Introduction
Thyroid cancer ranks among the malignancies with the
most rapidly increasing incidence. In the United States, its
age-standardized incidence rate nearly tripled from 1975–1979 to
2015–2019. This trend has been particularly prominent among female
patients in large urban areas, with the incidence among female
patients rising from 7.8 to 29.5/100,000 during the same period,
making it one of the most common cancer types in female patients
(1,2). Although early-stage thyroid cancer is
generally associated with favorable outcomes following timely
diagnosis and treatment, the 5-year relative survival rate is
~98.5%, advanced disease accompanied by local invasion and distant
metastasis remains a key challenge, often leading to poor prognosis
(3). Thus, elucidating the
molecular mechanisms underlying thyroid cancer progression and
identifying relevant biomarkers are key for the enhancement of
diagnostic accuracy and therapeutic strategies.
Folate receptor γ (FOLR3) is a key member of the
folate receptor family. FOLR3 belongs to a soluble receptor subtype
that serves a key role in the uptake and utilization of
extracellular folic acid (4).
Folate serves a dual role in cancer. On the one hand, folate may
help prevent the development of certain cancer types. Previous
studies have suggested that high folate intake is associated with a
reduced risk of breast, colorectal and ovarian cancer types, making
it a candidate for targeted cancer therapy (5–8). By
contrast, folate may also promote cancer progression, for example,
certain studies indicate that it might accelerate the development
of prostate cancer (9,10). Dietary folate promotes
hepatocellular carcinoma progression by hijacking methionine
metabolism through VCIP135-mediated stabilization of methionine
adenosyltransferase IIα, thereby coupling one-carbon and methionine
metabolic flux to fuel tumor growth (11). The abnormal expression of FOLR3 may
contribute to tumorigenesis and progression by interfering with
folate metabolism, affecting DNA synthesis, methylation and cell
proliferation. Furthermore, FOLR3 can exert antimicrobial and
antitumor effects by sequestering natural folate, thereby depriving
bacteria and tumor cells of this vital nutrient (12,13).
Although evidence suggests that FOLR3 is involved in cancer
pathogenesis, its specific function in thyroid cancer remains
poorly understood (14–17). Therefore, investigating the
expression level of FOLR3 in thyroid cancer and its association
with clinicopathological features may offer novel biomarkers for
early diagnosis and prognosis evaluation of thyroid cancer.
Materials and methods
Bioinformatic analysis
Gene expression data for thyroid cancer were
obtained from The Cancer Genome Atlas (TCGA;
portal.gdc.cancer.gov/) database and the Gene Expression Omnibus
(GEO; ncbi.nlm.nih.gov/geo/) dataset GSE250521 (18). From TCGA, the Thyroid Carcinoma
project dataset was downloaded via the Genomic Data Commons Data
Portal, which includes RNA-Seq data from tumor and adjacent normal
tissue. The combined dataset included clinical information for 572
individuals, comprising of 513 thyroid cancer cases and 59 normal
controls. Differential gene expression analysis was performed using
the ‘limma’ package in the R software (version 3.64.1; R
Development Core Team) (19), with
a significance threshold set at adjusted P<0.05 and
|log2FC|>2.5. Genes with log2FC >2.5
were considered upregulated, those with log2FC <-2.5
were classified as downregulated and genes with values between −2.5
and 2.5 were regarded as unchanged. Functional enrichment analyses,
including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analyses were performed using the
‘clusterProfiler’ package (version 4.16.0) in R (20). Univariate Cox regression analysis
and multivariate Cox regression analysis were performed using the
‘survival’ (version 3.8–3) and ‘survminer’ packages (version 0.4.9)
in R (21). Furthermore, FOLR3
expression in thyroid cancer was evaluated using the GEPIA2.0
(gepia.cancer-pku.cn) and University of Alabama at Birmingham
Cancer data analysis Portal (ualcan.path.uab.edu/index.html)
databases. For immune/stromal infiltration analysis, Cell-type
Identification by Estimating Relative Subsets of RNA Transcripts
(CIBERSORT) estimated immune cell proportions via deconvolution,
while ESTIMATE calculated immune and stromal scores from signature
gene sets (22,23). Together, they provide a
comprehensive tumor microenvironment (TME) composition profile.
The single-cell gene expression matrix was
preprocessed using the ‘Seurat’ R package (version 4.4.0) to ensure
data quality and prepare for downstream analyses (24). Cell filtering was performed by
retaining cells that met the following criteria: Detection of ≥500
genes, presence in ≥3 cells with non-zero unique molecular
identifier (UMI) counts and a mitochondrial UMI ratio <25%.
Genes detected in <3 cells were excluded. Highly variable genes
were selected for dimensionality reduction via principal component
analysis and the top 30 principal components were used for Uniform
Manifold Approximation and Projection to visualize high-dimensional
gene expression patterns in two dimensions. The Harmony algorithm
was applied to correct for batch effects and minimize technical
variability (25). Cell clustering
was performed using the ‘FindClusters’ function with a resolution
parameter of 0.7 to identify distinct cell populations.
Differential expression analysis across cell subpopulations was
conducted with the ‘FindAllMarkers’ function using the Wilcoxon
rank-sum test and P-values were adjusted for multiple analysis with
the false discovery rate (FDR) correction. Cell-cell communication
analysis was carried out with the ‘CellChat’ package (version
1.6.1) to predict ligand-receptor interactions (26). Folate metabolism-related gene sets
were obtained from the Molecular Signatures database and pathway
activity scores were calculated using the ‘AddModuleScore’ function
(27). Cells were stratified into
high- and low-score groups based on the median pathway score, with
a cutoff of 0.108. Myeloid cell subpopulations were annotated using
the same workflow and analytical methods.
Clinical data
The present study included a total of 293 patients
with thyroid cancer who were treated at Chifeng Hospital of Inner
Mongolia (Chifeng, China) between January 2021 and June 2024. The
cohort comprised 51 male and 242 women, with ages ranging from 19
to 78 years. According to the histopathological diagnosis, there
were 287 cases of papillary thyroid carcinoma (PTC), 3 cases of
follicular thyroid carcinoma and 3 cases of anaplastic thyroid
carcinoma. The collected clinical and pathological parameters
included sex, age, capsule invasion, perineural invasion, striated
muscle cell infiltration, pathological tumor (pT) stage, tumor
location, tumor focality, histological type, vascular infiltration
and pathological lymph node (pN) stage, assessed according to the
American Joint Committee on Cancer TNM staging system (8th edition)
(28).
Construction of high-throughput
thyroid tumor tissue microarray
Tissue samples were sourced from the surgical
resection specimens of the 293 thyroid cancer patients. A manual
tissue microarrayer was used to mark the tumor regions on large
tissue sections. A 1 mm diameter core was extracted from each donor
block and transferred into a recipient paraffin block to construct
a high-throughput thyroid tumor tissue microarray. For each case,
one core was taken from the tumor invasion front and one from
adjacent normal thyroid tissue.
Immunohistochemical staining
(IHC)
Formalin-fixed, paraffin-embedded tissue sections
(thickness, 4 µm) were used. Tissue samples were fixed in 10%
neutral-buffered formalin at room temperature for 24–48 h. Antigen
retrieval was performed using EnVision two-step method (95–100°C,
15–20 min). Sections were dewaxed, rehydrated through a descending
alcohol series (100, 95, 70% ethanol) to distilled water, and
endogenous peroxidase activity was blocked with 3% hydrogen
peroxide for 10 min at room temperature. Non-specific binding was
blocked with 5% normal goat serum for 1 h at room temperature.
Normal serum was incubated with rabbit anti-human FOLR3 antibody
(1:100; cat. no. DF9519; Affinity Biosciences) overnight at 4°C.
After washing, EnVision HRP-conjugated goat anti-rabbit secondary
antibody (1:100; Wuhan Proteintech, SA00001-2) was added for 30 min
at room temperature, and DAB staining was used for color
development. Hematoxylin re-dyeing for 2 min at room temperature.
Phosphate buffer, isotype rabbit IgG (1:100, Wuhan Boster
Biological Technology, Ltd.) instead of primary antibody were used
as a negative control. Images were acquired using a light
microscope (Olympus Corporation) at ×200 magnification. The
percentage of cells positive for FOLR3 in thyroid cancer tissue vs.
paired normal tissue was identified by QuPath software (version
0.4.4, QuPath) (29). The criteria
for cytosol staining intensity was as follows: Negative, <0.20;
1, 0.2–0.39; 2, 0.40–0.6 and 3+, >0.6 (30). IHC results were evaluated using
H-score method (31).
Cell culture
Human thyroid carcinoma TPC-1 cells (Abcells; cat.
no. AC153) were cultured in complete medium (Seven Biotech) at 37°C
in a 5% CO2 incubator (Thermo Fisher Scientific, Inc.).
Cells were passaged every 2–3 days to maintain exponential
growth.
Transfection
TPC-1 cells were seeded in 6-well plates and grown
to ~70% confluence. The human FOLR3 overexpression plasmid
(pcDNA3.1-hFOLR3-3XFLAG) and corresponding empty vector control
(pcDNA3.1) were synthesized by Beijing Likeli Biotechnology Co.,
Ltd.). Cells were transfected with 2.5 µg FOLR3 overexpression or
the empty control plasmid using 4 µl Lipo8000™ transfection reagent
(Beyotime Biotechnology, C0533), according to the manufacturer's
instructions. The transfection complexes were added at 37°C. The
culture medium was not replaced post-transfection. Transfected
cells were harvested for subsequent experimentation 48 h
post-transfection. Transfected cells were divided into two groups:
Flag-FOLR3 group (FOLR3 overexpression) and the Flag-negative
control (NC) group.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from Flag-FOLR3 and NC
groups using the RNAprep pure cell kit (Tiangen Biotech Co., Ltd.)
and reverse transcribed into cDNA with the PrimeScript™ RT reagent
kit (Takara Bio, Inc.). RT was performed at 37°C for 15 min,
followed by enzyme inactivation at 85°C for 5 sec. RT-qPCR was
using the Beyofast SYBR™ Green qPCR mix (Beyotime Biotechnology) on
a Fluorescent Quantitative PCR Instrument (Bio-Rad). Thermocycling
conditions were as follows: initial denaturation at 95°C for 2 min,
followed by 40 cycles of denaturation at 95°C for 15 s and
annealing/extension at 60°C for 30 sec. The relative expression
level of FOLR3 and folate metabolism-related genes was normalized
to that of GAPDH, which served as the reference gene. All primers
were designed and synthesized by Beyotime Biotechnology and
quantification was performed using the 2−ΔΔCq method
(data not shown; Table I) (32). Each experiment was performed three
times in duplicate.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Primer sequences
(5′-3′ |
|---|
| FOLR3 | F:
CGCAAAGAGCGCATTCTGAAC |
|
| R:
CTGGGCTGAGTCAAACCACA |
| GAPDH | F:
ACGGATTTGGTCGTATGGG |
|
| R:
CGCTCCTGGAAGATGGTGAT |
| TYMS | F:
TGATGGTGTCAATCACTCTTTGC |
|
| R:
TGGGGCAGAATACAGAGATATGG |
| MTHFR | F:
GAGCGGCATGAGAGACTCC |
|
| R:
CCGGTCAAACCTTGAGATGAG |
| SLC19A1 | F:
CTCAGCTTCGTGTCGGTGT |
|
| R:
AGCGAGATGTAGTTGAGCGTG |
| PCFT | F:
GGCCCAGGGTTATGCCAAC |
|
| R:
GGACAATGGATCGGTGGTGAC |
| Cyclin D1 | F:
CAATGACCCCGCACGATTTC |
|
| R:
CATGGAGGGCGGATTGGAA |
| Bcl-2 | F:
GGTGGGGTCATGTGTGTGG |
|
| R:
CGGTTCAGGTACTCAGTCATCC |
| c-FOS | F:
GGGGCAAGGTGGAACAGTTAT |
|
| R:
CCGCTTGGAGTGTATCAGTCA |
| c-Jun | F:
TCCAAGTGCCGAAAAAGGAAG |
|
| R:
CGAGTTCTGAGCTTTCAAGGT |
| VEGF | F:
AGGGCAGAATCATCACGAAGT |
|
| R:
AGGGTCTCGATTGGATGGCA |
| MCL1 | F:
GTGCCTTTGTGGCTAAACACT |
|
| R:
AGTCCCGTTTTGTCCTTACGA |
Protein isolation and western
blotting
TPC-1 cells were lysed using RIPA buffer (Beyotime
Biotechnology) supplemented with PMSF (Beyotime Biotechnology) and
homogenized mechanically until complete lysis was achieved. The
lysate was centrifuged at 12,000 × g for 5 min at 4°C to collect
the supernatant. Protein concentration was determined using the
Enhanced BCA Protein Assay Kit. Proteins (10 µg per lane) were
separated by 10% SDS-PAGE and transferred to PVDF membrane. The
membrane was blocked with 5% non-fat milk in TBST at room
temperature for 1 h, the corresponding FOLR3 (1:2,000; Affinity
Biosciences; cat. no. DF9519) and GAPDH antibody (1:2,000; Wuhan
Boster Biological Technology, Ltd, BA1050) was added overnight at
4°C. The membrane was washed and then the HRP-conjugated goat
anti-rabbit secondary antibody (1:5,000; Wuhan Boster Biological
Technology, Ltd, BM3874) was added for 1 h at room temperature. The
indicator proteins were assessed using a hypersensitive ECL
chemiluminescence kit (Seven Biotech) and the relative expression
level of FOLR3 was normalized to GAPDH. Each experiment was
performed three times in duplicate.
Scratch assay
After transfection, cells from the Flag-FOLR3 and NC
groups were seeded at 1×106 cells per well into 6-well
plate until they reached 100% confluence. A scratch was made in
each well using a sterile 200 µl pipette tip. Cells were washed and
cultured in RPMI-1640 (Beijing Solarbio Science & Technology
Co., Ltd.). Wound healing was observed and captured at 0, 24 and 48
h using an inverted microscope (Olympus Corporation). The migration
distance was quantified using ImageJ software (version 1.53f;
National Institutes of Health). Each experiment was performed three
times in duplicate.
Cell Counting Kit-8 (CCK-8) assay
Transfected cells were seeded into 96-well plates.
After culturing for 0, 24, 48, 72 and 96 h, enhanced CCK-8
(Beyotime Biotechnology) was added to each well. Following
incubation for 1.5 h, the optical density value at 450 nm was
measured using a microplate reader (Thermo Fisher Scientific,
Inc.), Each experiment was performed three times in duplicate.
Clone formation assay
Transfected cells were seeded at 1,000 cells/well
into 6-well plates and cultured for 14 days in a CO2
incubator at 37°C. Cells were fixed at room temperature with 4%
paraformaldehyde (Beyotime Biotechnology) for 20 min and stained
with 0.1% crystal violet solution (Beyotime Biotechnology) at room
temperature for another 20 min. Each experiment was performed three
times in duplicate.
Transwell invasion assay
The upper chamber of Transwell plates were
pre-coated with Matrigel™ Basement Membrane Matrix (Beyotime
Biotechnology) and incubated for 2 h at 37°C. A cell suspension
containing 5×104 cells in 200 µl serum-free RPMI-1640
medium (Beijing Solarbio Science & Technology Co., Ltd.). was
added to the upper chamber and 600 µl TPC-1 complete medium
(Abcells) containing 10% FBS was placed in the lower chamber.
Following incubation for 24 h at 37°C in a 5% CO2
incubator, cells that had invaded through the membrane were fixed
with 4% paraformaldehyde at room temperature for 20 min, stained
with 0.1% crystal violet at room temperature for 20 min and counted
under a microscope. Each experiment was performed three times in
duplicate.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism (version 8.0.2; Dotmatics), ImageJ (version 1.53f; National
Institutes of Health) and SPSS software (version 23.0; IBM Corp.),
and R software (version 4.2.3). Data from the IHC analysis are
presented as mean ± SD. P<0.05 was considered statistically
significant. In bioinformatic analyses, significance thresholds
were set at P<0.05 and a false discovery rate (FDR) <0.05.
For functional experiments, group comparisons were conducted as
follows: the CCK-8 cell proliferation, scratch wound healing,
colony formation and Transwell invasion assays were analyzed using
unpaired Student's t-tests. Comparisons of FOLR3 expression between
paired tumor and adjacent normal tissue in IHC were analyzed using
paired Student's t-test. The associations between FOLR3 expression
and clinicopathological features (including age, sex, tumor
location, pT and pN stage, perineural and vascular invasion) were
evaluated using the χ2 or Fisher's exact test, as
appropriate. Independent prognostic factors were identified using
multivariate Cox regression analysis. All bioinformatic statistical
computations, data processing and visualizations were performed
using R software.
Results
Bioinformatics analysis of FOLR3
expression in thyroid cancer and normal adjacent tissue
Expression profiling analysis of thyroid cancer data
from TCGA identified 520 differentially expressed genes (DEGs),
including 235 up- and 285 downregulated genes. The distribution of
these DEGs is visually represented in the volcano plot (Fig. 1A). GO enrichment analysis identified
24 enriched terms in the three major categories: Biological
process, cellular component and molecular function, while KEGG
pathway analysis revealed 10 significantly enriched pathways
(P<0.01; Fig. 1B and C). GO
enrichment analysis was primarily associated with ‘neural
synapse-related functions’, ‘extracellular matrix remodeling’ and
‘ion channel activity’. KEGG pathway analysis highlighted processes
such as ‘cell adhesion molecules’, ‘tyrosine metabolism’, ‘protein
digestion and absorption’ and ‘complement and coagulation
cascades’.
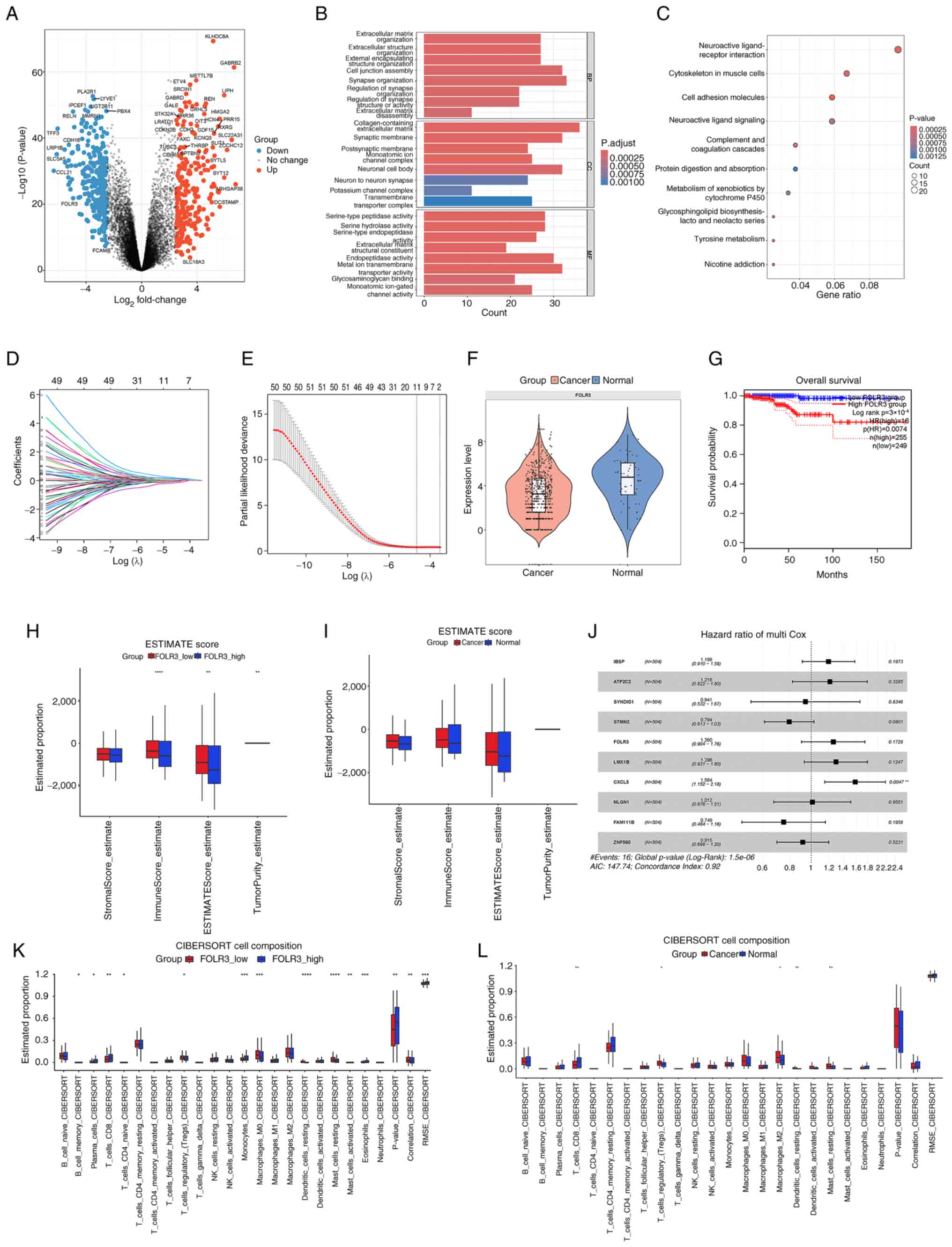 | Figure 1.Analysis of FOLR3 expression and its
impact in thyroid cancer. (A) Volcano plot of DEGs (red,
upregulated; blue, downregulated). (B) GO enrichment analysis
demonstrating three major functional categories. (C) KEGG pathway
enrichment of DEGs. (D) LASSO regression model for feature
selection. (E) A 10-fold cross-validation curve for the LASSO
model. (F) FOLR3 expression levels in thyroid cancer vs. normal
tissue. (G) Survival analysis based on FOLR3 expression. (H)
ESTIMATE scores comparing high vs. low FOLR3 expression groups. (I)
ESTIMATE scores in thyroid cancer vs. normal tissues. (J)
Univariate Cox regression analysis of prognostic factors. (K)
CIBERSORT analysis of immune infiltration in high vs. low FOLR3
expression groups. (L) CIBERSORT analysis of immune infiltration in
thyroid cancer vs. normal tissues. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. FOLR3, folate receptor γ; GO,
Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes;
LASSO, least absolute shrinkage and selection operator; CIBERSORT,
Cell-type Identification by Estimating Relative Subsets of RNA
Transcripts; DEGs, differentially expressed genes. |
Furthermore, a least absolute shrinkage and
selection operator (LASSO) regression model, developed using
machine learning techniques was employed to identify genes
associated with the progression and prognosis of thyroid cancer
(Fig. 1D). The resulting LASSO
regression model and the results of cross-validation results
selected 10 relevant genes (integrin-binding sialoprotein, ATPase
secretory pathway Ca2+ transporting 2, synapse
differentiation induced gene 1, stathmin-2, FOLR3, LIM homeobox
transcription factor 1-β, C-X-C motif chemokine ligand 5,
neuroligin-1, family with sequence similarity 111, member B and
zinc finger protein 560; Fig. 1E).
FOLR3 was selected as a research focus as it is one of the
significantly downregulated genes in thyroid cancer, and the LASSO
regression model identified it as one of the ten key genes
associated with disease progression and prognosis.
The present study results demonstrated that FOLR3
expression was significantly downregulated in thyroid cancer
tissues compared with normal tissues (P<0.001; Fig. 1F). Survival analysis indicated that
high expression level of FOLR3 was associated with significantly
worse overall survival in patients with thyroid cancer (P=0.0074;
Fig. 1G). Multivariate Cox
regression analysis identified CXCL5 as a significant independent
prognostic biomarker for thyroid cancer (P=0.0047; Fig. 1J). Comparative assessment of TME
profiles revealed distinct immune and stromal features associated
with FOLR3 expression levels and tissue status in thyroid cancer.
ESTIMATE algorithm revealed low FOLR3 expression was associated
with lower tumor purity (P<0.001) and higher immune scores (both
P<0.001) and ESTIMATE scores (P=0.0011; Fig. 1H). Similarly, thyroid cancer tissues
exhibited markedly lower immune and stromal infiltration overall
compared with normal tissues (Fig.
1I). CIBERSORT-based immune deconvolution further illustrated
divergent microenvironmental landscapes. High FOLR3 expression was
associated with increased infiltration of antitumor immune subsets,
including plasma cells (P=0.0415), CD8+ T cells
(P=0.0027). By contrast, low FOLR3 expression favored an
immunosuppressive milieu, characterized by elevated naïve
CD4+ memory T cells (P=0.043), M0 macrophages (P=0.0005)
and activated mast cells (P=0.0078) (Fig. 1K). Thyroid cancer tissues displayed
enriched infiltration of immunosuppressive cells, such as resting
Tregs (P=0.023), M2 macrophages (P=0.431), resting dendritic cells
(P=0.0042) and resting mast cells (P=0.0011), compared
with normal tissues, indicating an overall immunosuppressive TME
(P<0.001) (Fig. 1L). These
results collectively indicate that FOLR3 expression is closely
associated with immune-active microenvironment phenotypes, while
its downregulation may contribute to an immunosuppressive TME in
thyroid cancer.
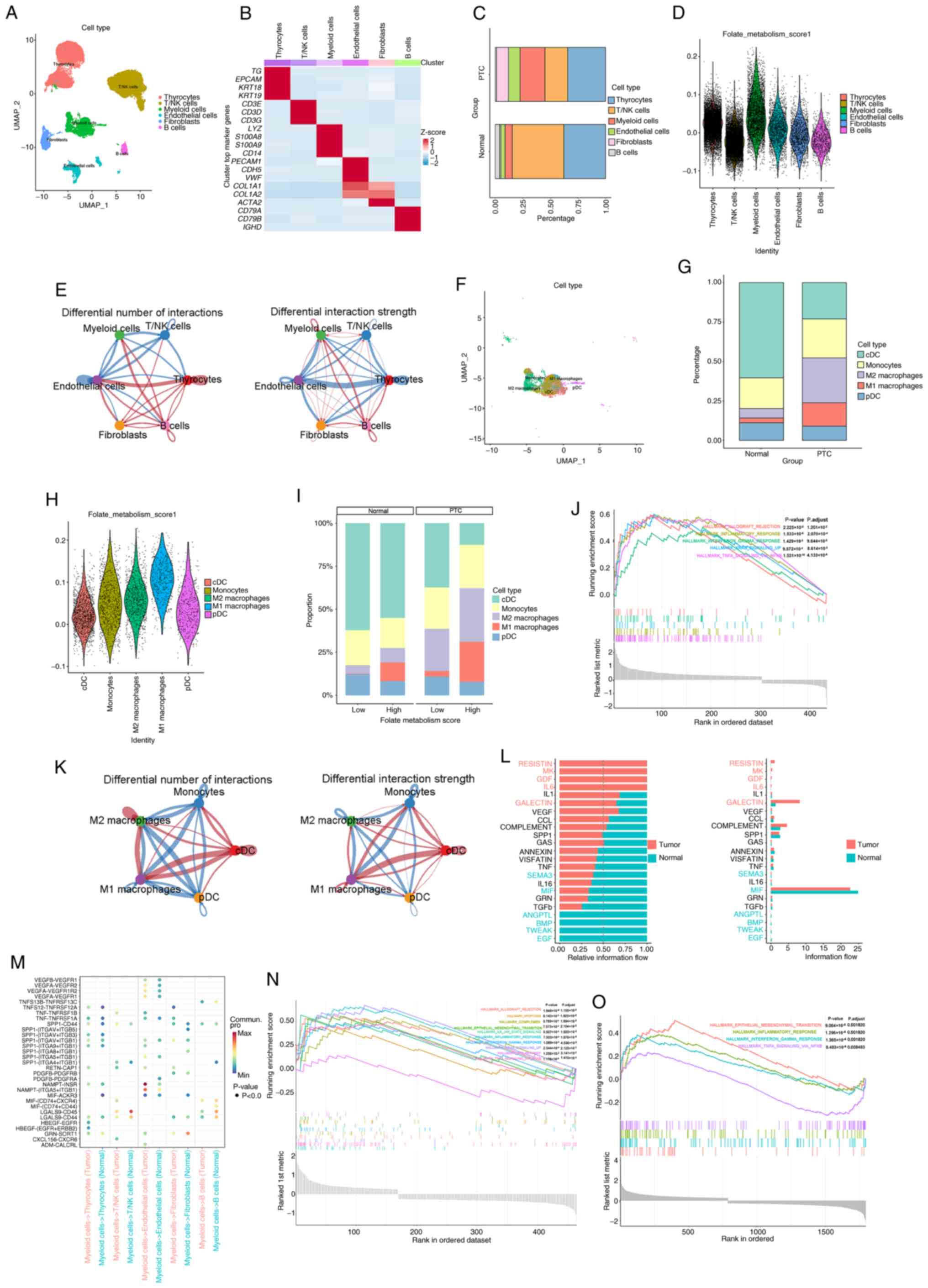
Remodeling of the TME in PTC:
Metabolic reprogramming, immune activation and altered cellular
communication driven by folate-high M1 macrophages
The present study comprehensively characterized the
TME of PTC through integrated analysis of single-cell
transcriptomic data and functional enrichment. The PTC dataset
GSE250521 was downloaded from the GEO database. After quality
control and normalization, the dataset contained 33,505 cells and
31,454 genes. Manual annotation of the main cell subsets revealed
six distinct populations: Thyrocytes [thyroglobulin (TG),
epithelial cell adhesion molecule, keratin (KRT) 18 and KRT19],
fibroblasts (collagen type I α 1 chain, collagen type I α 2 chain,
collagen type III α 1 chain and actin, α 2, smooth muscle),
endothelial cells (platelet-endothelial cell adhesion molecule 1,
CD34, cadherin 5 and von Willebrand factor), T/natural killer (NK)
cells (CD3D, CD3E, CD3G and CD247), B cells (CD79A, CD79B,
immunoglobulin heavy constant µ and immunoglobulin heavy constant
Δ) and myeloid cells (lysozyme, S100 calcium binding protein A8,
S100 calcium binding protein A9 and CD14) (Fig. 2A and B).
Key findings revealed extensive remodeling of the
TME in PTC, beginning with a marked shift in cellular composition:
The present study observed a marked decrease in thyrocytes and a
concurrent increase in myeloid cells, indicative of enhanced
stromal and immune infiltration accompanied by loss of functional
thyroid epithelium (Fig. 2C).
Further analysis identified elevated folate metabolism activity as
a hallmark of PTC, particularly within myeloid cells. Notably, M1
macrophages not only demonstrated the highest folate metabolic
score among all subsets but also constituted the majority of cells
within the high folate metabolism group (Fig. 2D, H and I). Sub-clustering of
myeloid cells confirmed the expansion of M1 macrophages in PTC
(Fig. 2F and G) and these cells
exhibited pronounced enrichment of immune-inflammatory pathways,
including TNF-α, IFN-γ, KRAS and NF-κB signaling, allograft
rejection, epithelial-mesenchymal transition and complement
activation (Fig. 2J, N and O). Cell
communication analysis revealed that M1 macrophages undergo the
most notable alterations in both the number and strength of
interactions, notably with T/NK cells and engage key
ligand-receptor axes such as secreted phosphoprotein 1-CD44,
TNF-TNF receptor superfamily, macrophage migration inhibitory
factor-(CD74+/CD44/C-X-C motif chemokine receptor 4) and
IL6-IL6 receptor, which are key in inflammation, adhesion and
immune regulation (Fig. 2E, K and
M). In addition, signaling network inference highlighted
‘RESISTIN’, ‘MK’ and ‘GDF’ pathways as key mediators of
intercellular crosstalk within the TME (Fig. 2L). Collectively, these results
underscore the dual role of M1 macrophages in PTC, mechanistically
associating folate metabolism with pro-tumorigenic inflammation and
immune modulation and identifying them as pivotal regulators of TME
remodeling.
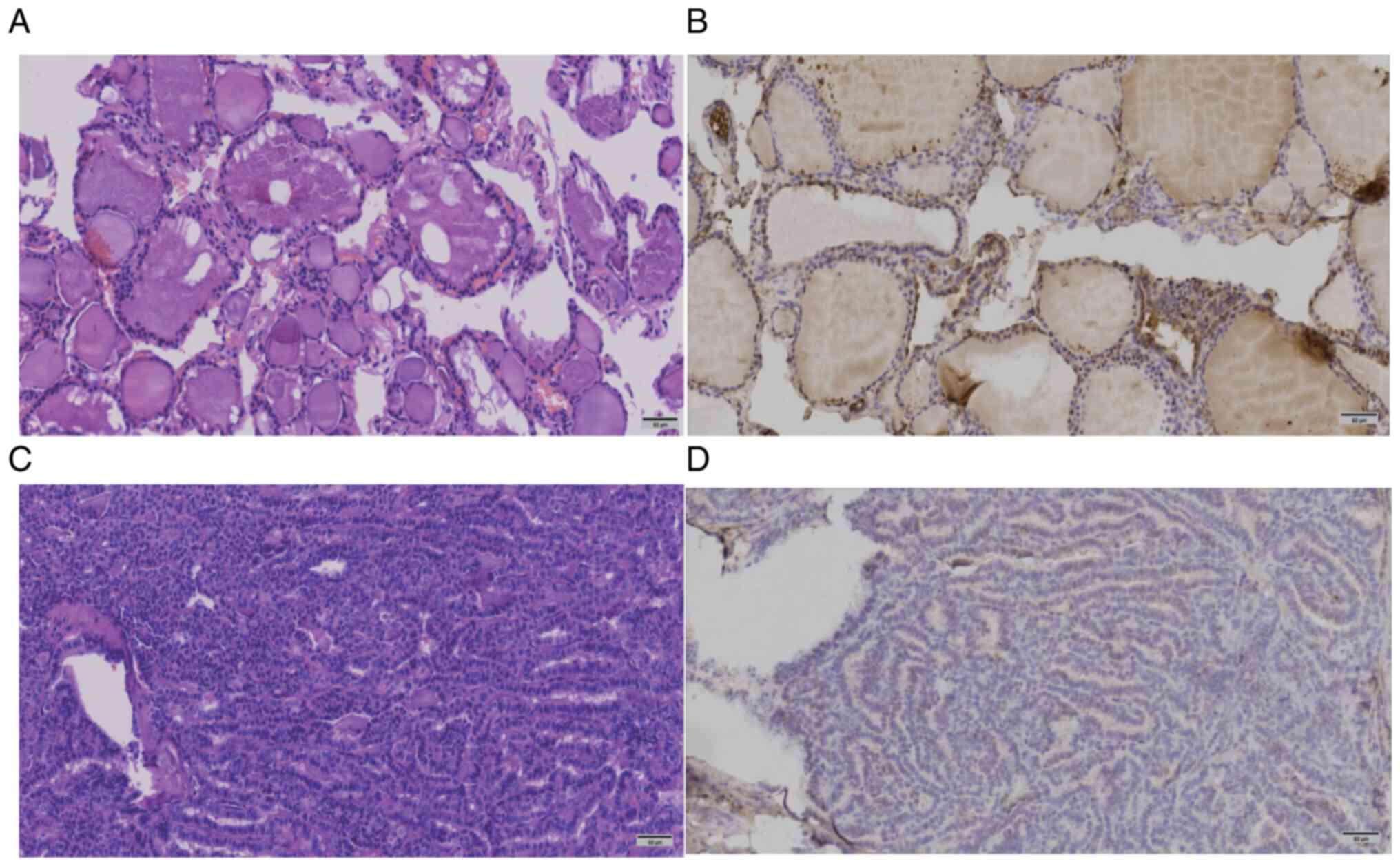
Expression of FOLR3 protein in thyroid
cancer and adjacent normal tissue
IHC revealed that FOLR3 expression was significantly
higher in normal thyroid tissues (H-score, 129.93±50.18) compared
with thyroid cancer tissues (H-score, 99.68±47.67) (Table II; Fig.
3). A paired t-test confirmed that this difference was
statistically significant (P<0.0001; Table II).
 | Table II.H-scores of the patients (n=293). |
Table II.
H-scores of the patients (n=293).
| Patient no. | Normal thyroid
tissues | Thyroid cancer
tissues |
|---|
| 1 | 164.55 | 146.37 |
| 2 | 146.00 | 136.58 |
| 3 | 133.02 | 129.21 |
| 4 | 247.31 | 195.57 |
| 5 | 148.17 | 87.76 |
| 6 | 160.78 | 138.36 |
| 7 | 114.05 | 53.49 |
| 8 | 211.47 | 154.97 |
| 9 | 193.39 | 153.36 |
| 10 | 154.94 | 145.04 |
| 11 | 150.71 | 143.13 |
| 12 | 183.65 | 98.63 |
| 13 | 171.79 | 153.39 |
| 14 | 151.49 | 146.88 |
| 15 | 201.25 | 154.38 |
| 16 | 186.21 | 151.62 |
| 17 | 116.70 | 104.32 |
| 18 | 152.38 | 126.82 |
| 19 | 162.38 | 148.82 |
| 20 | 171.70 | 61.32 |
| 21 | 125.82 | 72.99 |
| 22 | 215.35 | 172.70 |
| 23 | 219.23 | 147.34 |
| 24 | 171.05 | 156.03 |
| 25 | 196.15 | 161.93 |
| 26 | 184.15 | 111.95 |
| 27 | 181.24 | 139.55 |
| 28 | 133.28 | 126.86 |
| 29 | 204.96 | 149.66 |
| 30 | 143.99 | 132.18 |
| 31 | 174.22 | 145.96 |
| 32 | 156.80 | 139.17 |
| 33 | 86.04 | 54.02 |
| 34 | 108.69 | 109.10 |
| 35 | 119.19 | 82.42 |
| 36 | 109.45 | 77.76 |
| 37 | 108.59 | 57.18 |
| 38 | 99.96 | 51.16 |
| 39 | 86.40 | 40.47 |
| 40 | 96.69 | 46.96 |
| 41 | 122.83 | 108.76 |
| 42 | 158.93 | 113.31 |
| 43 | 140.82 | 118.09 |
| 44 | 105.50 | 63.45 |
| 45 | 131.70 | 111.66 |
| 46 | 139.03 | 86.48 |
| 47 | 94.88 | 68.67 |
| 48 | 56.94 | 39.40 |
| 49 | 85.17 | 48.56 |
| 50 | 11.55 | 8.62 |
| 51 | 135.13 | 132.38 |
| 52 | 147.64 | 137.57 |
| 53 | 127.17 | 119.15 |
| 54 | 92.66 | 103.77 |
| 55 | 134.85 | 84.52 |
| 56 | 96.37 | 39.76 |
| 57 | 54.15 | 42.18 |
| 58 | 65.47 | 13.09 |
| 59 | 63.74 | 1.07 |
| 60 | 5.88 | 0.18 |
| 61 | 113.42 | 91.81 |
| 62 | 210.17 | 64.71 |
| 63 | 190.18 | 122.17 |
| 64 | 88.44 | 70.45 |
| 65 | 116.35 | 157.87 |
| 66 | 76.91 | 56.08 |
| 67 | 159.75 | 68.34 |
| 68 | 156.54 | 119.36 |
| 69 | 160.64 | 136.02 |
| 70 | 59.53 | 55.13 |
| 71 | 81.79 | 78.75 |
| 72 | 93.73 | 50.22 |
| 73 | 130.00 | 39.09 |
| 74 | 87.28 | 41.11 |
| 75 | 216.87 | 103.31 |
| 76 | 98.67 | 74.03 |
| 77 | 99.00 | 73.30 |
| 78 | 90.94 | 79.29 |
| 79 | 164.41 | 93.89 |
| 80 | 167.83 | 166.5 |
| 81 | 137.57 | 69.51 |
| 82 | 111.62 | 49.47 |
| 83 | 201.42 | 134.27 |
| 84 | 163.93 | 118.29 |
| 85 | 80.09 | 61.97 |
| 86 | 192.61 | 88.74 |
| 87 | 170.62 | 152.33 |
| 88 | 191.49 | 50.53 |
| 89 | 119.56 | 119.22 |
| 90 | 67.67 | 45.43 |
| 91 | 67.53 | 51.54 |
| 92 | 106.34 | 6.95 |
| 93 | 120.59 | 35.01 |
| 94 | 61.15 | 38.78 |
| 95 | 120.41 | 89.81 |
| 96 | 50.26 | 16.43 |
| 97 | 62.52 | 42.29 |
| 98 | 48.86 | 25.90 |
| 99 | 35.35 | 24.36 |
| 100 | 113.82 | 113.06 |
| 101 | 73.47 | 64.73 |
| 102 | 81.64 | 76.59 |
| 103 | 61.18 | 14.07 |
| 104 | 74.41 | 28.97 |
| 105 | 112.23 | 58.89 |
| 106 | 75.94 | 45.13 |
| 107 | 116.07 | 93.56 |
| 108 | 99.80 | 58.09 |
| 109 | 92.19 | 22.33 |
| 110 | 42.86 | 12.04 |
| 111 | 10.30 | 11.21 |
| 112 | 124.12 | 50.37 |
| 113 | 85.12 | 42.66 |
| 114 | 83.75 | 74.22 |
| 115 | 32.02 | 14.77 |
| 116 | 84.79 | 90.94 |
| 117 | 73.23 | 96.52 |
| 118 | 1.33 | 8.67 |
| 119 | 117.84 | 96.20 |
| 120 | 66.44 | 56.91 |
| 121 | 95.26 | 97.71 |
| 122 | 127.92 | 76.75 |
| 123 | 73.33 | 79.05 |
| 124 | 56.06 | 45.97 |
| 125 | 117.35 | 76.76 |
| 126 | 100.10 | 44.13 |
| 127 | 126.63 | 110.81 |
| 128 | 131.09 | 100.99 |
| 129 | 30.80 | 28.38 |
| 130 | 101.37 | 82.66 |
| 131 | 64.22 | 55.62 |
| 132 | 51.98 | 35.75 |
| 133 | 172.32 | 64.74 |
| 134 | 67.35 | 60.24 |
| 135 | 58.16 | 41.55 |
| 136 | 33.55 | 33.89 |
| 137 | 85.79 | 60.32 |
| 138 | 26.20 | 5.91 |
| 139 | 34.32 | 10.65 |
| 140 | 59.71 | 42.04 |
| 141 | 188.62 | 104.21 |
| 142 | 45.80 | 31.95 |
| 143 | 147.59 | 75.87 |
| 144 | 70.59 | 38.08 |
| 145 | 84.66 | 59.77 |
| 146 | 108.32 | 87.49 |
| 147 | 71.66 | 57.89 |
| 148 | 80.62 | 134.46 |
| 149 | 137.00 | 116.60 |
| 150 | 140.84 | 139.98 |
| 151 | 134.13 | 81.44 |
| 152 | 135.82 | 131.41 |
| 153 | 185.20 | 161.26 |
| 154 | 158.57 | 147.50 |
| 155 | 147.48 | 132.21 |
| 156 | 180.52 | 95.11 |
| 157 | 78.38 | 66.41 |
| 158 | 161.33 | 154.09 |
| 159 | 111.38 | 106.17 |
| 160 | 184.15 | 152.06 |
| 161 | 148.90 | 142.03 |
| 162 | 190.60 | 167.59 |
| 163 | 194.39 | 152.33 |
| 164 | 166.85 | 154.07 |
| 165 | 158.80 | 141.93 |
| 166 | 188.04 | 187.25 |
| 167 | 143.03 | 89.98 |
| 168 | 162.27 | 148.97 |
| 169 | 168.05 | 176.93 |
| 170 | 127.95 | 117.53 |
| 171 | 147.88 | 116.69 |
| 172 | 112.18 | 102.63 |
| 173 | 164.60 | 29.74 |
| 174 | 124.81 | 106.56 |
| 175 | 104.00 | 71.29 |
| 176 | 80.54 | 58.33 |
| 177 | 39.82 | 18.88 |
| 178 | 100.87 | 44.71 |
| 179 | 96.21 | 69.35 |
| 180 | 133.71 | 71.09 |
| 181 | 69.68 | 51.30 |
| 182 | 106.75 | 86.33 |
| 183 | 131.84 | 64.39 |
| 184 | 71.89 | 23.83 |
| 185 | 132.16 | 39.71 |
| 186 | 129.86 | 45.19 |
| 187 | 52.52 | 49.63 |
| 188 | 86.69 | 99.01 |
| 189 | 96.55 | 66.90 |
| 190 | 39.05 | 5.49 |
| 191 | 57.76 | 52.49 |
| 192 | 78.92 | 66.92 |
| 193 | 53.93 | 49.64 |
| 194 | 114.65 | 100.65 |
| 195 | 84.82 | 65.47 |
| 196 | 111.75 | 101.47 |
| 197 | 80.33 | 94.78 |
| 198 | 108.18 | 73.45 |
| 199 | 76.27 | 77.60 |
| 200 | 112.31 | 68.72 |
| 201 | 80.01 | 58.27 |
| 202 | 89.42 | 72.15 |
| 203 | 96.58 | 79.57 |
| 204 | 102.38 | 69.58 |
| 205 | 97.33 | 70.65 |
| 206 | 93.90 | 76.41 |
| 207 | 117.50 | 60.98 |
| 208 | 23.55 | 3.58 |
| 209 | 157.45 | 107.52 |
| 210 | 81.05 | 60.24 |
| 211 | 137.02 | 116.14 |
| 212 | 118.78 | 79.55 |
| 213 | 133.41 | 133.66 |
| 214 | 164.76 | 116.36 |
| 215 | 180.06 | 182.92 |
| 216 | 166.59 | 118.32 |
| 217 | 170.52 | 170.16 |
| 218 | 193.79 | 163.37 |
| 219 | 193.36 | 158.70 |
| 220 | 187.24 | 170.76 |
| 221 | 183.74 | 145.64 |
| 222 | 182.94 | 172.33 |
| 223 | 131.85 | 89.28 |
| 224 | 148.43 | 77.22 |
| 225 | 166.30 | 160.53 |
| 226 | 150.27 | 142.50 |
| 227 | 135.90 | 81.77 |
| 228 | 146.95 | 133.12 |
| 229 | 151.41 | 150.12 |
| 230 | 143.85 | 133.38 |
| 231 | 99.93 | 43.97 |
| 232 | 151.49 | 121.59 |
| 233 | 144.99 | 124.87 |
| 234 | 142.00 | 122.68 |
| 235 | 165.83 | 140.79 |
| 236 | 208.44 | 187.60 |
| 237 | 178.17 | 163.92 |
| 238 | 168.91 | 141.85 |
| 239 | 165.80 | 145.45 |
| 240 | 136.95 | 139.40 |
| 241 | 147.78 | 135.69 |
| 242 | 105.73 | 86.53 |
| 243 | 73.78 | 0.95 |
| 244 | 97.68 | 47.81 |
| 245 | 163.53 | 57.35 |
| 246 | 166.40 | 107.74 |
| 247 | 129.13 | 108.89 |
| 248 | 137.95 | 112.03 |
| 249 | 143.59 | 109.13 |
| 250 | 137.34 | 132.70 |
| 251 | 175.34 | 147.97 |
| 252 | 184.94 | 172.82 |
| 253 | 156.07 | 143.52 |
| 254 | 189.84 | 188.35 |
| 255 | 183.57 | 169.42 |
| 256 | 168.51 | 138.99 |
| 257 | 127.93 | 118.84 |
| 258 | 158.56 | 143.35 |
| 259 | 123.80 | 105.50 |
| 260 | 194.29 | 135.53 |
| 261 | 159.92 | 99.10 |
| 262 | 131.32 | 120.29 |
| 263 | 48.63 | 30.04 |
| 264 | 164.66 | 127.32 |
| 265 | 127.74 | 77.67 |
| 266 | 101.59 | 88.66 |
| 267 | 128.55 | 117.84 |
| 268 | 100.90 | 57.06 |
| 269 | 134.81 | 87.24 |
| 270 | 134.47 | 110.14 |
| 271 | 118.35 | 109.63 |
| 272 | 131.51 | 111.68 |
| 273 | 126.47 | 120.74 |
| 274 | 243.09 | 132.76 |
| 275 | 138.70 | 112.34 |
| 276 | 215.87 | 91.34 |
| 277 | 163.43 | 113.18 |
| 278 | 192.07 | 133.95 |
| 279 | 177.21 | 123.38 |
| 280 | 174.17 | 112.87 |
| 281 | 152.79 | 121.19 |
| 282 | 130.25 | 125.07 |
| 283 | 182.06 | 170.84 |
| 284 | 163.29 | 152.19 |
| 285 | 142.50 | 133.29 |
| 286 | 135.76 | 112.89 |
| 287 | 162.53 | 132.69 |
| 288 | 164.47 | 133.78 |
| 289 | 114.49 | 115.11 |
| 290 | 183.97 | 136.89 |
| 291 | 158.09 | 88.39 |
| 292 | 98.89 | 63.07 |
| 293 | 160.68 | 47.88 |
Association analysis of FOLR3 and
clinicopathological factors in thyroid cancer
The expression levels of FOLR3 were significantly
associated with capsule invasion, neurofibrillary invasion,
striated muscle cell invasion and pT stage (P<0.05) and there
was no significant association with sex, age, tumor location, tumor
focality, histological type, vascular invasion or pN stage
(P<0.05) (Table III). FOLR3
was commonly downregulated or absent in cases with capsule,
rhabdomyocyte invasion or larger tumor burden, suggesting that
FOLR3 could serve as an indicator of focal invasion in thyroid
cancer.
 | Table III.Association between FOLR3 expression
and clinicopathological parameters in patients with thyroid cancer
(n=293). |
Table III.
Association between FOLR3 expression
and clinicopathological parameters in patients with thyroid cancer
(n=293).
|
|
| FOLR3
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | No. of cases | Low, n | High, n | P-value |
|---|
| Sex |
|
|
| 0.465 |
|
Male | 51 | 29 | 22 |
|
|
Female | 242 | 124 | 118 |
|
| Age, years |
|
|
| 0.165 |
|
≤65 | 282 | 145 | 137 |
|
|
>65 | 11 | 8 | 3 |
|
| Tumor location |
|
|
| 0.473 |
|
Unilateral | 255 | 129 | 126 |
|
|
Bilateral | 38 | 18 | 20 |
|
| Histological
type |
|
|
| 0.832 |
|
Papillary carcinoma | 287 | 145 | 142 |
|
|
Follicular carcinoma | 3 | 1 | 2 |
|
|
Anaplastic cancer | 3 | 1 | 2 |
|
| Tumor focality |
|
|
| 0.446 |
| Single
lesion | 165 | 80 | 85 |
|
|
Multiple lesions | 128 | 67 | 61 |
|
| Capsule
invasion |
|
|
| 0.042a |
| No | 102 | 45 | 57 |
|
|
Yes | 191 | 108 | 83 |
|
| Rhabdomyocyte
invasion |
|
|
| 0.006a |
|
Yes | 285 | 145 | 140 |
|
| No | 8 | 8 | 0 |
|
| pT stage |
|
|
| 0.047a |
| 1 | 272 | 141 | 131 |
|
| 2 | 14 | 5 | 9 |
|
| 3 | 5 | 5 | 0 |
|
| 4 | 2 | 2 | 0 |
|
| pN stage |
|
|
| 0.864 |
| 0 | 168 | 87 | 81 |
|
| 1 | 125 | 66 | 59 |
|
| Vascular
invasion |
|
|
| 0.123 |
| No | 287 | 148 | 139 |
|
|
Yes | 6 | 5 | 1 |
|
| Perineural
invasion |
|
|
| 0.025a |
| No | 284 | 145 | 139 |
|
|
Yes | 9 | 8 | 1 |
|
FOLR3 inhibits proliferation, invasion
and migration of thyroid cancer TPC-1 cells
The western blotting analysis (Fig. 4A and B) confirmed successful
overexpression of FOLR3 protein in TPC-1 cells of the Flag-FOLR3
group compared with the NC (P<0.01). RT-qPCR results
demonstrated a significant upregulation of FOLR3 mRNA expression in
the Flag-FOLR3 group (P<0.0001; Fig.
4C). Colony formation assays demonstrated the clonogenic
ability of Flag-FOLR3 group was significantly lower compared with
that of NC group (P<0.05), indicating that FOLR3 overexpression
impaired the proliferative capacity of TPC-1 cells (Fig. 4E and F). The results of the scratch
assay demonstrated cell migration was significantly attenuated in
the Flag-FOLR3 group compared with the NC group at both 24
(P<0.05) and 48 h (P<0.001), confirming that FOLR3
overexpression suppresses the migratory ability of TPC-1 cells
(Fig. 4G and H). Similarly,
Transwell invasion assays indicated that the invasive capacity of
TPC-1 cells was significantly reduced in the Flag-FOLR3 group after
24 h (P<0.01), indicating that FOLR3 overexpression
significantly inhibits cell invasion (Fig. 4I and J). CCK-8 assays indicated that
TPC-1 cell proliferation was significantly inhibited at 48, 72 and
96 h after FOLR3 overexpression (P<0.05; Fig. 4D).
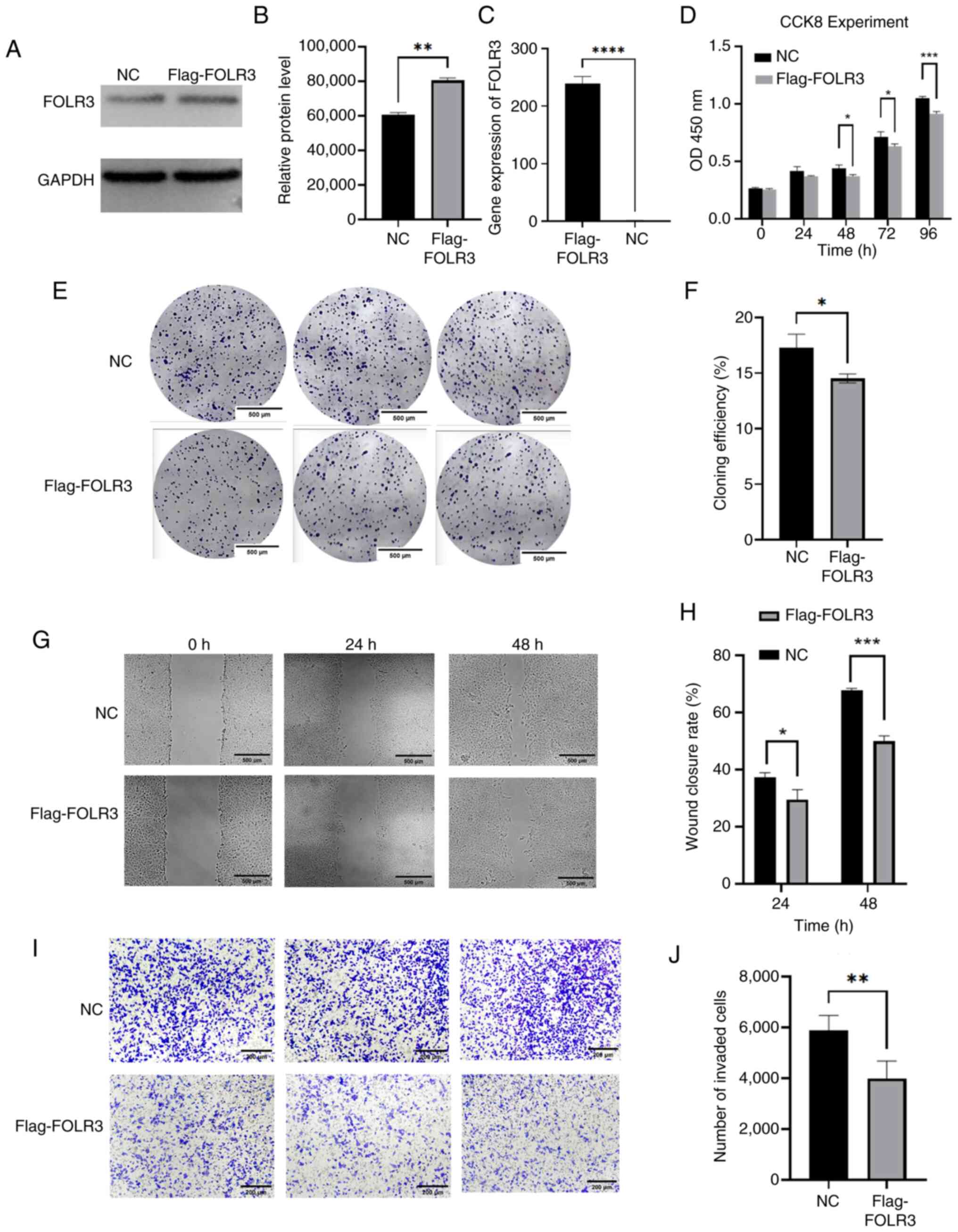 | Figure 4.Functional characterization of FOLR3
in thyroid cancer cells in vitro. (A) Protein expression
levels of FOLR3 assessed using western blotting and (B)
semi-quantification. (C) Relative mRNA relative expression levels
of FOLR3 by reverse transcription-quantitative PCR. (D) CCK-8 for
0, 24, 48, 72 and 96 h. (E) Colony forming ability evaluated using
colony formation assay and (F) quantification (magnification, ×4).
(G) Cell migratory capacity analyzed using a wound healing assay
and (H) quantification (magnification, ×20). (I) Invasive potential
determined using Transwell invasion assay and (J) quantification
(magnification, ×10). *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. FOLR3, folate receptor γ; CCK-8, Cell Counting
Kit-8; NC, negative control. |
FOLR3 overexpression suppresses the
progression of thyroid cancer by inactivating the STAT3/MAPK
signaling pathways and subsequently reprogramming folate
metabolism
To elucidate the molecular mechanism underlying the
tumor-suppressive role of FOLR3, the present study first
investigated its impact on folate metabolism using qPCR analysis.
Notably, FOLR3 overexpression significantly downregulated the mRNA
level of the folate transporter gene solute carrier family 19
member 1 and upregulated that of solute carrier family 46 member 1
(P<0.05). Concurrently, expression levels of key folate
metabolic enzyme genes, methylenetetrahydrofolate reductase and
thymidylate synthase, were significantly downregulated (P<0.05).
These results indicate that FOLR3 overexpression induces notable
reprogramming of folate metabolism in thyroid cancer cells
(Fig. 5A), which may contribute to
the inhibition of tumor progression.
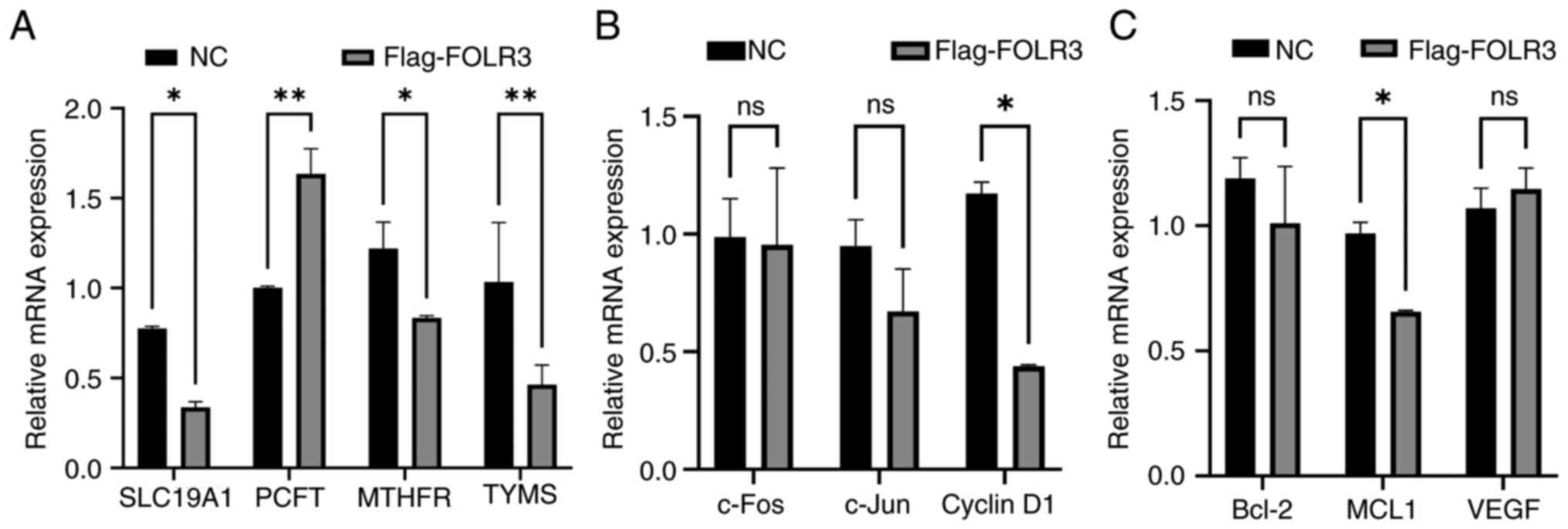 | Figure 5.FOLR3 overexpression promotes folate
metabolism and activates oncogenic signaling pathways in thyroid
cancer cells. (A) mRNA expression levels of folate
metabolism-related genes (SLC19A1, PCFT, MTHFR and TYMS) after
FOLR3 overexpression. (B) mRNA expression levels of MAPK pathway
downstream genes (c-Fos, c-Jun and cyclin D1) after FOLR3
overexpression. (C) mRNA expression levels of STAT3 pathway
downstream genes after FOLR3 overexpression. *P<0.05 and
**P<0.01. ns, not significant; FOLR3, folate receptor γ; TYMS,
thymidylate synthase; MTHFR, methylenetetrahydrofolate reductase;
SLC19A1, solute carrier family 19 member 1; PCFT, proton-coupled
folate transporter; c-FOS, Fos proto-oncogenes, AP-1 transcription
factor subunit; MCL1, myeloid cell leukemia-1; NC, negative
control. |
The present study further examined whether FOLR3
modulates oncogenic signaling pathways by assessing the expression
levels of downstream effectors of STAT3 and MAPK signaling. RT-qPCR
revealed that overexpression of FOLR3 significantly downregulated
both mRNA and protein levels of myeloid cell leukemia-1 (MCL1), an
anti-apoptotic gene downstream of STAT3 (P<0.05). Similarly,
expression levels of cyclin D1, a key cell cycle regulator mediated
by the MAPK pathway, were also significantly suppressed (P<0.05;
Fig. 5B and C). These findings
demonstrated that FOLR3 overexpression effectively inhibits the
activity of both STAT3 and MAPK signaling pathways. Collectively,
the present study results suggest that FOLR3 constrains thyroid
cancer development through dual mechanisms: Inactivation of STAT3
and MAPK signaling pathways and concomitant reprogramming of folate
metabolism.
Discussion
Thyroid cancer is one of the fastest-growing
malignancies in recent years, with a notably higher incidence among
women (33,34). Despite the notable improvements in
early diagnosis and treatment, patients with advanced thyroid
cancer still face poor prognosis due to tumor invasiveness and
metastasis (35,36). Therefore, understanding the
molecular mechanisms underlying thyroid cancer and identifying
novel biomarkers are key to the improvement of diagnosis, treatment
and prognosis. To the best of our knowledge, the present study
investigated the expression level of FOLR3 in thyroid cancer and
its association with clinicopathological features, for the first
time. FOLR3, a member of the folate receptor family, has been
implicated in various cancer types (37–39).
In tongue cancer, FOLR3 is an upregulated immune-related target
associated with patient prognosis (37). Similarly, its expression is
upregulated in metastatic uterine leiomyosarcoma compared to
primary tumors, suggesting a potential role in promoting metastatic
disease (38). Furthermore,
transcriptomic analysis of peripheral blood mononuclear cells from
patients with head and neck squamous cell carcinoma revealed FOLR3
as one of the differentially expressed genes, indicating its
systemic involvement in cancer-related immune responses (39). The present bioinformatics analysis
and experimental data revealed that FOLR3 is significantly
downregulated in thyroid cancer tissues compared with normal tissue
and its high expression was associated with worse overall survival.
The seemingly contradictory link between FOLR3 expression and
clinical prognosis may be explained by its cell type-specific
functions within the tumor microenvironment. While FOLR3 is
typically downregulated in cancer cells (a tumor-suppressive
effect), its high overall levels in tumors mainly comes from
tumor-infiltrating immune cells such as macrophages. This
immune-derived FOLR3 enrichment is indicative of an
immunosuppressive tumor microenvironment that promotes disease
progression and is associated with poorer patient survival.
Therefore, FOLR3 exhibits a dual function, acting as a putative
tumor suppressor within cancer cells while serving as a biomarker
and functional driver of an immune-favorable, pro-tumorigenic niche
(12,37–41).
Therefore, FOLR3 has the potential for targeted therapy.
Single-cell transcriptomic analysis revealed a
significant expansion of M1 macrophages with heightened folate
metabolism in the TME of PTC. These folate-high M1 macrophages
drive TME remodeling and promote pro-tumor inflammatory responses
through activation of immune-inflammatory pathways and enhanced
intercellular communication. Mechanistically, FOLR3 suppresses
thyroid cancer growth by inactivating the STAT3 and MAPK signaling
pathways through downregulation of their key downstream oncogenes,
MCL1 and cyclin D1, ultimately leading to reprogramming of folate
metabolism. Notably, FOLR3 has also been reported to influence
other cancer types. Recent studies have reported that FOLR3
undergoes hypomethylation in the blood of patients with non-small
cell lung cancer and lung adenocarcinoma, suggesting that FOLR3
hypomethylation could serve as an early diagnostic marker for these
cancers (12,40). Furthermore, high expression of FOLR3
in patients with laryngeal squamous cell carcinoma after
chemotherapy has been reported to influence subsequent treatment
strategies (41). These findings
across cancer types reinforce the biological and clinical relevance
of FOLR3 and support the present study results highlighting its
importance in thyroid cancer.
The human body expresses three isoforms of folate
receptors (FOLRs): FOLR1, FOLR2 and FOLR3, which have distinct
distributions, structures and functions. Both FOLR1 and FOLR2 are
glycosylphosphatidylinositol (GPI)-anchored membrane proteins that
lack transmembrane domains, whereas FOLR3 is secreted into
extracellular compartments, such as blood and breast milk, in a
soluble form due to the absence of a GPI-anchoring signal (42,43).
FOLR1 mainly facilitates folate endocytosis to support classical
folate-dependent proliferative pathways. Although FOLR1 is
upregulated in various types of cancers including ovarian cancer,
Non-Small Cell Lung cancer and rectal cancer, and is generally
associates with accelerated tumor progression and poor prognosis,
its specific role and mechanisms in thyroid cancer remain to be
elucidated in future studies (44–47).
Currently, most folate receptor-targeted drugs and diagnostic
technologies focus on FOLR1. High expression level of
membrane-bound FOLR1 is a recognized predictor of targeted therapy
sensitivity, while soluble FOLR3 levels may be associated with
treatment resistance (45,48). The present study identified reduced
expression levels of FOLR3 in thyroid cancer, which appears to
specifically reflect malignant progression. FOLR3 may influence
carcinogenesis through the folate metabolism-signaling axis and
modulate local folate availability via epigenetic regulation of
soluble folate pools. Further research could explore ‘metabolic
compensation’ strategies using FOLR3 for aggressive thyroid cancer
with poor prognosis, highlighting its therapeutic potential. Since
FOLR3 is secreted into the bloodstream, it may serve as a
non-invasive biomarker in liquid biopsies. For instance,
hypomethylation of FOLR3 in the blood of patients with lung cancer
suggests its utility as an early diagnostic marker (12).
However, whether FOLR3 can be used for thyroid
cancer diagnosis via liquid biopsy requires further validation.
Theoretically, blood-based FOLR3 levels may be associated with
tumor burden, metabolic activity or disease stage. While FOLR3
alone may not be sufficient for diagnosis, its combination with
other markers, such as CA-125, human epididymis protein 4 or
circulating tumor DNA mutations, could significantly enhance
diagnostic accuracy and sensitivity (49–51).
Although the present study provided evidence supporting the
tumor-suppressive role of FOLR3 in thyroid cancer, several
limitations should be acknowledged. The clinical samples, despite
having detailed pathological annotations, were obtained from a
single center with a limited cohort size (n=293). Validation in a
larger, multi-center cohort is warranted to enhance the statistical
power and generalizability of the association between FOLR3
expression and clinicopathological characteristics. Furthermore,
the lack of in vivo validation using animal models, such as
xenograft or orthotopic models, restricts the comprehensive
confirmation of its tumor-suppressive function within a more
complex physiological context. The present study primarily focused
on the role of FOLR3 in thyroid cancer itself, while its
interactions with the tumor immune microenvironment, stromal
components and other folate metabolism-related genes have not been
extensively investigated. In the future, FOLR3 may have the
potential to serve as a diagnostic biomarker and a novel
therapeutic target in thyroid cancer; however, further research is
warranted to explore its clinical applicability in both treatment
and prognosis assessment.
Acknowledgements
The authors would like to thank Mr Xiaokai Liu (Chi
Feng No. 2 Senior High School, Chifeng, China) for their assistance
in polishing the English language and grammar of the
manuscript.
Funding
The present study was supported by the Chifeng Municipal Natural
Science Foundation (grant no. SZR2023063).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JC, YL and LT conceptualized the present study; JC,
YL and XD devised the methodology. JC and XD; JC, YL and XD
validated the data. JC, LH, YL and LT analyzed data. JC, YL and BL
performed the experiments. YL and LT prepared the resources. JC, YL
and XD curated the data. JC, YL and XD visualized the data. JC, YL
and XD prepared the original draft of the manuscript. JC, YL, LH
and LT reviewed and edited the manuscript. LT supervised the
present study. YL and LT contributed to project administration. All
authors read and approved the final manuscript. JC and YL confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Scientific
Research Ethics Committee of Chifeng City Hospital (approval no.
CK2023037; approval on 20 April 2023; Chifeng, China). The present
study is retrospective in nature, and the informed consent
requirement was waived by the Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence tools were used to improve the readability and
language of the manuscript, and subsequently, the authors revised
and edited the content produced by the artificial intelligence
tools as necessary, taking full responsibility for the ultimate
content of the present manuscript.
References
|
1
|
Chen MM, Luu M, Sacks WL, Orloff L,
Wallner LP, Clair JM, Pitt SC, Ho AS and Zumsteg ZS: Trends in
incidence, metastasis, and mortality from thyroid cancer in the USA
from 1975 to 2019: A population-based study of age, period, and
cohort effects. Lancet Diabetes Endocrinol. 13:188–195. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI
|
|
3
|
Boucai L, Zafereo M and Cabanillas ME:
Thyroid cancer: A review. JAMA. 331:425–435. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shen F, Wu M, Ross JF, Miller D and Ratnam
M: Folate receptor type gamma is primarily a secretory protein due
to lack of an efficient signal for glycosylphosphatidylinositol
modification: Protein characterization and cell type specificity.
Biochemistry. 34:5660–5665. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang HC, Huo YN and Lee WS: Folic acid
prevents the progesterone-promoted proliferation and migration in
breast cancer cell lines. Eur J Nutr. 59:2333–2344. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moazzen S, Dolatkhah R, Tabrizi JS,
Shaarbafi J, Alizadeh BZ, de Bock GH and Dastgiri S: Folic acid
intake and folate status and colorectal cancer risk: A systematic
review and meta-analysis. Clin Nutr. 37:1926–1934. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sadeghi Jam Z, Tafvizi F, Khodarahmi P,
Jafari P and Baghbani-Arani F: Cisplatin-loaded UiO-66-NH2
functionalized with folic acid enhances apoptotic activity and
antiproliferative effects in MDA-MB-231 breast and A2780 ovarian
cancer cells: An in vitro study. Heliyon. 11:e426852025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Crider KS, Yang TP, Berry RJ and Bailey
LB: Folate and DNA methylation: A review of molecular mechanisms
and the evidence for Folate's role. Adv Nutr. 3:21–38. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Petersen LF, Brockton NT, Bakkar A, Liu S,
Wen J, Weljie AM and Bismar TA: Elevated physiological levels of
folic acid can increase in vitro growth and invasiveness of
prostate cancer cells. BJU Int. 109:788–795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ullevig SL, Bacich DJ, Gutierrez JM,
Balarin A, Lobitz CA, O'Keefe DS and Liss MA: Feasibility of
dietary folic acid reduction intervention for men on active
surveillance for prostate cancer. Clin Nutr ESPEN. 44:270–275.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li JT, Yang H, Lei MZ, Zhu WP, Su Y, Li
KY, Zhu WY, Wang J, Zhang L, Qu J, et al: Dietary folate drives
methionine metabolism to promote cancer development by stabilizing
MAT IIA. Signal Transduct Target Ther. 7:1922022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qu Y, Zhang X, Qiao R, Di F, Song Y, Wang
J, Ji L, Zhang J, Gu W, Fang Y, et al: Blood FOLR3 methylation
dysregulations and heterogeneity in non-small lung cancer highlight
its strong associations with lung squamous carcinoma. Respir Res.
25:592024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Holm J and Hansen SI: Characterization of
soluble folate receptors (folate binding proteins) in humans.
Biological roles and clinical potentials in infection and
malignancy. Biochim Biophys Acta Proteins Proteom. 1868:1404662020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu MH, Luo JD, Wang WC, Chang TH, Hwang
WL, Lee KH, Liu SY, Yang JW, Chiou CT, Chang CH and Chiang WF: Risk
analysis of malignant potential of oral verrucous hyperplasia: A
follow-up study of 269 patients and copy number variation analysis.
Head Neck. 40:1046–1056. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hansen MF, Greibe E, Skovbjerg S, Rohde S,
Kristensen AC, Jensen TR, Stentoft C, Kjær KH, Kronborg CS and
Martensen PM: Folic acid mediates activation of the pro-oncogene
STAT3 via the Folate Receptor alpha. Cell Signal. 27:1356–1368.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma Q, Geng K, Xiao P and Zeng L:
Identification and prognostic value exploration of radiotherapy
sensitivity-Associated genes in Non-Small-cell lung cancer. Biomed
Res Int. 2021:59638682021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zee RY, Rose L, Chasman DI and Ridker PM:
Genetic variation of fifteen folate metabolic pathway associated
gene loci and the risk of incident head and neck carcinoma: The
Women's Genome Health Study. Clin Chim Acta. 418:33–36. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liao T, Zeng Y, Xu W, Shi X, Shen C, Du Y,
Zhang M, Zhang Y, Li L, Ding P, et al: A spatially resolved
transcriptome landscape during thyroid cancer progression. Cell Rep
Med. 6:1020432025. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ritchie ME, Phipson B and Wu D, Ritchie
ME, Phipson B and Wu D: Limma powers differential expression
analyses for RNA-sequencing and microarray studies. Nucleic Acids
Res. 43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2:1001412021.PubMed/NCBI
|
|
21
|
Li X, Zhang L, Liu C, He Y, Li X, Xu Y, Gu
C, Wang X, Wang S, Zhang J and Liu J: Construction of mitochondrial
quality regulation genes-related prognostic model based on
bulk-RNA-seq analysis in multiple myeloma. Biofactors.
51:e21352025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang X, Sun Y, Song J, Huang Y, Shi H,
Qian A, Cao Y, Zhou Y and Wang Q: Prognostic value of fatty acid
metabolism-related genes in colorectal cancer and their potential
implications for immunotherapy. Front Immunol. 14:13014522023.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ning X, Li R, Zhang B, Wang Y, Zhou Z, Ji
Z, Lyu X and Chen Z: Immune score indicator for the survival of
melanoma patients based on tumor microenvironment. Int J Gen Med.
14:10397–10416. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao AY, Unterman A, Abu Hussein NS,
Sharma P, Nikola F, Flint J, Yan X, Adams TS, Justet A, Sumida TS,
et al: Single-cell analysis reveals novel immune perturbations in
fibrotic hypersensitivity pneumonitis. Am J Respir Crit Care Med.
210:1252–1266. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Li F, Xu L, Shi X, Xue H, Liu J, Bai
S, Wu Y, Yang Z, Xue F, et al: Single cell analyses reveal the PD-1
blockade response-related immune features in hepatocellular
carcinoma. Theranostics. 14:3526–3547. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin S, Guerrero-Juarez CF, Zhang L, Chang
I, Ramos R, Kuan CH, Myung P, Plikus MV and Nie Q: Inference and
analysis of cell-cell communication using CellChat. Nat Commun.
12:10882021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang A, Sun Z, Hong H, Yang Y, Chen J,
Gao Z and Gu J: Novel hypoxia- and lactate metabolism-related
molecular subtyping and prognostic signature for colorectal cancer.
J Transl Med. 22:5872024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marrero JA, Kulik LM, Sirlin CB, Zhu AX,
Finn RS, Abecassis MM, Roberts LR and Heimbach JK: Diagnosis,
staging, and management of hepatocellular carcinoma: 2018 practice
guidance by the American association for the study of liver
diseases. Hepatology. 68:723–750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Humphries MP, Maxwell P and Salto-Tellez
M: QuPath: The global impact of an open source digital pathology
system. Comput Struct Biotechnol J. 19:852–859. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wen Z, Luo D, Wang S, Rong R, Evers BM,
Jia L, Fang Y, Daoud EV, Yang S, Gu Z, et al: Deep Learning-Based
H-Score quantification of immunohistochemistry-stained images. Mod
Pathol. 37:1003982024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siena S, Raghav K, Masuishi T, Yamaguchi
K, Nishina T, Elez E, Rodriguez J, Chau I, Di Bartolomeo M,
Kawakami H, et al: HER2-related biomarkers predict clinical
outcomes with trastuzumab deruxtecan treatment in patients with
HER2-expressing metastatic colorectal cancer: Biomarker analyses of
DESTINY-CRC01. Nat Commun. 15:102132024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li M, Hu M, Jiang L, Pei J and Zhu C:
Trends in cancer incidence and potential associated factors in
China. JAMA Netw Open. 7:e24403812024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miranda-Filho A, Lortet-Tieulent J, Bray
F, Cao B, Franceschi S, Vaccarella S and Dal Maso L: Thyroid cancer
incidence trends by histology in 25 countries: A population-based
study. Lancet Diabetes Endocrinol. 9:225–234. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tao Z, Ding Z, Guo B, Fan Y and Deng X:
Influence factors and survival outcomes of different invasion sites
in locally advanced thyroid cancer and new site-based risk
stratification system. Endocrine. 88:501–510. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu X, Deng Q, Gao X, He L, Hu D and Yang
L: A prognostic nomogram for distant metastasis in thyroid cancer
patients without lymph node metastasis. Front Endocrinol
(Lausanne). 16:15237852025. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lv X and Yu X: Signatures and prognostic
values of related immune targets in tongue cancer. Front Surg.
9:9523892023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Davidson B, Abeler VM, Førsund M, Holth A,
Yang Y, Kobayashi Y, Chen L, Kristensen GB, Shih IeM and Wang TL:
Gene expression signatures of primary and metastatic uterine
leiomyosarcoma. Hum Pathol. 45:691–700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bin-Alee F, Arayataweegool A,
Buranapraditkun S, Mahattanasakul P, Tangjaturonrasme N, Hirankarn
N, Mutirangura A and Kitkumthorn N: Transcriptomic analysis of
peripheral blood mononuclear cells in head and neck squamous cell
carcinoma patients. Oral Dis. 27:1394–1402. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qiao R, Di F, Wang J, Wei Y, Xu T, Dai L,
Gu W, Han B and Yang R: Identification of FUT7 hypomethylation as
the blood biomarker in the prediction of early-stage lung cancer. J
Genet Genomics. 50:573–581. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li L, Wang R, He S, Shen X, Kong F, Li S,
Zhao H, Lian M and Fang J: The identification of induction
chemo-sensitivity genes of laryngeal squamous cell carcinoma and
their clinical utilization. Eur Arch Otorhinolaryngol.
275:2773–2781. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
O'Byrne MR, Au KS, Morrison AC, Lin JI,
Fletcher JM, Ostermaier KK, Tyerman GH, Doebel S and Northrup H:
Association of folate receptor (FOLR1, FOLR2, FOLR3) and reduced
folate carrier (SLC19A1) genes with meningomyelocele. Birth Defects
Res A Clin Mol Teratol. 88:689–694. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Grarup N, Sulem P, Sandholt CH,
Thorleifsson G, Ahluwalia TS, Steinthorsdottir V, Bjarnason H,
Gudbjartsson DF, Magnusson OT, Sparsø T, et al: Genetic
architecture of vitamin B12 and folate levels uncovered applying
deeply sequenced large datasets. PLoS Genet. 9:e10035302013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nawaz FZ and Kipreos ET: Emerging roles
for folate receptor FOLR1 in signaling and cancer. Trends
Endocrinol Metab. 33:159–174. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mai J, Wu L, Yang L, Sun T, Liu X, Yin R,
Jiang Y, Li J and Li Q: Therapeutic strategies targeting folate
receptor α for ovarian cancer. Front Immunol. 14:12545322023.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Matsunaga Y, Yamaoka T, Ohba M, Miura S,
Masuda H, Sangai T, Takimoto M, Nakamura S and Tsurutani J: Novel
Anti-FOLR1 Antibody-drug conjugate MORAb-202 in breast cancer and
non-Small cell lung cancer cells. Antibodies (Basel). 10:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen CI, Li WS, Chen HP, Liu KW, Tsai CJ,
Hung WJ and Yang CC: High expression of folate receptor alpha
(FOLR1) is associated with aggressive tumor behavior, poor response
to chemoradiotherapy, and worse survival in rectal cancer. Technol
Cancer Res Treat. 21:153303382211417952022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Varaganti P, Buddolla V, Lakshmi BA and
Kim YJ: Recent advances in using folate receptor 1 (FOLR1) for
cancer diagnosis and treatment, with an emphasis on cancers that
affect women. Life Sci. 326:121802M2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Medina JE, Annapragada AV, Lof P, Short S,
Bartolomucci AL, Mathios D, Koul S, Niknafs N, Noë M, Foda ZH, et
al: Early detection of ovarian cancer Using Cell-Free DNA
fragmentomes and protein biomarkers. Cancer Discov. 15:105–118.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Muinao T, Deka Boruah HP and Pal M:
Multi-biomarker panel signature as the key to diagnosis of ovarian
cancer. Heliyon. 5:e028262019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Heitz F, Lakis S, Harter P, Heikaus S,
Sehouli J, Talwar J, Menon R, Ataseven B, Bertrand M, Schneider S,
et al: Cell-free tumor DNA, CA125 and HE4 for the objective
assessment of tumor burden in patients with advanced high-grade
serous ovarian cancer. PLoS One. 17:e02627702022. View Article : Google Scholar : PubMed/NCBI
|