Introduction
Cervical cancer (CC) is the fourth most common
cancer in female patients worldwide, with an estimated incidence of
662,301 novel cases and 348,874 deaths in 2022 (1,2). It
has been reported that ~90% of CC deaths occur in low- and
middle-income countries and are primarily associated with delayed
diagnosis and failure to deliver guideline-concordant treatment in
the advanced stages of CC (3).
According to the International Federation of
Gynecology and Obstetrics classification system, the treatment of
invasive CC depends on clinical evaluation and histopathological
characteristics of tumors (4).
However, some patients do not respond to the treatment chosen based
on these criteria, highlighting the need to find predictive
biomarkers to lower the risk of disease recurrence or metastasis
(5). Furthermore, the
identification of genetic and epigenetic alterations associated
with therapy resistance may increase survival in patients with
locally advanced (LA)CC (5).
SET domain-containing protein 2 (SETD2) is a tumor
suppressor associated with the trimethylation of lysine 36 on
histone 3 (H3K36me3), a histone mark primarily associated with
actively transcribed regions. Loss of function of SETD2 due to
deletion or mutation is frequently detected in hematological and
solid tumors, such as renal, lung, colon and breast cancer and CC
(6). Furthermore, SETD2 mutations
are associated with acquired chemoradiotherapy (CRT) resistance and
poor survival due to alterations in mechanisms associated with DNA
damage repair and tumor heterogeneity mediated by chromatin
instability (7).
The increased risk of CRT resistance associated with
the loss of SETD2 is associated with the increased activity of the
hedgehog (Hh) and Wnt/β-catenin pathways in patients with brain
cancer and osteosarcoma, respectively (7,8).
Additionally, abnormal Hh signaling is detected in multiple human
malignant tumors (9). In CC,
alterations in Hh signaling are frequently associated with an
increased risk of developing CRT, which may be associated with
tumor hypoxia (10–13). Furthermore, increased expression of
the glioma-associated oncogene homolog 1 (GLI1) protein, a
transcriptional activator of the Hh pathway, is associated with
chemoresistance in gynecological cancer (14). To the best of our knowledge,
however, the relationship between SETD2 and GLI1 protein levels has
not been investigated and their potential role as prognostic
markers in patients with LACC remains unknown.
The present study explored the relationship between
SETD2 and GLI1 based on their localization and protein levels using
immunohistochemical assays. In addition, the immunoreactivity
scores (IS) of SETD2 and GLI1, as well as the pathological
characteristics of tumors and survival indicators, were evaluated
to determine their potential utility as prognostic biomarkers in
patients with LACC.
Materials and methods
Patients and sample collection
The present retrospective study included 84
patients, aged 25–81 years, with histological results of squamous
cell cervical carcinoma (SCC), adenocarcinoma (ADC) and
adenosquamous cell carcinoma (ASC) who received treatment between
January 2016 and December 2018 at the National Cancer Institute
(México City, México). The inclusion criteria were as follows: i)
Patients with a diagnosis of LACC [stage IB1-IVA; International
Federation of Gynecology and Obstetrics (FIGO) 2009] (15); ii) patients who underwent biopsy;
iii) patients who did not receive treatment before biopsy
(chemotherapy, radiotherapy, immunotherapy or hormonal therapy) and
iv) patients who received concurrent treatment with cisplatin 40
mg/m2 weekly with external beam radiation therapy of 45
Gy. Patients with incomplete treatment schemes or other types of
primary cancer were excluded from the present study.
Treatment response was evaluated according to the
Response Evaluation Criteria In Solid Tumors (RECIST; version 1.1)
(16). Patients with complete and
partial responses were defined as responders, whereas those with
stable or progressive disease were designated as
non-responders.
Sociodemographic and clinicopathological data were
collected from the medical files. Smoking was defined as
consumption of ≥2 cigarettes/week for ≥1 year at any point in their
lifetime; alcohol consumption was defined as alcohol intake >1
drink/day on average.
Immunohistochemistry (IHC)
IHC staining was performed using formalin-fixed
tissue as described previously (17). Formalin-fixed paraffin-embedded
tumor blocks were obtained from the Institutional Pathology Tissue
Bank between January 2024 and March 2024. Polyclonal antibodies
directed at the C-terminus of SETD2 (cat. no. HPA042451; 1:50;
Millipore Sigma) and GLI-1/GLI1 (cat no. C1; 1:50; Santa Cruz
Biotechnology, Inc.) were used for the IHC assays. The conditions
and antibody concentrations were previously validated in colon
adenocarcinoma tissue for GLI1 and in the human small intestine for
the SETD2 protein.
Two observers specializing in gynecological oncology
at the National Institute of Cancer independently evaluated and
scored immunoreactivity in a blinded manner. SETD2 and GLI1
expression were analyzed according to a previously described IS
(18). The percentage of
tumor-positive cells was graded as follows: 0, <5%; 1, 6–25%; 2,
26–50%; 3, 51–75% and 4, 76–100%. Positive cells (5%) were used as
the cut-off to define negative tumors. The intensity of the
immunoreactivity was scored as follows: 1, weak; 2, moderate and 3,
strong. The percentage of positive cells and intensity values were
multiplied to obtain the IS. Localization was categorized as
membranous, cytoplasmic, nuclear or a combination.
DNA extraction and human
papillomavirus (HPV) genotyping
Genomic DNA was extracted from the tumor tissue
using the QIAamp DNA FFPE tissue kit (cat. no. 56404, Qiagen GmbH)
according to the manufacturer's instructions. The integrity of
extracted DNA was evaluated by PCR amplification of the β-globin
gene using specific primers (forward: 5′GAAGAGCCAAGGACAGGTAC3′;
reverse: 5′CAACTTCATCCACGTTCACC3′ as described previously (19). Primers were synthesized at
Integrated DNA Technologies (San Diego, Ca, USA).
HPV sequences were detected and typed using the E6
nested multiplex PCR protocol (Table
SI) (20). PCR products were
analyzed by electrophoresis on 2% agarose gels stained with GelRed
(Biotium, Inc.) and visualized using the iBright FL1500
documentation system (Thermo Fisher Scientific, Inc.). The HPV type
was determined by assessing the size of the amplified fragments.
DNA samples from HPV-positive HeLa and CaSki cell lines (American
Type Culture Collection), cultured in DMEM-F12 medium (GIBCO-BRL)
with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific,
Inc.) and incubated at 37°C in a humidified environment with 5%
CO2 was used as positive controls, while a mixture
without DNA was used as a negative control.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 22; IBM Corp.). Descriptive statistics are
presented as mean ± SD of ≥2 independent experimental repeats for
normally distributed values and median (25th and 75th percentiles)
for skewed variables. The unpaired Student's t-test, Mann-Whitney U
and Kruskal-Wallis tests were used to assess quantitative
variables. Pearson's correlation coefficient was used to evaluate
the correlation between SETD2 and GLI1 levels. Fisher's exact test
was used to evaluate differences between categorical variables.
Overall survival (OS) and disease-free survival (DFS) were
estimated using the Kaplan-Meier method according to IS categories.
Cox proportional hazard regression models were used to estimate the
hazard ratios (HR) and 95% CIs of the variables with OS and DFS.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characteristics of the study
population
The overall sociodemographic characteristics of the
patients are shown in Table SII.
The median age was 46 years (range, 25–81 years). According to the
BMI, 52.4% (44/84) of participants were overweight or obese.
Alcohol and tobacco consumption were reported in 15.5% (13/84) and
27.4% (23/84) of patients, respectively. Hormonal contraception was
reported in 20.2% (17/84) of patients. According to
histopathological classification, 89.3% (75/84) of patients had
SCC, 7.1% (6/84) had ADC and 3.6% (3/84) had ASC (Table SIII). Based on the RECIST criteria,
the overall response rate was 92.9% (78/84). Additional
clinicopathological characteristics of the patients, such as the
FIGO classification, lymph node status and tumor differentiation,
are provided in Table SIII.
The distribution of the sociodemographic and
histopathological characteristics, categorized by SCC and non-SCC
(ADC + ASC) groups, revealed a significant difference in tumor
differentiation (Table SIV). Most
patients in the SCC group had moderately differentiated tumors
(68.0%; 51/75), whereas those in the non-SCC group primarily had
poorly differentiated tumors (55.6%; 5/9).
Expression and localization of SETD2
and GLI1
The overall positivity of SETD2 and GLI1 proteins
was detected in 90.5% (76/84) and 82.1% (69/84) of tumors,
respectively (Table SV; Fig. S1). Samples positive for SETD2
revealed a consistent nuclear (n) staining pattern, whereas GLI1
protein localization indicated nGLI1 and both n and cytoplasmic
(c)GLI1 staining in 59.4% (41/69) and 40.6% (28/69) of samples,
respectively. According to the histological classification, the
positivity and localization of SETD2 and GLI1 were not
significantly different between the SCC and non-SCC groups
(Table SV).
Expression of SETD2 and GLI1 was compared using IS
values. Analysis of SETD2 and GLI1 IS values according to the
sociodemographic and histopathological characteristics of the
patients revealed that increased nGLI1 levels were significantly
associated with a history of hormonal contraceptive use (Table I). Tumors positive for HPV18 or
other genotypes demonstrated significantly increased levels of
nSETD2 compared with those positive for HPV16 (Table I). Differences in clinical outcomes
were not significantly associated with changes in nGLI1, ncGLI1 or
nSETD2 levels. However, an increasing trend was observed in ncGLI1
levels in overweight/obese patients (Table I).
 | Table I.GLI1 and SETD2 cellular localization
in patients with locally advanced cervical cancer. |
Table I.
GLI1 and SETD2 cellular localization
in patients with locally advanced cervical cancer.
|
| Cellular
localization |
|---|
|
|
|
|---|
| Variable | nGLI1 (n=41) | P-value | ncGLI1 (n=29) | P-value | nSETH2 (n=76) | P-value |
|---|
| BMIa, kg/m2) |
|
|
|
|
|
|
| Normal
(<25.0) | 8 (2–9) | 0.793 | 8 (8–12) | 0.098 | 4 (2–8) | 0.838 |
|
Overweight (≥25.0) | 8 (6–8) |
| 12 (8–12) |
| 2 (1–8) |
|
| Tobacco
consumptiona |
|
|
|
|
|
|
| No | 7 (4–9) | 0.772 | 12 (8–12) | 0.533 | 4 (2–8) | 0.286 |
|
Yes | 7 (2–8) |
| 8 (8–12) |
| 4 (1–6) |
|
| Alcohol
consumptiona |
|
|
|
|
|
|
| No | 8 (4–8) | 0.957 | 8 (8–12) | 0.842 | 4 (2–8) | 0.681 |
|
Yes | 6 (4–9) |
| 10 (8–12) |
| 3 (1–8) |
|
| Hormonal
contraceptiona |
|
|
|
|
|
|
| No | 8 (6–8) | 0.029c | 8 (4–12) | 0.365 | 4 (3–5) | 0.957 |
|
Yes | 9 (8–9) |
| 8 (8–12) |
| 4 (1–8) |
|
| FIGO
classificationb |
|
|
|
|
|
|
| II | 8 (6–8) | 0.247 | 8 (8–12) | >0.999 | 4 (1–8) | 0.400 |
|
III | 8 (4–8) |
| 10 (8–12) |
| 4 (1–6) |
|
| IV | 5 (1–8) |
| - |
| 4 (3–8) |
|
| Tumor
differentiationb |
|
|
|
|
|
|
|
Well | 6 (4–12) | 0.897 | 6 (4–8) | 0.149 | 5 (2–11) | 0.232 |
|
Moderately | 8 (6–8) |
| 12 (8–12) |
| 3 (1–6) |
|
|
Poorly | 7 (5–8) |
| 8 (6–10) |
| 5 (4–8) |
|
| Lymph node
statusa |
|
|
|
|
|
|
| N0 | 8 (6–9) | 0.327 | 8 (8–12) | 0.872 | 4 (1–8) | 0.968 |
| N1 | 6 (4–8) |
| 10 (8–12) |
| 4 (2–8) |
|
| HPV
genotypeb |
|
|
|
|
|
|
| 16 | 8 (4–8) | 0.734 | 10 (6–12) | 0.587 | 3 (1–4) | 0.031c |
| 18 | 5 (4–8) |
| 8 (4–8) |
| 5 (3–8) |
|
|
Other | 8 (6–9) |
| 8 (8–8) |
| 8 (1–9) |
|
| Clinical
responsea |
|
|
|
|
|
|
|
Responder | 6 (4–8) | 0.289 | 12 (8–12) | 0.416 | 4 (1–6) | 0.694 |
|
Non-responder | 8 (8–12) |
| 10 (8–12) |
| 7 (2–8) |
|
Relationship between SETD2 and GLI1
expression levels
The relationship between SETD2 and GLI1 expression
was evaluated based on tumor positivity and IS values. The
distribution of samples positive for nSETD2 revealed no
statistically significant differences between the nGLI1 (50.0%;
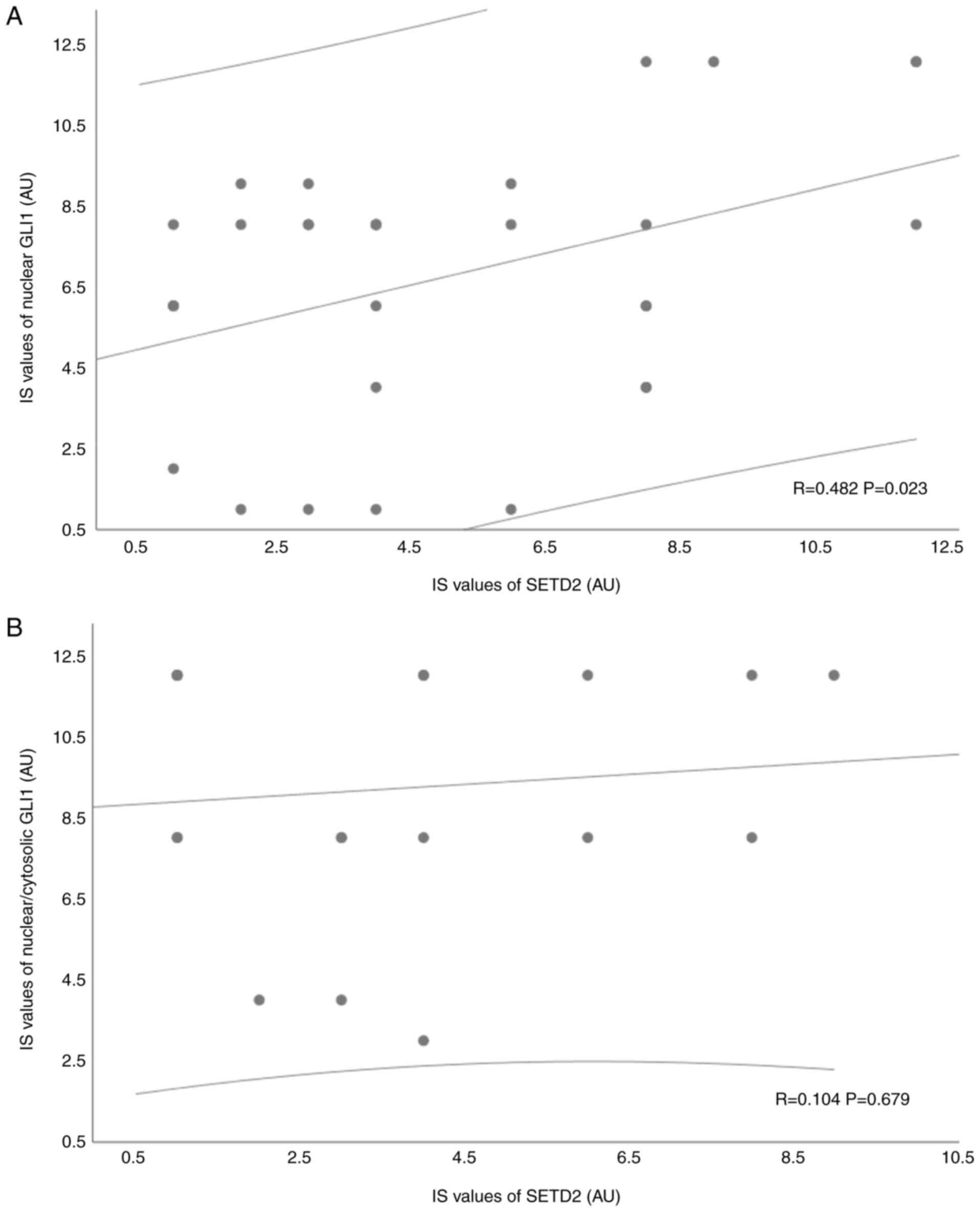
38/76) and ncGLI1 (31.6%; 24/76) groups (Table SVI). However, a positive
correlation was detected between nSETD2 and nGLI1 levels based on
IS values (R=0.428; Fig. 1A). No
significant association was found between nSETD2 and ncGLI1
expression (Fig. 1B).
Prognostic utility of SETD2 and GLI1
IS values
The median follow-up period was 53 months (range,
1–120 months). Patients positive for GLI1 or SETD2 demonstrated no
significant differences in OS and DFS rates compared with those
negative for these proteins (Fig.
S2). The predictive values of SETD2 and GLI1 for OS and DFS
were evaluated by categorizing the IS values according to cell
positivity >50% in combination with moderate or strong
immunoreactivity (IS >6; Figs
S3 and S4). Increased GLI1 and
SETD2 expression (IS >6) was observed in 65.2% (45/69) and 26.3%
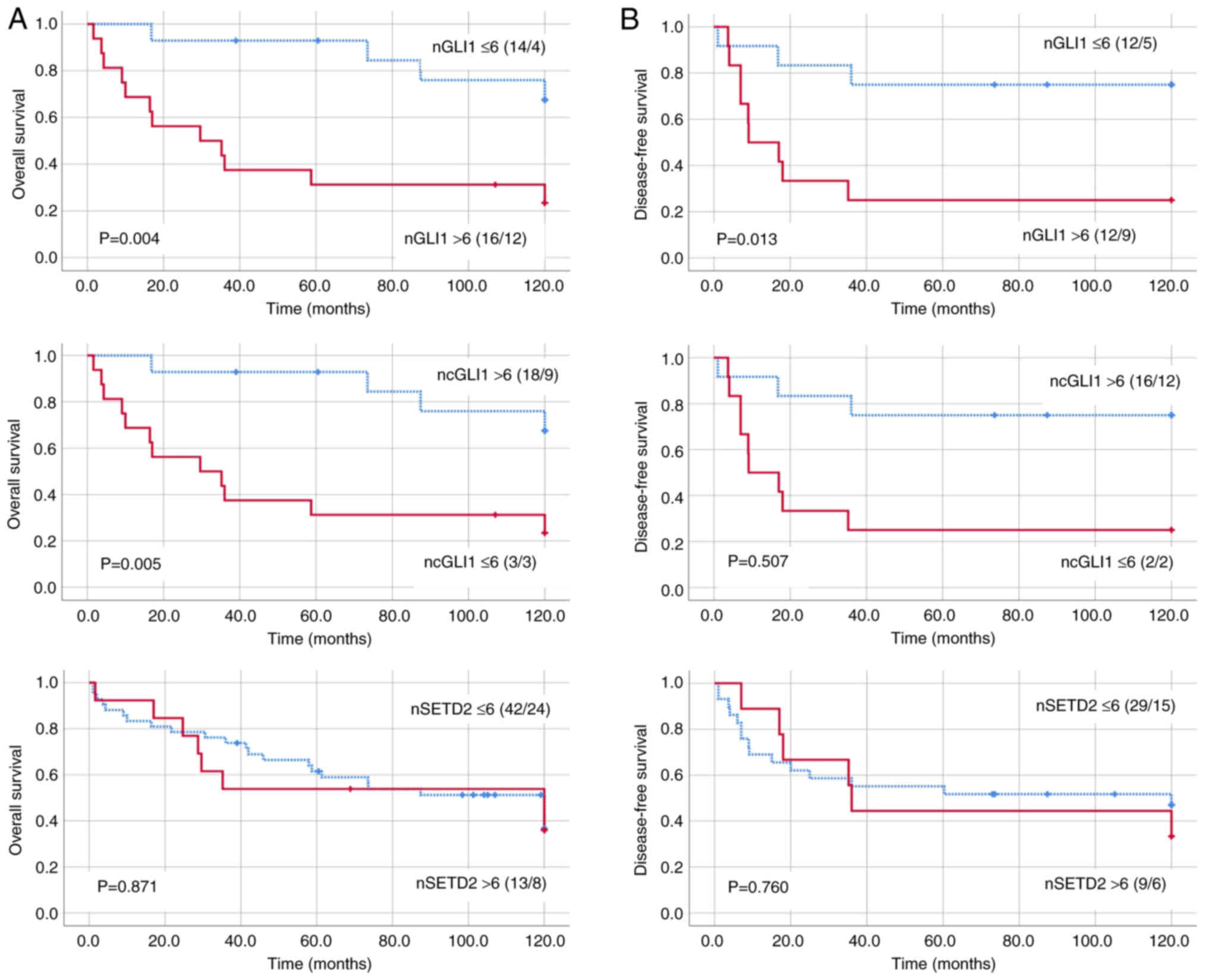
(20/76) of cases, respectively. Patients with nGLI1 IS >6 had
significantly worse OS and DFS rates compared with those with nGLI1
IS ≤6 (Fig. 2). Conversely,
patients with ncGLI1 IS >6 exhibited improved OS compared with
those with ncGLI1 IS ≤6. No significant differences were observed
in the OS or DFS between the categories for nSETD2.
The univariate analysis revealed an increased
probability of lower OS associated with nGLI IS >6 (HR, 1.50;
95% CI, 1.44–14.13) and ncGLI1 IS ≤6 (HR, 1.86; 95% CI,
1.42–29.55), whereas a lower probability of DFS was significantly
correlated with nGLI IS >6 (HR, 1.53; 95% CI, 1.23–17.49).
However, no significant association between nGLI1 and ncGLI1 and
clinicopathological characteristics was detected in the
multivariate analysis (Table
II).
 | Table II.Uni- and multivariate Cox regression
analysis of survival rates in patients with locally advanced
cervical cancer. |
Table II.
Uni- and multivariate Cox regression
analysis of survival rates in patients with locally advanced
cervical cancer.
| A, Overall
survival |
|---|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| nGLI IS ≤6 | Reference |
|
|
|
| 0.437 |
| nGLI IS >6 | 1.507 | 1.44–14.13 | 0.010 | 0.953 | 0.23–28.70 |
|
| ncGLI IS >6 | Reference |
|
|
|
| 0.988 |
| ncGLI1 IS ≤6 | 1.869 | 1.42–29.55 | 0.016 | 7.15 | 0.00–16.27 |
|
| Hormonal
contraception |
|
|
|
|
|
|
| No | Reference |
|
|
|
|
|
|
Yes | 0.431 | 0.57–4.14 | 0.394 |
|
|
|
| BMI,
kg/m2) |
|
|
|
|
|
|
| Normal
(<25.0) | Reference |
|
|
|
|
|
|
Overweight/obese (≥25.0) | 0.155 | 0.60–2.25 | 0.645 |
|
|
|
| Histological
subtype |
|
|
|
|
|
|
|
SCC | Reference |
|
|
|
|
|
|
Non-SCC | −0.089 | 0.32–2.56 | 0.866 |
|
|
|
| FIGO
classification |
|
|
|
|
|
|
| II | Reference |
|
|
|
|
|
|
III | 0.344 | 0.72–2.74 | 0.311 |
|
|
|
| IV | 0.129 | 0.47–2.70 | 0.771 |
|
|
|
| Lymph node
status |
|
|
|
|
|
|
| N0 | Reference |
|
|
|
|
|
| N1 | −0.145 | 0.42–1.78 | 0.694 |
|
|
|
|
| B, Disease-free
survival |
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| nGLI IS ≤6 | Reference |
|
|
|
| 0.514 |
| nGLI IS >6 | 1.535 | 1.23–17.49 | 0.023 | 1.032 | 0.12–61.92 |
|
| ncGLI1 IS
>6 | Reference |
|
|
|
|
|
| ncGLI1 IS ≤6 | 0.750 | 0.21–20.49 | 0.517 |
|
|
|
| Hormonal
contraception |
|
|
|
|
|
|
| No | Reference |
|
|
|
|
|
|
Yes | 0.017 | 0.34–3.02 | 0.976 |
|
|
|
| BMI,
kg/m2 |
|
|
|
|
|
|
| Normal
(<25.0) | Reference |
|
|
|
|
|
|
Overweight/obese (≥25.0) | −0.061 | 0.44–2.01 | 0.874 |
|
|
|
| Histological
subtype |
|
|
|
|
|
|
|
SCC | Reference |
|
|
|
|
|
|
Non-SCC | 0.496 | 0.62–4.28 | 0.311 |
|
|
|
| FIGO
classification |
|
|
|
|
|
|
| II | Reference |
|
|
|
|
|
|
III | 0.152 | 0.52–2.60 | 0.710 |
|
|
|
| IV | 0.405 | 0.57–3.87 | 0.404 |
|
|
|
| Lymph node
status |
|
|
|
|
|
|
| N0 | Reference |
|
|
|
|
|
| N1 | 0.395 | 0.67–3.28 | 0.330 |
|
|
|
Discussion
The present study demonstrated the utility of IS in
evaluating the prognostic impact of SETD2 and GLI1 expression in
patients with LACC. Evaluation of IS values according to cellular
localization revealed a significant correlation between increased
levels of nSETD2 and nGLI1 proteins. Furthermore, increased nGLI1
levels were associated with oral estrogen consumption. The present
results demonstrated the prognostic value of changes in the
expression of nGLI1 and ncGLI1 on OS and DFS and their clinical
value in predicting a higher likelihood of mortality.
Previous studies have suggested that chemotherapy
resistance associated with upregulation or mutation of SETD2 in
several malignancies, such as prostate, renal cancer, and gastric
cancer, may be associated with alterations in Wnt/β-catenin and Hh
signaling pathways (21–23). Furthermore, abnormal function of
SETD2 and GLI1 is associated with CRT resistance in advanced CC
(10,13,24).
Therefore, this interaction has been proposed as a potential
candidate for synthetic lethality in patients with a high
probability of recurrence following chemoradiation (5). However, the causal relationship
between alterations in SETD2 and GLI1 during cervical
carcinogenesis remains unknown. In the overall study cohort, a
significant association between increased nSETD2 and nGLI1 was
reported, which indicated that their interactions were associated
with nuclear accumulation. Consistent with previous studies
(11,13), the frequency of increased GLI1
expression (65.2%; 45/69) was notably higher compared with SETD2
upregulation (26.3%; 20/76). Although increased GLI1 expression is
associated with epidermoid tumors (25), no significant differences were
detected in GLI1 positivity according to histological
classification in the present study. These results suggested that
abnormal SETD2 expression was associated with increased nuclear
accumulation and transcriptional activity of GLI1. Further studies
are warranted to determine the mechanisms underlying this
interaction.
The abnormal functionality of SETD2 in CC is
associated with the maintenance of the productive replicative cycle
during HPV infection, particularly in high-risk genotypes (26). According to Gautam et al
(26), upregulation of SETD2 in
cervical epithelial cells is associated with the ability of E7
oncoproteins to prolong protein half-life, which is key to sustain
H3K36me3 levels in the early region of the viral genome. The
present results demonstrated that tumors positive for HPV18 and
other HPV genotypes (HPV6, 11, 31, 43, 42, 45 and 58) significantly
increased nSETD2 levels compared with samples positive for the
HPV16 genotype. This is consistent with previous findings that
cells harboring HPV31 maintain higher levels of SETD2 compared with
those with HPV16 infection (26).
Thus, these findings support the hypothesis that HPV oncoproteins
differentially modify the activity of target proteins during
malignant transformation depending on the cellular environment.
Further studies are required to elucidate whether differences in
SETD2 protein levels associated with HPV genotypes affect
therapeutic strategies that target SETD2 in cancer.
The long-term consumption of oral contraceptives
(OCs) is associated with an increased risk of developing CC,
particularly glandular origin and HPV-positive tumors (HR,
1.77–3.3; CI 95%, 1.4–2.24) (27,28).
Although findings regarding the role of steroid hormones in CC are
controversial, the deleterious effects are primarily associated
with enhanced transcriptional activity of HPV oncogenes (28,29).
Similarly, the risk of invasive CC increases according to the
duration of OC consumption (odds ratio, 1.90; 95% CI, 1.69–2.13),
in addition to being associated with a lower OS time (30,31).
The role of OCs in regulating non-HPV proteins implicated in
cervical carcinogenesis and treatment responses remains poorly
understood. In the present study, significantly higher nGLI1 levels
were observed in patients who used OCs. This finding is consistent
with those of previous studies reporting crosstalk between androgen
stimulation and Hh signaling in prostate cancer, particularly the
association between epithelial-mesenchymal transition and GLI1
upregulation (32,33). Furthermore, estrogenic stimulation
in gynecological cancer is associated with the upregulation and
nuclear translocation of GLI1 via the inhibition of glycogen
synthase kinase-3β (34). Thus, the
present results supported the hypothesis that long-term OC use
contributes to CC progression by stimulating upregulation and
nuclear translocation of GLI1.
Although 90.5% (76/84) of patients with LACC were
SETD2-positive, exhibiting increased expression in 26.3% of cases,
no notable associations were found between SETD2 and OS and DFS. In
malignant neoplasia such as lung and endometrial cancer and CC, the
abnormal function of SETD2 is associated with mutations that modify
gene expression patterns, increasing the risk of CRT resistance and
decreasing survival rates (4,5,19).
According to The Cancer Genome Atlas Program, endocervical ADC
presents the highest rate of SETD2 mutations (7%) (6). In the present study, SETD2 expression
exhibited no difference between histological groups, which may be
associated with the lower proportion of tumors of glandular origin.
Further studies are warranted to clarify the association between
the expression and the mutation rate of SETD2. Rate of SETD2
mutations may increase following treatment with neoadjuvant
chemotherapy (platinum + paclitaxel) in patients with LACC and is
associated with a lack of response (19). This suggests it is necessary to
integrate both expression and mutational analyses when determining
the prognostic and predictive value of SETD2 in LACC.
The Hh signaling cascade serves a key role in the
proliferation, metastasis, recurrence, invasion and CRT resistance
of CC (10). Nevertheless, the
prognostic and predictive value depend on the target protein
analyzed and the criteria employed for evaluation (13,18,35).
Consistent with previous studies, the present study analyzed the
prognostic utility of GLI1 by categorizing the IS; by contrast with
other studies, the present study evaluated the differences
according to cellular localization (18,36).
This revealed that increased nGLI levels may predict a higher
probability of mortality, associated with lower OS and DFS. By
contrast, an increased probability of OS was associated with
increased ncGLI1 levels. These results suggested that the
prognostic and predictive utility of GLI1 depends on the analysis
of proteins according to their cellular localization. Furthermore,
this supports the hypothesis that the role of GLI1 in CRT
resistance is determined by its nuclear translocation, which may
markedly affect the probability of survival (11).
To the best of our knowledge, the present study is
the first to investigate the association between the expression and
localization of SETD2 and GLI1 proteins and their role in the
outcome of patients with LACC. A prior study on members of the Hh
pathway in CC established an increase in their expression using
IHC. However, the aforementioned analyses did not include
associations with pathological characteristics or clinical outcomes
(37). Furthermore, epigenetic
dysregulation is associated with several types of cancer including
breast, liver cancer, prostate cancer, and small-cell bladder
cancer (38). Histone
methyltransferases are frequently mutated or deleted in human
tumors (39,40); however, whether alterations in
histone methyltransferases are associated with disease progression
and response to treatment remains unknown. Despite the positive
outcomes, the present study had limitations. The retrospective
study design, with a limited sample size, based on FFPE samples,
limited access to fresh biological material for functional assays
of GLI1 and SETD2 proteins. However, the consistency of sensitivity
analysis results suggested that sample size was not a notable
limitation.
The outcomes in patients with LACC are poor because
they present high rates of resistance to treatment (41). Therefore, the identification of
patients at increased risk of recurrence who may benefit from more
aggressive treatment strategies and the identification of novel
therapeutic targets are key. The interactions of the Hh pathway in
tumor cells are complex and the present study provided insight into
the positive correlation between the expression of SETD2 and the
transcription factor GLI1, which may represent a novel mechanism of
epigenetic regulation involved in the clinical outcome of patients
with LACC. The present data suggested that GLI1 was a valid and
effective therapeutic target in patients with LACC who undergo
chemoradiation.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Alejandro
Lopez-Saavedra (Advanced Microscopy Applications Unit, Mexico City,
Mexico) for technical assistance with capturing the images.
Funding
The present study was supported by Consejo Nacional de Ciencia y
Tecnología (grant CF-2019-263979), Proyecto Nacional de
Investigación e Incidencia-7 Virus y Cancer (grant no. 303044),
Instituto Nacional de Cancerología (grant nos. 015/039/IBI,
CEI/998/15, 018/051/IBI and CEI/1294/18) and Consejo de Ciencia y
Tecnología del estado de Tabasco (grant nos. PRODECTI-2023-01/090
and PRODECTI-REICTI-012).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ML conceived the study. ACo and EC confirm the
authenticity of all the raw data. ACo, EC and ML wrote the
manuscript. ACo and ML acquired funding. ACo conceptualized the
study. GM, LC and ACa designed the methodology. AA and MR
constructed figures. JC and LC supervised the study. JC, AA, EC and
MR analyzed data. ACo, EC and ML edited the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics and
Scientific Institutional Review Board (approval no.
INCAN/CEI577/15) of the National Cancer Institute (Mexico City,
México), and written informed consent was obtained from all
participating patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Ferlay J, Ervik M, Lam F, Laversanne M,
Colombet M, Mery L, et al: Global Cancer Observatory: Cancer Today.
Lyon, France, International Agency for Research on Cancer.
2024.https://gco.iarc.who.int/today
|
|
3
|
Hull R, Mbele M, Makhafola T, Hicks C,
Wang S, Reis R, Mehrotra R, Mkhize-Kwitshana Z, Kibiki G, Bates DO
and Dlamini Z: Cervical cancer in low and middle-income countries
(Review). Oncol Lett. 20:2058–2074. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sehnal B, Kmoníčková E, Sláma J, Tomancová
V and Zikán M: Current FIGO staging for carcinoma of the cervix
uteri and treatment of particular stages. Klin Onkol. 32:224–231.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lizano M, Carrillo-García A, De La
Cruz-Hernández E, Castro-Muñoz LJ and Contreras-Paredes A:
Promising predictive molecular biomarkers for cervical cancer
(Review). Int J Mol Med. 53:502024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu M, Zhao B, Liu M, Wu L, Li Y, Zhai Y
and Shen X: Pan-cancer analysis of SETD2 mutation and its
association with the efficacy of immunotherapy. NPJ Precis Oncol.
5:512021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng Z, Zhang J, Li J, Li Y, Huang Z, Han
L, Xie C and Gong Y: SETD2 regulates gene transcription patterns
and is associated with radiosensitivity in lung adenocarcinoma.
Front Genet. 13:9356012022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Viaene AN, Santi M, Rosenbaum J, Li MM,
Surrey LF and Nasrallah MP: SETD2 mutations in primary central
nervous system tumors. Acta Neuropathol Commun. 6:1232018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing J, Wu Z, Wang J, Luo G, Lin H, Fan Y
and Zhou C: Hedgehog signaling in tissue homeostasis, cancers and
targeted therapies. Signal Transduct Target Ther. 8:3152023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu C and Wang R: The roles of hedgehog
signaling pathway in radioresistance of cervical cancer. Dose
Response. 17:15593258198852932019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Z, Huang C, Li R, Li H, Lu H and Lin Z:
PRKCI mediates radiosensitivity via the Hedgehog/GLI1 pathway in
cervical cancer. Front Oncol. 12:8871392022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang C, Lu H, Li J, Xie X, Fan L, Wang D,
Tan W, Wang Y, Lin Z and Yao T: SOX2 regulates radioresistance in
cervical cancer via the hedgehog signaling pathway. Gynecol Oncol.
151:533–541. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chaudary N, Pintilie M, Hedley D, Fyles
AW, Milosevic M, Clarke B, Hill RP and Mackay H: Hedgehog pathway
signaling in cervical carcinoma and outcome after chemoradiation.
Cancer. 118:3105–3115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chai JY, Sugumar V, Alshanon AF, Wong WF,
Fung SY and Looi CY: Defining the Role of GLI/Hedgehog signaling in
Chemoresistance: Implications in therapeutic approaches. Cancers
(Basel). 13:47462021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gennigens C, De Cuypere M, Hermesse J,
Kridelka F and Jerusalem G: Optimal treatment in locally advanced
cervical cancer. Expert Rev Anticancer Ther. 21:657–671. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vázquez-Ulloa E, Ramos-Cruz AC, Prada D,
Avilés-Salas A, Chávez-Blanco AD, Herrera LA, Lizano M and
Contreras-Paredes A: Loss of nuclear NOTCH1, but not its negative
regulator NUMB, is an independent predictor of cervical malignancy.
Oncotarget. 9:18916–18928. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bohr Mordhorst L, Ahlin C and Sorbe B:
Prognostic impact of the expression of Hedgehog proteins in
cervical carcinoma FIGO stages I–IV treated with radiotherapy or
chemoradiotherapy. Gynecol Oncol. 135:305–311. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lizano M, De la Cruz-Hernández E,
Carrillo-García A, García-Carrancá A, Ponce de Leon-Rosales S,
Dueñas-González A, Hernández-Hernández DM and Mohar A: Distribution
of HPV16 and 18 intratypic variants in normal cytology,
intraepithelial lesions, and cervical cancer in a Mexican
population. Gynecol Oncol. 102:230–235. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sotlar K, Diemer D, Dethleffs A, Hack Y,
Stubner A, Vollmer N, Menton S, Menton M, Dietz K, Wallwiener D, et
al: Detection and typing of human papillomavirus by e6 nested
multiplex PCR. J Clin Microbiol. 42:3176–3184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar A, Kumari N, Gupta V and Prasad R:
Renal cell carcinoma: Molecular aspects. Indian J Clin Biochem.
33:246–254. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang C, He C, Wu Z, Li F and Xiao J:
Histone methyltransferase SETD2 regulates osteosarcoma cell growth
and chemosensitivity by suppressing Wnt/β-catenin signaling.
Biochem Biophys Res Commun. 502:382–388. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen R, Zhao WQ, Fang C, Yang X and Ji M:
Histone methyltransferase SETD2: A potential tumor suppressor in
solid cancers. J Cancer. 11:3349–3356. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li W, Huang Y, Xiao M, Zhao J, Du S, Wang
Z, Hu S, Yang L and Cai J: PBRM1 presents a potential ctDNA marker
to monitor response to neoadjuvant chemotherapy in cervical cancer.
iScience. 27:1091602024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vishnoi K, Mahata S, Tyagi A, Pandey A,
Verma G, Jadli M, Singh T, Singh SM and Bharti AC: Cross-talk
between Human Papillomavirus Oncoproteins and hedgehog signaling
synergistically promotes stemness in cervical cancer cells. Sci
Rep. 6:343772016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gautam D, Johnson BA, Mac M and Moody CA:
SETD2-dependent H3K36me3 plays a critical role in epigenetic
regulation of the HPV31 life cycle. PLoS Pathog. 14:e10073672018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Asthana S, Busa V and Labani S: Oral
contraceptives use and risk of cervical cancer-A systematic review
& meta-analysis. Eur J Obstet Gynecol Reprod Biol. 247:163–175.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gadducci A, Cosio S and Fruzzetti F:
Estro-progestin contraceptives and risk of cervical cancer: A
debated issue. Anticancer Res. 40:5995–6002. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wei X, Hong L, Liang H, Ren K, Man W, Zhao
Y and Guo P: Estrogen and viral infection. Front Immunol.
16:15567282025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gashu C, Tasfa B, Alemu C and Kassa Y:
Assessing survival time of outpatients with cervical cancer: At the
university of Gondar referral hospital using the Bayesian approach.
BMC Womens Health. 23:592023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
International Collaboration of
Epidemiological Studies of Cervical Cancer, . Appleby P, Beral V,
Berrington de González A, Colin D, Franceschi S, Goodhill A, Green
J, Peto J, Plummer M and Sweetland S: Cervical cancer and hormonal
contraceptives: Collaborative reanalysis of individual data for
16,573 women with cervical cancer and 35,509 women without cervical
cancer from 24 epidemiological studies. Lancet. 370:1609–1621.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamamichi F, Shigemura K, Behnsawy HM,
Meligy FY, Huang WC, Li X, Yamanaka K, Hanioka K, Miyake H, Tanaka
K, et al: Sonic hedgehog and androgen signaling in tumor and
stromal compartments drives epithelial-mesenchymal transition in
prostate cancer. Scand J Urol. 48:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lubik AA, Nouri M, Truong S, Ghaffari M,
Adomat HH, Corey E, Cox ME, Li N, Guns ES, Yenki P, et al:
Paracrine sonic hedgehog signaling contributes significantly to
acquired steroidogenesis in the prostate tumor microenvironment.
Int J Cancer. 140:358–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kaushal JB, Sankhwar P, Kumari S, Popli P,
Shukla V, Hussain MK, Hajela K and Dwivedi A: The regulation of
Hh/Gli1 signaling cascade involves Gsk3β-mediated mechanism in
estrogen-derived endometrial hyperplasia. Sci Rep. 7:65572017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cui Y, Cui CA, Yang ZT, Ni WD, Jin Y and
Xuan YH: Gli1 expression in cancer stem-like cells predicts poor
prognosis in patients with lung squamous cell carcinoma. Exp Mol
Pathol. 102:347–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sinicrope FA, Ruan SB, Cleary KR, Stephens
LC, Lee JJ and Levin B: Bcl-2 and p53 oncoprotein expression during
colorectal tumorigenesis. Cancer Res. 55:237–241. 1995.PubMed/NCBI
|
|
37
|
Xuan YH, Jung HS, Choi YL, Shin YK, Kim
HJ, Kim KH, Kim WJ, Lee YJ and Kim SH: Enhanced expression of
hedgehog signaling molecules in squamous cell carcinoma of uterine
cervix and its precursor lesions. Mod Pathol. 19:1139–1147. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu X, Zhao H, Wang R, Chen Y, Ouyang X, Li
W, Sun Y and Peng A: Cancer epigenetics: From laboratory studies
and clinical trials to precision medicine. Cell Death Discov.
10:282024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tinsley E, Bredin P, Toomey S, Hennessy BT
and Furney SJ: KMT2C and KMT2D aberrations in breast cancer. Trends
Cancer. 10:519–530. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Na F, Pan X, Chen J, Chen X, Wang M, Chi
P, You L, Zhang L, Zhong A, Zhao L, et al: KMT2C deficiency
promotes small cell lung cancer metastasis through DNMT3A-mediated
epigenetic reprogramming. Nat Cancer. 3:753–767. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Niu Y, Du C, Zhou Y, Zhang M, Guo Q and
Zhou H: A comparative analysis of survival outcomes and adverse
effects between preoperative brachytherapy with radical surgery and
concurrent chemoradiotherapy in patients with locally advanced
cervical cancer. Front Oncol. 15:15117482025. View Article : Google Scholar : PubMed/NCBI
|