow cytometer
(Mindray). Data analysis was conducted with MR Flow software
(version 3.0.2, Mindray) to quantify apoptotic cells. Flow
cytometry parameters were set to detect FITC (Ex 488 nm/Em 530 nm)
and PI (Ex 488 nm/Em 617 nm). Compensation values were adjusted
using the single-stained controls: 5–10% from FITC to PI channel
and 1–3% from PI to FITC channel to correct for fluorescence
spillover. Data analysis distinguished live cells (Annexin
V−/PI−), early apoptotic (Annexin
V+/PI−) and late apoptotic/necrotic
(double-positive) populations and total apoptosis rate (%) was
calculated as follows: (Early+late apoptotic cells) / total cells ×
100%. All steps were performed under light-protected conditions and
detection was completed within 1 h after staining to ensure
reliability.
Western blot analysis
Transfected cells were harvested by scraping in
ice-cold PBS, followed by two washes to remove residual medium.
Cell pellets were lysed in RIPA buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) supplemented with protease/phosphatase
inhibitors cocktail (1X final concentration; cat. no. 78440; Thermo
Fisher Scientific, Inc.) on ice for 30 min with intermittent
vortexing (every 10 min). Lysates were clarified by centrifugation
at 14,000 × g for 15 min at 4°C, and supernatants were transferred
to pre-chilled 1.5 ml tubes. Protein concentration was determined
using a BCA assay kit (cat. no. 23225; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions, with BSA (cat.
no. G3522; GBCBIO Technologies, Inc.) as a standard. Samples were
adjusted to 2 µg/µl with lysis buffer, mixed with 5X SDS loading
buffer to a final 1X concentration and denatured at 95°C for 5
min.
Proteins were separated by SDS-PAGE gels (1.5 mm
thick) with 4% stacking gels. Samples (20–40 µg protein per lane,
quantified by BCA assay) were electrophoresed at 120 V for 1.5 h in
standard running buffer. Following separation, the proteins were
transferred to polyvinylidene fluoride membranes (0.45 µm) using
wet transfer at 300 mA for 90 min in transfer buffer containing 20%
methanol. Transfer efficiency was verified by Ponceau S staining (5
min) and destaining.
Membranes were blocked with 5% (w/v) non-fat dry
milk in TBS with Tween-20 [TBST; 20 mM Tris-HCl (pH 7.5), 150 mM
NaCl and 0.1% Tween-20] at room temperature for 1 h with gentle
agitation. The following primary antibodies were diluted in
antibody dilution buffer (5% BSA in TBST): Anti-caspase-3 (cat. no.
25128-1-AP; 1:1,000; Proteintech Group, Inc.), anti-cleaved-poly
(adenosine diphosphate-ribose) polymerase (PARP; cat. no.
bsm-52408R; 1:1,000; BIOSS), anti-YAP1 (cat. no. bs-3605R; 1:2,000;
BIOSS), anti-Sox2 (cat. no. bs-0523R; 1:500; BIOSS) and
anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; cat. no.
60004-1-Ig; 1:5,000; Proteintech Group, Inc.). Membranes were
incubated with primary antibodies overnight at 4°C, followed by
three 10-min washes in TBST. Horseradish peroxidase-conjugated goat
anti-rabbit (cat. no. A0208; 1:5,000) and goat anti-mouse (cat. no.
A0216; 1:5,000) IgG H&L secondary antibodies (both from
Beyotime Institute of Biotechnology) were applied for 1 h at room
temperature, followed by three additional TBST washes.
Chemiluminescent signals were detected using ECL
substrate (cat. no. WBKLS0500; MilliporeSigma) and were captured
with a ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc.).
Exposure times were optimized for each target protein (10 sec-10
min) to avoid signal saturation, with multiple exposures collected
for dynamic range verification. Band intensities were
semi-quantified using ImageLab software (version 6.1; Bio-Rad
Laboratories, Inc.).
CDDP sensitivity assay
Ovarian cancer cells were divided into four groups
(siNC, siLINC00473, siNC+30 µM CDDP and siLINC00473+30 µM CDDP),
which were co-treated with CDDP (cat. no. 15663-27-1;
MedChemExpress) for 48 h post-transfection and analyzed for cell
viability using the CCK-8 assay. Briefly, culture medium was
discarded, the cells were washed with PBS and fresh medium
containing 10% CCK-8 reagent was add to each well and incubated at
37°C in a humidified atmosphere with 5% CO2 in the dark
for 2 h. Lastly, the absorbance was measured at 450 nm using a
microplate reader and the relative survival rate (% of control
group) was calculated. Three independent replicates were performed
to evaluate the cytotoxic effects of CDDP and the regulatory effect
of LINC00473 silencing on drug sensitivity.
Statistical analysis
Relative cell proliferation was expressed as the
mean OD450 (blank-corrected) at each time point. For
CCK-8 relative cell viability (normalized to the siNC control), a
two-way ANOVA evaluated the effects of siRNA treatment (siNC vs.
siLINC00473) and CDDP exposure (0 vs. 30 µM), including their
interaction. Data were normalized to the 24 h time point (set to
100%) to compare proliferation rates across the groups. All
experiments were performed with three independent biological
replicates. Data are presented as mean±SD from three replicates.
Bar graphs display the mean±SD with individual data points and
significant differences are marked as *P<0.05, **P<0.01,
***P<0.001. Statistical analyses were performed using GraphPad
Prism (version 20.0; Dotmatics) with α=0.05 (two-tailed). Prior to
selecting the statistical test, the normality of the data was
assessed using Q-Q plot analysis. One-way/two-way ANOVA (with
Tukey's post hoc test) was applied for data that conformed to a
normal distribution, while the Kruskal-Wallis test was used for
non-normal data. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression level of the lncRNA
LINC00473
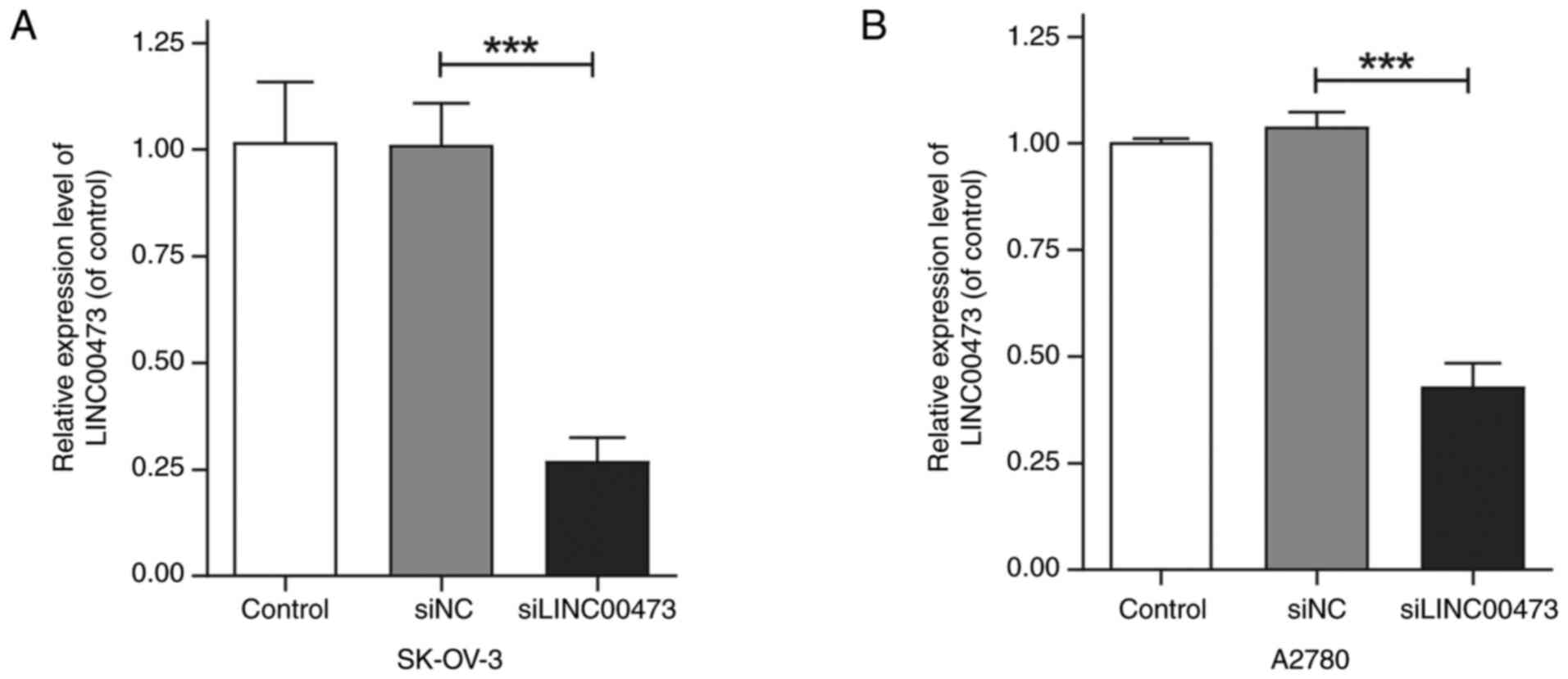
To investigate the knockdown efficiency of siRNA
targeting the lncRNA LINC00473, SK-OV-3 and A2780 ovarian cancer
cell lines were transfected with siLINC00473 or siNC using a
Lipofectamine-based transfection method. qPCR analysis revealed a
significant reduction in LINC00473 expression in both cell lines
transfected with siLINC00473. Compared with that in the siNC and
blank control groups, LINC00473 expression was decreased by
27.01±4.70% (P<0.01, Fig. 1A) in
SK-OV-3 cells and 42.96±4.39% (P<0.01, Fig. 1B) in A2780 cells, with statistically
significant differences. Notably, the knockdown efficiency
demonstrated no notable variation between the two cell lines,
indicating the broad applicability of siRNA-mediated LINC00473
suppression.
Cell proliferation and apoptosis
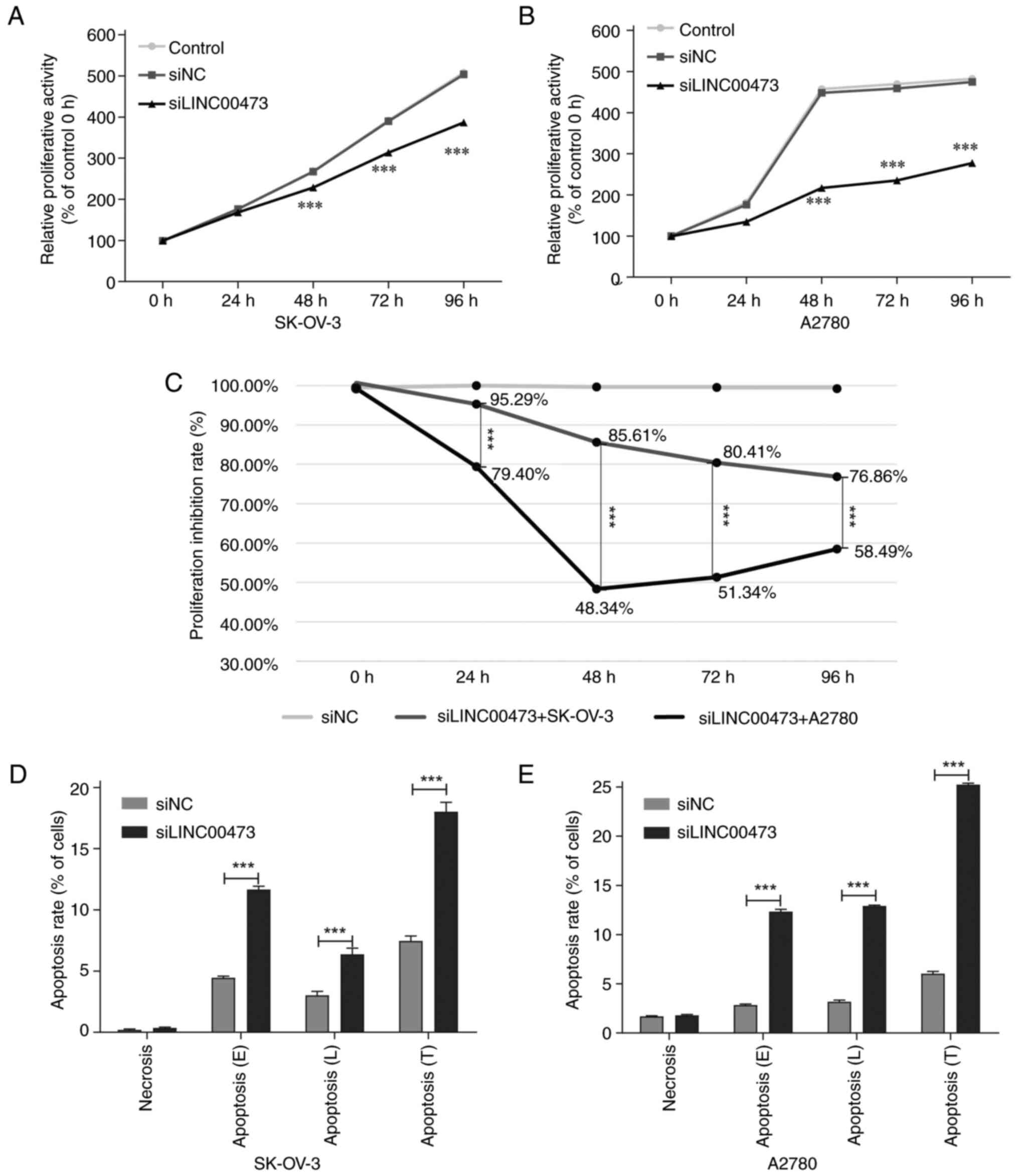
Time-course analysis demonstrated that siLINC00473
transfection induced progressive and duration-dependent suppression
of the proliferative activity of both SK-OV-3 and A2780 ovarian
cancer cells. Significant inhibition emerged at 48 h
post-transfection compared with in the siNC and untreated control
groups (Fig. 2A and B). At this
time point, SK-OV-3 cells retained 85.61% relative proliferative
activity (14.39% suppression rate, P<0.001), while A2780 cells
exhibited markedly stronger suppression, with 48.34% residual
proliferative activity (51.66% suppression rate, P<0.001). By 72
h, SK-OV-3 suppression further intensified, with the suppression
rate increased to 19.59% (P<0.001), whereas A2780 cells
exhibited no additional suppression, stabilizing at 48.66%
suppression rate (P<0.001). At the 96 h endpoint, suppression
rate plateaued at 23.14% for SK-OV-3 (P<0.001) and 41.51% for
A2780 cells (P<0.001). These findings indicated the specific
anti-proliferative efficacy of siLINC00473 in ovarian cancer models
(Fig. 2C).
siLINC00473-induced apoptosis in ovarian cancer
cells exhibited time-dependent dynamics and was synchronized with
cell proliferation inhibition (Figs.
2D, S1A and S1B). During the 48 h early apoptotic
phase (E), the apoptosis rate of SK-OV-3 cells was 11.66±0.27%,
significantly higher compared with that of the siNC group
(4.45±0.14%; P<0.001), accompanied by a proliferation inhibition
rate of 14.39±1.23%. A2780 cells displayed a similar apoptotic
response (Figs. 2E, S1C and S1D), with an apoptosis rate of
12.33±0.23% (vs. siNC group, 2.84±0.11%; P<0.001), while its
proliferation inhibition rate reached 51.66%±0.68%. By the 72 h
late apoptotic phase (L), the apoptosis rate of SK-OV-3 decreased
to 6.36±0.52%, remaining significantly higher compared with that in
the siNC group (3.01±0.34%; P<0.001), yet its proliferation
inhibition rate increased to 19.59±1.85%. By contrast, A2780 cells
maintained a high apoptosis rate (12.90%±0.10%, vs. siNC group,
3.18±0.17%; P<0.001), although its proliferation inhibition rate
slightly declined to 48.66±4.13%. Total apoptosis rates (E+L; T)
revealed that SK-OV-3 cells reached 18.02±0.76% and A2780 cells
25.23±0.15%, both significantly higher compared with their
respective control groups (SK-OV-3 control, 7.45±0.41%; P<0.001;
A2780 control, 6.02±0.24%; P<0.001). While apoptosis and
proliferation inhibition rates in both cell lines increased over
time, A2780 cells demonstrated higher sensitivity to siLINC00473,
as evidenced by its elevated total apoptosis and proliferation
inhibition rates compared with SK-OV-3 cells.
Suppression of cell migration and
invasion
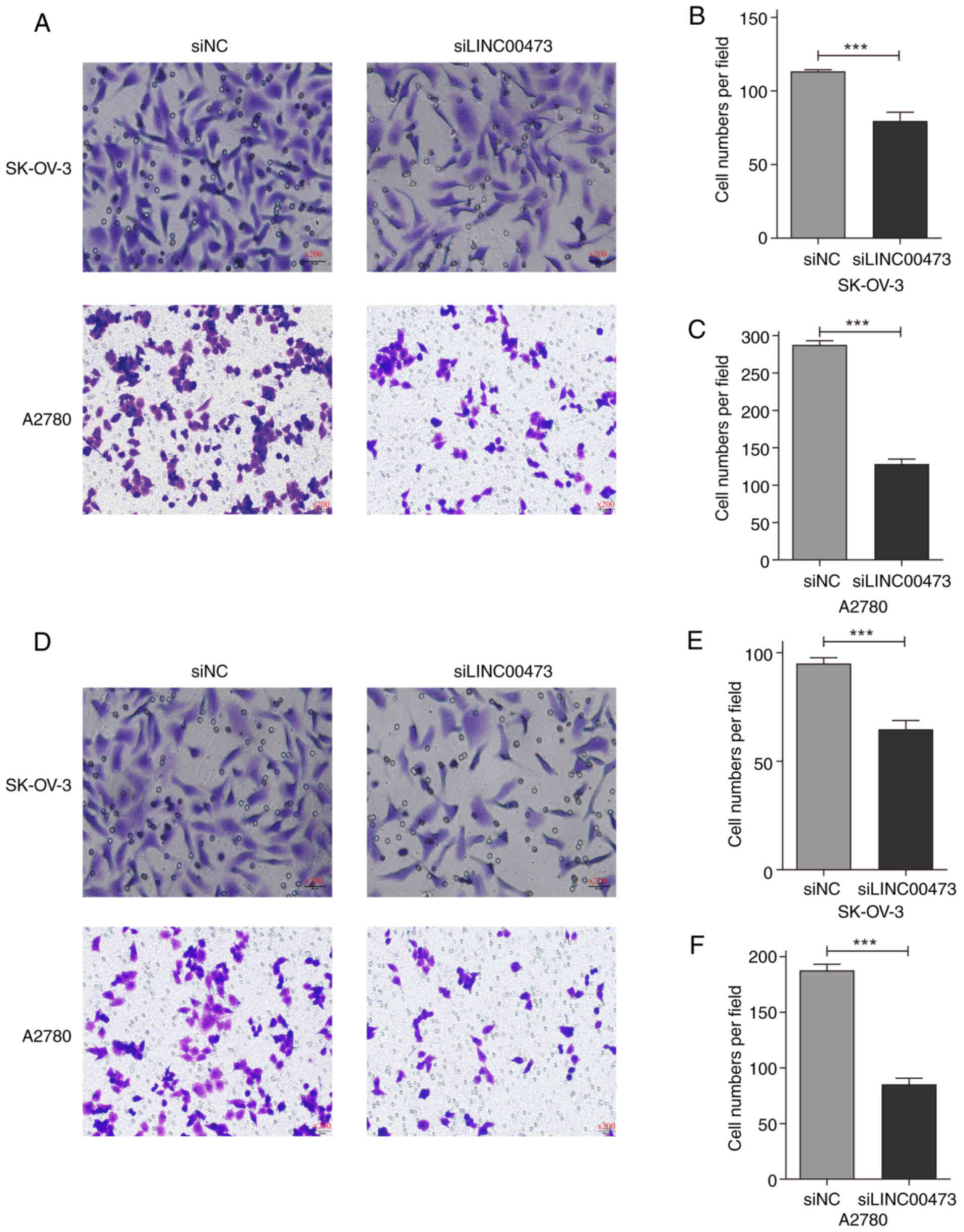
To investigate the regulatory role of LINC00473 in
ovarian cancer metastasis, the present study systematically
analyzed cell migration and invasion phenotypes using Transwell
assays. The results demonstrated that transfection with siLINC00473
significantly suppressed both migratory and invasive capacities in
the two ovarian cancer cell lines (Fig.
3).
In SK-OV-3 cells (Fig.
3B and E), the migratory cell count decreased from 113.3±1.2
cells/field in the siNC group to 79.7±5.9 cells/field (30.71%
inhibition; P<0.001), while the invasive cell count dropped from
95.0±2.6 to 64.7±4.0 cells/field (31.93% suppression; P<0.001).
A2780 cells exhibited even more pronounced inhibition (Fig. 3C and F): Migratory cell numbers
sharply declined from 287.7±5.5 to 128.3±6.5 cells/field (55.39%
reduction; P<0.001) and invasive cell counts decreased from
187.7±5.5 to 85.3±5.5 cells/field (54.53% attenuation;
P<0.001).
Microscopic imaging (×200 magnification) revealed a
marked reduction in membrane, traversing cell density in the
siLINC00473-transfected groups, accompanied by pseudopodia
retraction and a rounded cellular morphology (Fig. 3A and D). Statistical analyses
further confirmed that A2780 cells demonstrated a 25% higher
sensitivity to LINC00473 silencing compared with SK-OV-3 cells, as
evidenced by the differential inhibition rates in both migration
and invasion. These findings collectively highlighted the key role
of LINC00473 in regulating the metastatic behaviors of ovarian
cancer cells.
Regulation of apoptosis and oncogenic
proteins
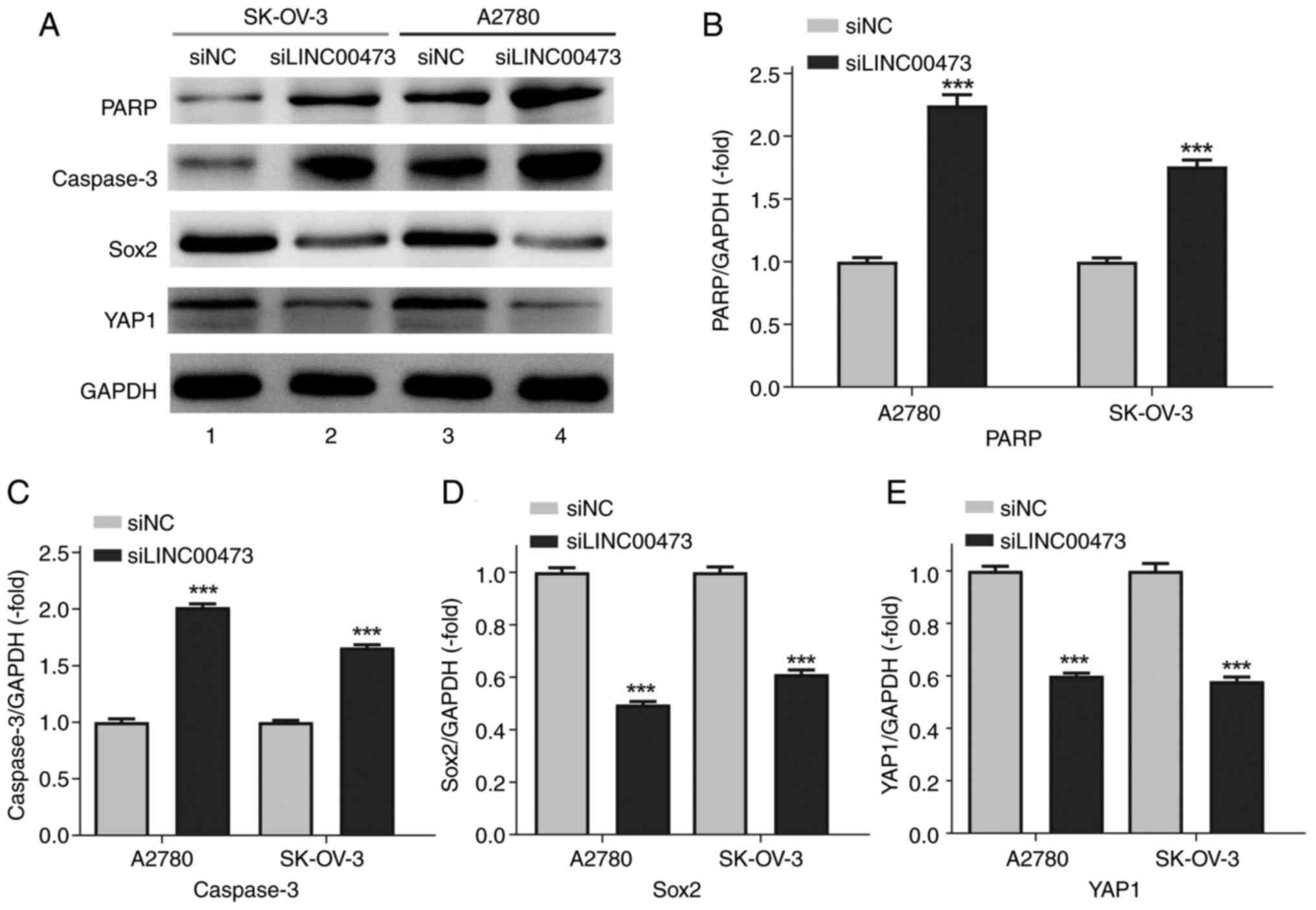
Western blot analysis (Fig. 4A) revealed that transfection with
siLINC00473 significantly upregulated the expression levels of
apoptosis-associated proteins PARP and caspase-3 (both P<0.001)
compared with those in the siNC group, while markedly suppressing
the protein levels of stemness and metastasis-related markers Sox2
and YAP1 (both P<0.001). Semi-quantitative normalization to
GAPDH (Fig. 4B-E) demonstrated that
the PARP/GAPDH ratio in SK-OV-3 and A2780 cells increased to
1.76±0.15-fold and 2.25±0.25-fold of the control group,
respectively. Similarly, caspase-3/GAPDH ratios rose to
1.66±0.08-fold (SK-OV-3) and 2.02±0.09-fold (A2780), indicating
robust activation of apoptotic pathways upon LINC00473 silencing
(Fig. 4B and C).
Concurrently, Sox2 and YAP1 expressions exhibited
consistent yet quantitatively distinct inhibition across the two
cell lines (Fig. 4D and E). In
SK-OV-3 cells, Sox2/GAPDH and YAP1/GAPDH ratios decreased to
0.61±0.05-fold and 0.58±0.05-fold of the control, respectively.
A2780 cells demonstrated more pronounced reductions, with Sox2 and
YAP1 levels dropping to 0.49±0.04-fold and 0.60±0.03-fold of the
control. These findings align with the aforementioned migration and
invasion assays. The coordinated suppression of stemness/metastasis
markers and activation of apoptosis further supports a dual
regulatory mechanism underlying the role of LINC00473 in ovarian
cancer progression.
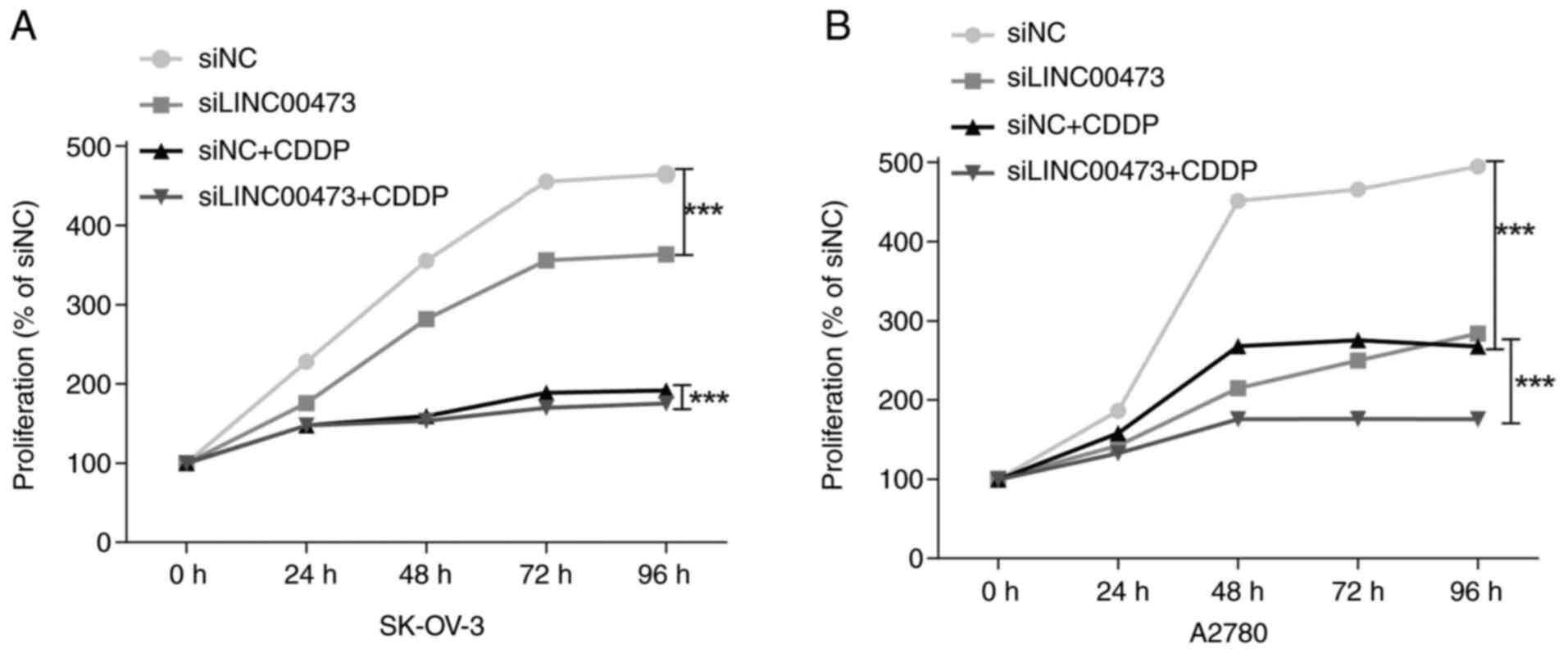
Enhanced CDDP sensitivity
Under CDDP treatment, both ovarian cancer cell lines
transfected with siLINC00473 exhibited significantly reduced
viability compared with that in the siNC group (both P<0.001),
with a time-dependent inhibitory effect. In SK-OV-3 cells (Fig. 5A), the relative proliferation rate
of the siLINC00473+CDDP combination group progressively declined
over time: 52.77±2.19% at 24 h, 43.19±0.39% at 48 h, 37.30±1.22% at
72 h, and 37.71±0.71% at 96 h, all significantly lower compared
with the siNC group (P<0.001). Although the siNC+CDDP group
indicated stronger antiproliferative effects compared with
siLINC00473 monotherapy, its impact remained weaker compared with
the combination group, while still being significantly suppressed
compared with siNC (P<0.001).
In A2780 cells (Fig.
5B), the inhibitory effects of the combined treatment were more
pronounced. Compared with in the siNC group, the siLINC00473+CDDP
group displayed a proliferation rate of 71.39±1.47% at 24 h,
followed by a sharp decline to 38.99±1.98% at 48 h and further
reductions to 37.82±0.35% at 72 h and 35.55±1.54% at 96 h
(P<0.001). Both the siNC+CDDP group and siLINC00473 monotherapy
group significantly suppressed proliferation (both P<0.001), but
their effects were less potent compared with the combination
treatment. These results indicated that LINC00473 silencing may
synergize with CDDP to induce time-enhanced antitumor activity,
with the inhibitory effects becoming particularly prominent after
48 h.
Discussion
LINC00473 is a lncRNA that is aberrantly upregulated
in various malignancies, including pancreatic cancer, liver cancer
and pituitary adenoma. LINC00473 expression level is markedly
associated with tumor invasion, metastasis and poor patient
prognosis, making it a potential pan-cancer biomarker (16,17).
Previous studies have reported that LINC00473 regulates tumor cell
malignancy through a competitive endogenous RNA (ceRNA) mechanism
by sponging microRNAs (miRNAs/miRs) (18), such as miR-502-3p (19), miR-424-5p (20) and miR-497-5p (21), thereby alleviating miRNA-mediated
suppression of downstream target genes. Furthermore, LINC00473
interacts with proteins such as phosphatidylethanolamine-binding
protein 1 to indirectly promote tumor proliferation by inhibiting
key signaling pathways (22–24).
The present study focused on ovarian cancer. Although not directly
involving miRNA research, silencing LINC00473 revealed its
biological functions: Suppression of LINC00473 significantly
inhibited ovarian cancer cell proliferation, migration and
invasion, which was closely associated with apoptosis activation
(PARP cleavage and caspase-3 activation) and downregulation of
CSC-related proteins (Sox2 and YAP1). Notably, LINC00473 silencing
significantly enhanced ovarian cancer cell sensitivity to CDDP,
suggesting its potential role in modulating pathways such as DNA
damage repair to influence chemotherapy efficacy.
Liposome transfection achieved significant silencing
of LINC00473 in SK-OV-3 and A2780 ovarian cancer cell lines, with
significant differences in silencing efficiency between SK-OV-3
(27.01%) and A2780 (42.96%). However, this efficiency contrasts
with higher lncRNA silencing rates reported in other studies. For
example, siRNA-mediated knockdown of lncRNA HULC in liver cancer
has been reported to achieve 52–70% efficiency (25) and siRNA targeting lncRNA HOTAIR in
SK-OV-3 demonstrated 51% knockdown efficiency (26), both markedly surpassing the 27.01%
observed in SK-OV-3 cells in the present study. This discrepancy
may stem from the use of lentiviral infection in the aforementioned
previous studies, which yield higher silencing efficiency compared
with liposome-based methods. Notably, the differential silencing
efficiency between SK-OV-3 and A2780 cells may arise from the
distinct molecular characteristics of the two cell lines. The
elevated expression levels of stemness markers such as CD133 in
A2780 4 cells could enhance cellular dependency on LINC00473,
thereby increasing susceptibility to silence interventions
(27).
Following transfection, siLINC00473 significantly
inhibited the proliferation of SK-OV-3 and A2780 cells. The rapid
response in A2780 cells (48 h inhibition rate, 51.67%) aligns with
previous reports of high sensitivity to lncRNA-dependent
proliferation in aggressive ovarian cancer cells (28–30),
while the later stagnation in inhibition may be associated with
limitations of in vitro culture conditions (such as nutrient
competition and microenvironment). By contrast, the delayed
inhibition in SK-OV-3 (23.14% at 96 h) may stem from its inherently
slower proliferation rate. The temporal synchronization between
apoptosis rates and proliferation inhibition suggests that
LINC00473 may coordinately regulate the proliferation-apoptosis
balance. The higher apoptosis rate in A2780 cells could be
associated with Bcl-2 family protein modulation, a phenomenon
previously observed in lncRNA CCAT1 silencing models (31). Notably, A2780 cells exhibited
stronger migration/inhibition suppression (~55%) compared with
SK-OV-3 cells (~30%), possibly due to a greater dependency on
epithelial-mesenchymal transition (EMT) phenotypes, analogous to
the mechanism by which lncRNA colon cancer-associated transcript 2
promotes EMT via the Wnt/β-catenin pathway (32).
The present study demonstrated that silencing
LINC00473 exerted a dual antitumor effect on ovarian cancer by
activating apoptotic pathways and suppressing proteins associated
with stemness and metastasis. Following LINC00473 knockdown,
cleaved-PARP were significantly upregulated, consistent with the
key roles of this proteins in regulating apoptosis (33). While activated caspase-3 was also
evaluated in the present study, it is important to clarify that the
level of cleaved caspase-3 was not actually assessed. Consequently,
the activation status of cleaved caspase-3 cannot be directly
determined, which represents a limitation of this study. Notably,
the upregulation of cleaved-PARP strongly suggests activation of
the apoptotic pathway, as this process is typically accompanied by
caspase-3 cleavage. The primary finding, that silencing LINC00473
promotes ovarian cancer cell apoptosis, still aligns with previous
studies associating LINC00473 to apoptotic regulation (20–22,34).
Simultaneously, the inhibition of Sox2 and YAP1
highlights the role of LINC00473 in modulating cancer stemness and
metastasis. Sox2 serves as a pivotal pluripotent factor
indispensable for sustaining the CSC population. It serves a key
role in driving chemotherapy resistance and relapse in ovarian
cancer (35). The present study
revealed that silencing LINC00473 resulted in a significant decline
in Sox2 levels, particularly in A2780 cells, where the levels were
0.49-fold lower compared with those in the control group. By
downregulating Sox2, LINC00473 may effectively diminish the CSC
subpopulation within ovarian tumors, leading to more efficient
suppression of tumor growth and progression. Furthermore, based on
the findings from the CDDP resistance study, it can be hypothesized
that LINC00473 modulates the sensitivity of ovarian cancer cells to
chemotherapeutic agents by regulating Sox2. When LINC00473 is
silenced and Sox2 levels decrease, tumor cells may potentially
become more vulnerable to the cytotoxic effects of chemotherapeutic
drugs, ultimately enhancing treatment outcomes. The Hippo pathway
effector, YAP1, contributes to metastatic dissemination by
promoting EMT and stromal remodeling (36). The present study demonstrated that
following LINC00473 knockdown, YAP1 underwent coordinated
downregulation, with levels decreasing by 0.58–0.60-fold. This
downregulation may impede EMT, curtail the migration and invasion
of ovarian cancer cells and thus, inhibit tumor metastasis.
Furthermore, it could disrupt the conducive tumor microenvironment
that facilitates cancer progression and metastasis, thereby
impacting the proliferation and survival of cancer cells.
The present study confirmed the synergistic
antitumor effect of LINC00473 silencing combined with CDDP
(siLINC00473+CDDP), which induced time-dependent proliferation
inhibition in both SK-OV-3 and A2780 cells, with significantly
enhanced efficacy after 48 h. These findings suggested that
targeting LINC00473 may overcome adaptive chemoresistance in tumor
cells, thereby sensitizing them to CDDP. The combination therapy
outperformed monotherapy (siLINC00473 or CDDP alone) in both cell
lines, particularly in A2780 cells, where the proliferation rate
dropped to 38.99±1.98% (vs. siNC group) after 48 h, indicating
time-accumulative synergy. This aligns with the clinical rationale
for chemotherapy cycle design, where prolonged drug exposure
amplifies therapeutic efficacy through cumulative molecular damage
(37). Similar synergy has been
reported in other lncRNA studies (38,39),
for example, silencing lncRNA HOTAIR has been shown to enhance
ovarian cancer cell sensitivity to paclitaxel via apoptosis pathway
activation (40). Although both
cell lines exhibited synergy, A2780 cells exhibited a more
pronounced response (96 h proliferation rate, 35.55±1.54% vs.
37.71±0.71% in SK-OV-3 cells). This discrepancy likely stems from
tumor heterogeneity: A2780 cells, derived from a chemotherapy-naïve
patient, exhibits lower intrinsic resistance, whereas SK-OV-3
cells, isolated from metastatic ascites, may possess stronger
microenvironmental adaptability (41). This underscores the need for
molecular subtyping to identify patient populations most likely to
benefit from such combinatorial strategies in future clinical
applications.
The present study revealed the key pro-tumorigenic
role of LINC00473 in ovarian cancer and its potential as a
therapeutic target. Experimental evidence demonstrated that
silencing LINC00473 suppressed malignant progression through dual
mechanisms: i) Activating the PARP/caspase-3 apoptosis pathway to
induce cell death; and ii) downregulating Sox2 and YAP1 to inhibit
CSC properties and metastasis-related phenotypes. Notably,
LINC00473 silencing exhibited significant synergistic effects with
CDDP treatment, particularly in chemotherapy-naïve A2780 cells,
which exhibit heightened sensitivity. The differential responses
across cell lines (including time-dependent proliferation
inhibition and varying levels of migration/invasion suppression)
reflect tumor heterogeneity and underscore the importance of
molecular subtyping for personalized therapy. These findings not
only validate the oncogenic function of LINC00473 but also reveal
its regulatory network independent of the ceRNA mechanism,
positioning it as a novel therapeutic target to inform clinical
strategies for ovarian cancer in the future.
Although the present in vitro experiments
have yielded valuable insights into the role of LINC00473 in
ovarian cancer, the lack of in vivo validation results stems
from limitations in project duration and funding. The in
vitro experimental environment, by its nature, cannot fully and
accurately replicate the intricate interplay among various factors
within the complex human physiological milieu. Notably, the
interference results of siLINC00473 have largely achieved the
anticipated verification effect. Due to this, it is imperative to
employ animal models in subsequent research to further investigate
and confirm the current findings. Based on the current research
findings, in future studies, knockdown/overexpression rescue
experiments in combination with pathway inhibitors should be
performed to thoroughly explore the direct interactions and
specific molecular mechanisms between Sox2, YAP1 and LINC00473 in
regulating the biological behavior of ovarian cancer cells, aiming
to clarify their regulatory relationships. Furthermore, it is
essential to perform a clinical evaluation of the therapeutic
potential of treatment methods targeting LINC00473.
In conclusion, LINC00473 has emerged as a pivotal
oncogenic lncRNA with multifaceted roles in ovarian cancer
progression and therapy resistance. Its upregulation may drive
tumor aggressiveness by orchestrating dual mechanisms: Apoptosis
evasion (via PARP/caspase-3 suppression) and CSC maintenance
(through Sox2/YAP1 upregulation). Silencing LINC00473 not only
inhibited proliferation, migration and invasion but also synergized
with CDDP, overcoming adaptive chemoresistance likely by impairing
DNA damage repair pathways. The divergent responses between SK-OV-3
(metastatic, slower-proliferating) and A2780 (chemotherapy-naïve,
aggressive) cells underscore the impact of tumor heterogeneity on
therapeutic outcomes, emphasizing the need for molecular subtyping
to tailor combinatorial strategies in the future.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Mr. Wenbo Zhao and
Mr. Chaoyang Chen (both affiliated with Xinjiang Dingju
Biotechnology Co., Ltd., Ürümqi, Xinjiang Uygur Autonomous Region,
China) for their technical support in data analysis.
Funding
The present study was supported by Special Programme for the
in-hospital Project of the People's Hospital of Xinjiang Uygur
Autonomous Region (grant no. 20210210).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CZ and KZ conceived and designed the study. CH and
MS collected the data. JM, WW and MG performed the analysis and
interpretation of the data. CZ drafted the manuscript. MG and KZ
revised the manuscript. WW and MG confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI
|
|
2
|
Lheureux S, Braunstein M and Oza AM:
Epithelial ovarian cancer: Evolution of management in the era of
precision medicine. CA Cancer J Clin. 69:280–304. 2019.PubMed/NCBI
|
|
3
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang X, Wang W, Zhu W, Dong J, Cheng Y,
Yin Z and Shen F: Mechanisms and functions of long non-coding RNAs
at multiple regulatory levels. Int J Mol Sci. 20:55732019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Slack FJ and Chinnaiyan AM: The Role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li L, Zhang X, Liu N, Chen X and Peng C:
LINC00473: A novel oncogenic long noncoding RNA in human cancers. J
Cell Physiol. 236:4174–4183. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang WZ, Zhou H and Yan Y: XIAP underlies
apoptosis resistance of renal cell carcinoma cells. Mol Med Rep.
17:125–130. 2018.PubMed/NCBI
|
|
8
|
Yang L, Xie HJ, Li YY, Wang X, Liu XX and
Mai J: Molecular mechanisms of platinum-based chemotherapy
resistance in ovarian cancer (review). Oncol Rep. 47:822022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zanconato F, Cordenonsi M and Piccolo S:
YAP/TAZ at the roots of cancer. Cancer Cell. 29:783–803. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Y, Li Z, Chen X and Zhang S: Long
non-coding RNAs: From disease code to drug role. Acta Pharm Sin B.
11:340–354. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mo J, Li B, Zhou Y, Xu Y, Jiang H, Cheng
X, Wu X and Zhang Y: LINC00473 promotes hepatocellular carcinoma
progression via acting as a ceRNA for microRNA-195 and increasing
HMGA2 expression. Biomed Pharmacother. 120:1094032019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang JY, Lu AQ and Chen LJ: LncRNAs in
ovarian cancer. Clin Chim Acta. 490:17–27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shankaraiah RC, Veronese A, Sabbioni S and
Negrini M: Non-coding RNAs in the reprogramming of glucose
metabolism in cancer. Cancer Lett. 419:167–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leeper NJ and Maegdefessel L: Non-coding
RNAs: Key regulators of smooth muscle cell fate in vascular
disease. Cardiovasc Res. 114:611–621. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Q, Wang G, Xu L, Yao Z and Song L:
Long non-coding RNA LINC00473 promotes glioma cells proliferation
and invasion by impairing miR-637/CDK6 axis. Artif Cells Nanomed
Biotechnol. 47:3896–3903. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu S, Fu W, Zhang L, Fu K, Hu J, Jia W
and Liu G: LINC00473 antagonizes the tumour suppressor miR-195 to
mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif.
51:e124162018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang H, Gao L, Chen Y, Zhang L, Bai Y,
Zhao C, Zhang L, Zuo L and Sun H: Identification of hub genes in
bladder transitional cell carcinoma through ceRNA network
construction integrated with gene network analysis. J Cell Mol Med.
28:e179792024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Qian Y, Zhang C, Wang W, Qiao Y,
Song H, Li L, Guo J, Lu D and Deng X: LncRNA LINC00473 is involved
in the progression of invasive pituitary adenoma by upregulating
KMT5A via ceRNA-mediated miR-502-3p evasion. Cell Death Dis.
12:5802021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang C and Yang T: Long Non-coding RNA
LINC00473 promotes breast cancer progression via miR-424-5p/CCNE1
pathway. Protein Pept Lett. 30:72–84. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu SH, Bo YH, Ma HC, Zhang HN and Shao MJ:
lncRNA LINC00473 promotes proliferation, migration, invasion and
inhibition of apoptosis of non-small cell lung cancer cells by
acting as a sponge of miR-497-5p. Oncol Lett. 21:4292021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu Y, Jiang Y, Wang Y, Zhao Z and Li T:
LINC00473 rescues human bone marrow mesenchymal stem cells from
apoptosis induced by dexamethasone through the PEBP1-mediated
Akt/Bad/Bcl-2 signaling pathway. Int J Mol Med. 47:171–182. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yeung K, Seitz T, Li S, Janosch P,
McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, et
al: Suppression of Raf-1 kinase activity and MAP kinase signalling
by RKIP. Nature. 401:173–177. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Noh HS, Hah YS, Zada S, Ha JH, Sim G,
Hwang JS, Lai TH, Nguyen HQ, Park JY, Kim HJ, et al: PEBP1, a RAF
kinase inhibitory protein, negatively regulates starvation-induced
autophagy by direct interaction with LC3. Autophagy. 12:2183–2196.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Song X, Wang X, Hu J and Jiang L:
Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in
human gastric cancer. J Med Biochem. 35:137–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Zhang Y, Shao Y, Yue X, Chu Y,
Yang C and Chen D: LncRNA HOTAIR down-expression inhibits the
invasion and tumorigenicity of epithelial ovarian cancer cells by
suppressing TGF-β1 and ZEB1. Discov Oncol. 14:2282023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Casagrande N, Borghese C, Agostini F,
Durante C, Mazzucato M, Colombatti A and Aldinucci D: In ovarian
cancer multicellular spheroids, platelet releasate promotes growth,
expansion of ALDH+ and CD133+ cancer stem cells, and protection
against the cytotoxic effects of cisplatin, carboplatin and
paclitaxel. Int J Mol Sci. 22:30192021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma N, Zhang M, Hu J, Wei Z and Zhang S:
Daphnetin induces ferroptosis in ovarian cancer by inhibiting
NAD(P)H:Quinone oxidoreductase 1 (NQO1). Phytomedicine.
132:1558762024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wojtowicz K and Nowicki M: The
characterization of the sensitive ovarian cancer cell lines A2780
and W1 in response to ovarian CAFs. Biochem Biophys Res Commun.
662:1–7. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, Guo Y, Xing Z, Gong T, Yang L, Yang
T, Chang B, Wang X, Yu B and Guo R: ABT-737 increases cisplatin
sensitivity through the ROS-ASK1-JNK MAPK signaling axis in human
ovarian cancer cisplatin-resistant A2780/DDP cells. Oncol Rep.
52:1222024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang DY, Li N and Cui YL: Long non-coding
RNA CCAT1 sponges miR-454 to promote chemoresistance of ovarian
cancer cells to cisplatin by regulation of surviving. Cancer Res
Treat. 52:798–814. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang B, Liu M, Zhuang R, Jiang J, Gao J,
Wang H, Chen H, Zhang Z, Kuang Y and Li P: Long non-coding RNA
CCAT2 promotes epithelial-mesenchymal transition involving
Wnt/β-catenin pathway in epithelial ovarian carcinoma cells. Oncol
Lett. 15:3369–3375. 2018.PubMed/NCBI
|
|
33
|
Wang H, Ge W, Jiang W, Li D and Ju X:
SRPK1-siRNA suppresses K562 cell growth and induces apoptosis via
the PARP-caspase3 pathway. Mol Med Rep. 17:2070–2076.
2018.PubMed/NCBI
|
|
34
|
Li S, Lv C, Li J, Xie T, Liu X, Zheng Z,
Qin Z, Hui X and Yu Y: LncRNA LINC00473 promoted colorectal cancer
cell proliferation and invasion by targeting miR-195 expression. Am
J Transl Res. 13:6066–6075. 2021.PubMed/NCBI
|
|
35
|
Ding LN, Yu YY, Ma CJ, Lei CJ and Zhang
HB: SOX2-associated signaling pathways regulate biological
phenotypes of cancers. Biomed Pharmacother. 160:1143362023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu C, Barger CJ and Karpf AR: FOXM1: A
multifunctional oncoprotein and emerging therapeutic target in
ovarian cancer. Cancers (Basel). 13:30652021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Disis ML, Taylor MH, Kelly K, Beck JT,
Gordon M, Moore KM, Patel MR, Chaves J, Park H, Mita AC, et al:
Efficacy and safety of Avelumab for patients with recurrent or
refractory ovarian cancer: Phase 1b results from the JAVELIN solid
tumor trial. JAMA Oncol. 5:393–401. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fu Y, Zhang Y, Cui J, Yang G, Peng S, Mi
W, Yin X, Yu Y, Jiang J, Liu Q, et al: SNP rs12982687 affects
binding capacity of lncRNA UCA1 with miR-873-5p: involvement in
smoking-triggered colorectal cancer progression. Cell Commun
Signal. 18:372020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Zhang H, Yin S, Yang Y, Yang H,
Yang J, Zhou Z, Li S, Ying G and Ba Y: lncRNA-encoded pep-AP
attenuates the pentose phosphate pathway and sensitizes colorectal
cancer cells to oxaliplatin. EMBO Rep. 23:e531402022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiang J, Wang S, Wang Z, Cai J, Han L, Xie
L, Han Q, Wang W, Zhang Y, He X and Yang C: HOTAIR promotes
paclitaxel resistance by regulating CHEK1 in ovarian cancer. Cancer
Chemother Pharmacol. 86:295–305. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kielbik M, Szulc-Kielbik I and Klink M:
Impact of selected signaling proteins on SNAIL 1 and SNAIL 2
expression in ovarian cancer cell lines in relation to cells'
cisplatin resistance and EMT markers level. Int J Mol Sci.
22:9802021. View Article : Google Scholar : PubMed/NCBI
|