Introduction
Adenoid cystic carcinoma (ACC), also known as
cylindroma, is a rare malignant tumour that accounts for 1% of all
head and neck cancer (HNSC) and 10% of salivary gland malignancies
(1). Salivary glands are the most
common site of ACC, but tumours may also occur in the lacrimal
glands (2), breast (3), nasal cavity and paranasal sinus
(4) vulva (5) and skin (6). ACC occurring in the vulvar region
predominantly arises from the Bartholin's gland. Although
characterized by indolent growth, this malignancy commonly exhibits
perineural invasion (7). The age of
onset is between 18 and 90 years; however, no notable differences
have been identified between the sexes (8,9). At
present, the common treatment is surgery with or without
radiotherapy, as no approved systemic therapy exists. ACC of the
head and neck progresses relatively slowly and indolently,
resulting in 5- and 10-year patient survival rates of ~85 and 67%,
respectively (8). However,
long-term outcomes have revealed a decline in the 20-year overall
survival rate of ~20% (10), which
is primarily associated with nerve invasion, local control failure
and distant metastasis (11). Due
to the complexity of its clinical biological behaviour and the
challenges in clinical treatment, ACC requires further
investigation.
NDC80 kinetochore complex component (NUF2) is the
gene encoding the protein cell division cycle associated 1, which
serves a key role in ensuring proper chromosomal segregation and is
a key component of the NDC80/NUF2 complex (12,13).
Previous studies have reported that the expression level of the
NUF2 gene increases in various cancer tissues and it is closely
associated with tumorigenesis and progression, including
hepatocellular carcinoma, clear cell renal cell carcinoma, gastric
and breast cancer (13–17). It has been reported that in ovarian
cancer, lung adenocarcinoma and breast cancer, downregulating the
expression level of the NUF2 gene not only inhibits cell
proliferation and colony formation, but also promotes apoptosis
(18–20). These studies support the possible
role of NUF2 in tumorigenesis.
Collectively, the high recurrence rate (50%)
(21) and distant metastasis of ACC
underscore the need for novel molecular targets. The established
role of NUF2 in proliferation and migration positions it as a
plausible contributor to ACC aggressiveness. However, to the best
of our knowledge, no studies have investigated the function of NUF2
in ACC, leaving its therapeutic potential unexplored. Therefore,
the aim of the present study was to screen the hub genes
distinguishing ACC from normal tissues by the application of
bioinformatics techniques. The present study identified the key
role of NUF2 in ACC by analysing public datasets and further
validated these findings using functional experiments.
Materials and methods
Patients and samples
ACC tissues and paired adjacent normal tissue
samples were obtained from patients with ACC undergoing surgery at
Suzhou Hospital, Affiliated Hospital of Medical School, Nanjing
University (Suzhou, China) between October 2021 and December 2024
for immunohistochemistry (IHC) and western blotting analysis. The
inclusion criteria were as follows: i) Age, 18–80 years; ii)
postoperative pathological confirmation of adenoid cystic
carcinoma; and iii) availability of complete clinical data. The
exclusion criterion was a history of prior radiotherapy or
chemotherapy. Due to the low incidence of ACC at Suzhou Hospital,
Affiliated Hospital of Medical School, Nanjing University, only 2
patients met the inclusion criteria during the present study
period. The present study was approved by the Ethics Committee of
Suzhou Hospital, Affiliated Hospital of Medical School, Nanjing
University (approval no. IRB2020094; Suzhou, China). Included
patients signed a consent form that authorized the use of their
tissues in the present study.
ACC dataset
In the present study, datasets were obtained from
the GEO data repository (https://www.ncbi.nlm.nih.gov/geo/). The GSE88804
dataset contained 13 surgical samples of ACC with 7 normal samples
(22). The GSE153002 dataset was
composed of 30 ACC samples and 7 normal samples (23). The GSE36820 dataset included 11 ACC
samples and 3 normal samples (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36820).
The GSE88804 and GSE153002 datasets were merged and normalized
utilizing the ‘sva’ R package (version no. 3.58.0; https://www.bioconductor.org/packages/release/bioc/html/sva.html;
R Development Core Team; Bioconductor) and the normalized data were
used for subsequent study. The GSE36820 dataset was utilized for
verification.
Differentially expressed genes
(DEGs)
The R package ‘limma’ (version 3.58.1; https://bioinf.wehi.edu.au/limma/; R Development
Core Team; Bioconductor) was used for the merged dataset. The
fold-change (FC) was calculated using false discovery and the
Benjamini-Hochberg method was used to adjust the original P-values
(24). Based on adjusted P<0.05
and log2FC >1, the present study identified the DEGs
in the merged dataset with the ‘limma’ package. Subsequently, a
volcano plot and heatmap were generated to visualize the
results.
Protein-protein interaction (PPI)
network construction and hub gene exploration
The Search Tool for Retrieval of Interacting
Genes/Proteins (STRING) database (https://cn.string-db.org/cgi/input?sessionId=bXmYsv7CnUrH&input_page_active_form=multiple_identifiers)
displayed the relationships between proteins in the form of network
graphs. In the present study, the DEGs were uploaded to the STRING
website to construct a PPI network for the prediction of vital
genes with a confidence score >0.4 and the organism was set to
Homo sapiens. The network data were exported and imported to
Cytoscape software (version 3.9.0) for analysis and visualization
of the molecular interaction diagrams. Subsequently, ‘cytoHubba’, a
plugin tool in Cytoscape (25), was
used to separately identify the first hub genes based on three
topological analysis methods, including degree, maximum clique
centrality (MCC) and maximum network connectivity (MNC). The
intersection of these genes were subsequently visualized.
Expression and validation analysis of
NUF2
Tumour Immune Estimation Resource (TIMER) 2.0 is a
database that utilizes RNA-sequencing (RNA-seq) expression profile
data to analyse differential gene expression in tumours (http://timer.cistrome.org/). The differences in NUF2
expression between normal and cancer tissues in different cancer
types were determined using the TIMER 2.0 database (26). In the present study, NUF2 was
inputted into the ‘Gene_DE’ module (https://compbio.cn/timer2/) of the TIMER 2.0 database
and the difference in NUF2 expression between normal and cancer
tissues from different tumours in The Cancer Genome Atlas database
was analysed (27).
The present study used box plots to depict the
differential expression level of NUF2 in normal and tumour tissues
using the combined dataset, after which the present study validated
the differential expression level of NUF2 in the GSE36820
dataset.
Functional enrichment analysis
Based on the median expression levels of NUF2 in
tumour tissues, the two groups were divided into high- and
low-expression groups and the differences were analysed using the
‘limma’ package. LogFC >1 and adjusted P<0.05 were set as the
cut-off point for the DEGs. Gene Ontology (GO) enrichment analysis
of the DEGs was performed with the ‘ClusterProfiler’ (version
4.10.0; http://yulab-smu.top/biomedical-knowledge-mining-book/)
and ‘org.Hs.eg.db’ (version 3.22; http://bioconductor.org/packages/org.Hs.eg.db)
package. Subsequently, the results were visualized using a bubble
diagram.
Immune infiltration analysis
TIMER is a web server used to analyse the level of
immune infiltration in different cancer types. In the TIMER
database, specific algorithms were used to analyse the immune
infiltration level. In the present study, the relationship between
NUF2 and immune cells in HNSC was discussed.
To assess differences in immune cell infiltration,
single sample Gene Set Enrichment Analysis (ssGSEA) was performed
on RNA-seq data from the NUF2 high- and low-expression groups.
IHC
The tissue samples were fixed in 10% neutral
formalin at room temperature for 24 h. IHC staining was performed
on 4-µm-thick paraffin-embedded tissue sections. The sections were
deparaffinized in xylene and rehydrated using a graded ethanol
series to water. Antigen retrieval was carried out in citrate
buffer (pH 6.0) at 121°C for 120 sec using a pressure cooker. To
quench endogenous peroxidase activity, the sections were incubated
in 3% hydrogen peroxide solution protected from light for 25 min at
room temperature. Non-specific binding sites were blocked with
rabbit serum (concentrated; cat. no. G1209; Wuhan Servicebio
Technology Co., Ltd.) for 30 min at room temperature. The sections
were then incubated with primary antibody [rabbit polyclonal
antibodies against NUF2 (1:500; cat. no. 15731-1-AP; Proteintech
Group, Inc.)] overnight at 4°C in a humidified chamber.
Subsequently, the sections were incubated with HRP-conjugated goat
anti-rabbit secondary antibody (1:200; cat. no. GB23303; Wuhan
Servicebio Technology Co., Ltd.) at room temperature for 50 min.
Colour development was performed using a DAB substrate kit
according to the manufacturer's instructions (cat. no. G1212; Wuhan
Servicebio Technology Co., Ltd.) at room temperature, followed by
counterstaining with haematoxylin for ~3 min. Lastly, the stained
sections were imaged under a light microscope. Each section was
evaluated by two blinded, independent pathologists. Discrepancies
between the two pathologists were resolved by a joint re-evaluation
and discussion until a consensus was reached. The percentages of
positive tumour cells were as follows: i) 1–25%; ii) 26–50%; iii)
51–75%; and iv) 76–100%. The immunoreaction intensity was
classified as 0 (negative), 1 (weak), 2 (moderate), 3 (strong). The
final staining scores were calculated as the sum of the intensity
and positive rates scores and was graded as follows: 0 score,
negative (−); 1–3 scores, weakly positive (+); 4–5 scores, moderate
positive (++) or 6–7 scores, strongly positive (+++) (28).
Western blotting analysis
Proteins were isolated from human ACC tissues and
paired adjacent normal tissues using RIPA buffer (Beyotime
Institute of Biotechnology). The protein concentration was
determined by the BCA method. Subsequently, 30 µg of protein was
loaded onto an 10% SDS-PAGE gel and then transferred to PVDF
membranes. The PVDF membranes were blocked with 5% milk at room
temperature for 1 h and then incubated with the primary antibody
against NUF2 (1:800; cat. no. 15731-1-AP; Proteintech Group, Inc.)
and GAPDH (1:10,000; cat. no. 10494-1-AP; Proteintech Group, Inc.)
overnight at 4°C. After washing with TBS-0.1% Tween 20, the
membranes were incubated with secondary antibodies (HRP-conjugated
Goat Anti-Rabbit IgG; 1:5,000; cat. no. SA00001-2; Proteintech
Group, Inc.) for 2 h at room temperature. Lastly, the bands were
detected using an ECL substrate (Beyotime Institute of
Biotechnology) according to the manufacturer's instructions and an
Amersham™ Imager 680 system (Cytiva).
Cell culture
The human salivary ACC (SACC)-83 cell line (cat. No.
FH0798; http://www.fudancell.com/sys-pd/?pid=3836) was
purchased from Shanghai Fuheng Biotechnology Co., Ltd. The SACC-83
cell line was cultivated in RPMI 1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 1% penicillin/streptomycin and
10% FBS (cat. no. FH100-900; Shanghai Fuheng Biotechnology Co.,
Ltd) at 37°C in a 5% CO2 atmosphere.
Small interfering RNA (siRNA) and
transfection
siRNAs targeting human the NUF2 gene and negative
control siRNA (si-NC) were purchased from Guangzhou RiboBio Co.,
Ltd. SACC-83 cells were transfected with si-NC (cat. no.
siN0000001-1-5) or siNUF2 (Table
SI) using Lipofectamine® 3000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
cells were seeded into 6-well plates at a density of
2×105 cells/well before transfection and cultured until
they reached a confluency of 60–70%. Transfected cells were
incubated at 37°C for 72 h and then were collected for Cell
Counting Kit-8 (CCK-8) assay and wound healing assay.
CCK-8
After 72 h target gene suppression, the cells were
seeded in 96-well plates at a density of 1.5×103
cells/well. A control group cultured with medium alone was
established in parallel. At 24, 48, 72 and 96 h, 10 µl of CCK-8
solution (MedChemExpress) was added to each well under
light-protected conditions. After a 2 h incubation, the absorbance
at 450 nm was measured with a spectrophotometer (Tecan
Biotechnology).
Wound healing assay
SACC-83 cells transfected with NUF2 siRNA were
seeded in 6-well plates at a density of 1.5×105
cells/well and cultured overnight to reach 80–90% confluence.
Linear wounds were generated using a 1,000 µl pipette tip held
orthogonal to the plate surface. After the cells were washed with
post-scratch PBS, cellular debris was removed and 2 ml of
serum-free RPMI-1640 was added to mitigate the effects on
proliferation. Cell migration was observed under an inverted
microscope at 0 and 24 h post-culture. Quantification was conducted
using ImageJ software. The wound area at 0 and 24 h was first
measured, after which the wound closure rate was calculated. Wound
closure was defined as the percentage reduction in wound area at 24
h relative to the initial wound area at 0 h. Each group included
three independent biological replicates, and all data were
presented as mean ± SD.
Statistical analysis
To evaluate immune infiltrates, TIMER 2.0 employs
partial Spearman's correlation, controlling for tumour purity, to
assess the association between the estimated levels of immune
infiltration and NUF2 expression. Data obtained from the CCK-8
assay were analysed using one-way ANOVA, followed by the Bonferroni
post-hoc test. The wound healing assay was analysed using an
unpaired t-test. All statistical tests were two-tailed. CCK-8 and
wound healing assay were performed in triplicate. The efficiency of
NUF2 knockdown by each specific siRNA (si-1, si-2, and si-3) was
evaluated via Western blot. Quantitative data were compared to the
negative control group (si-NC) using one-way ANOVA, followed by the
Bonferroni post-hoc test. Statistical analyses were performed with
GraphPad Prism (version 10; Dotmatics). For comparisons between two
groups of independent samples, such as tumour samples vs. normal
control samples from different individuals in the GEO datasets, the
Mann-Whitney U test was used. For comparisons involving paired
samples with sufficient sample size (n≥5), the Wilcoxon signed-rank
test was employed. P<0.05 was considered to indicate a
statistically significant difference.
Results
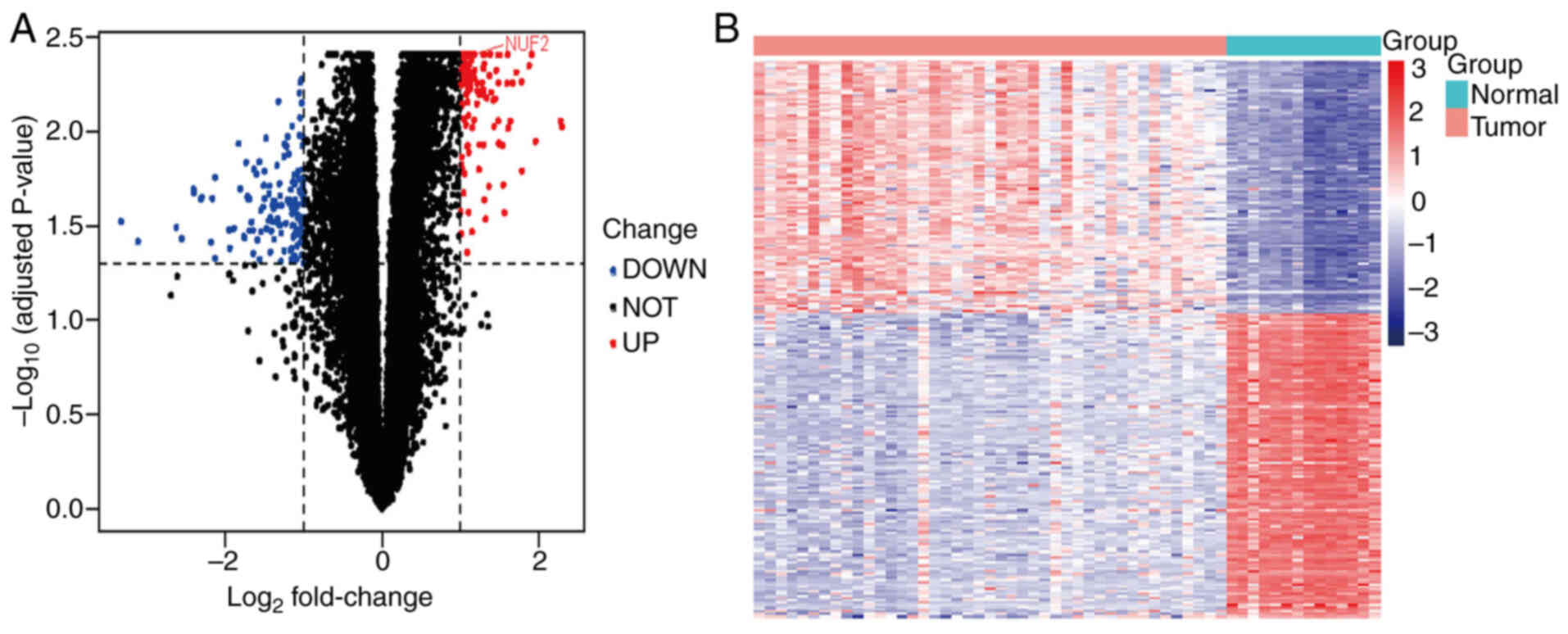
Screening of DEGs
In the present study, three gene expression
datasets, GSE88804, GSE153002 and GSE36820, were downloaded from
the GEO database. A total of 54 ACC with 17 normal salivary gland
tissues data were obtained. In the merged GSE88804 and GSE153002
datasets, the present study applied the rectified data for
differential expression analysis and obtained 248 DEGs, including
113 upregulated genes and 135 downregulated genes (Fig. 1A and B).
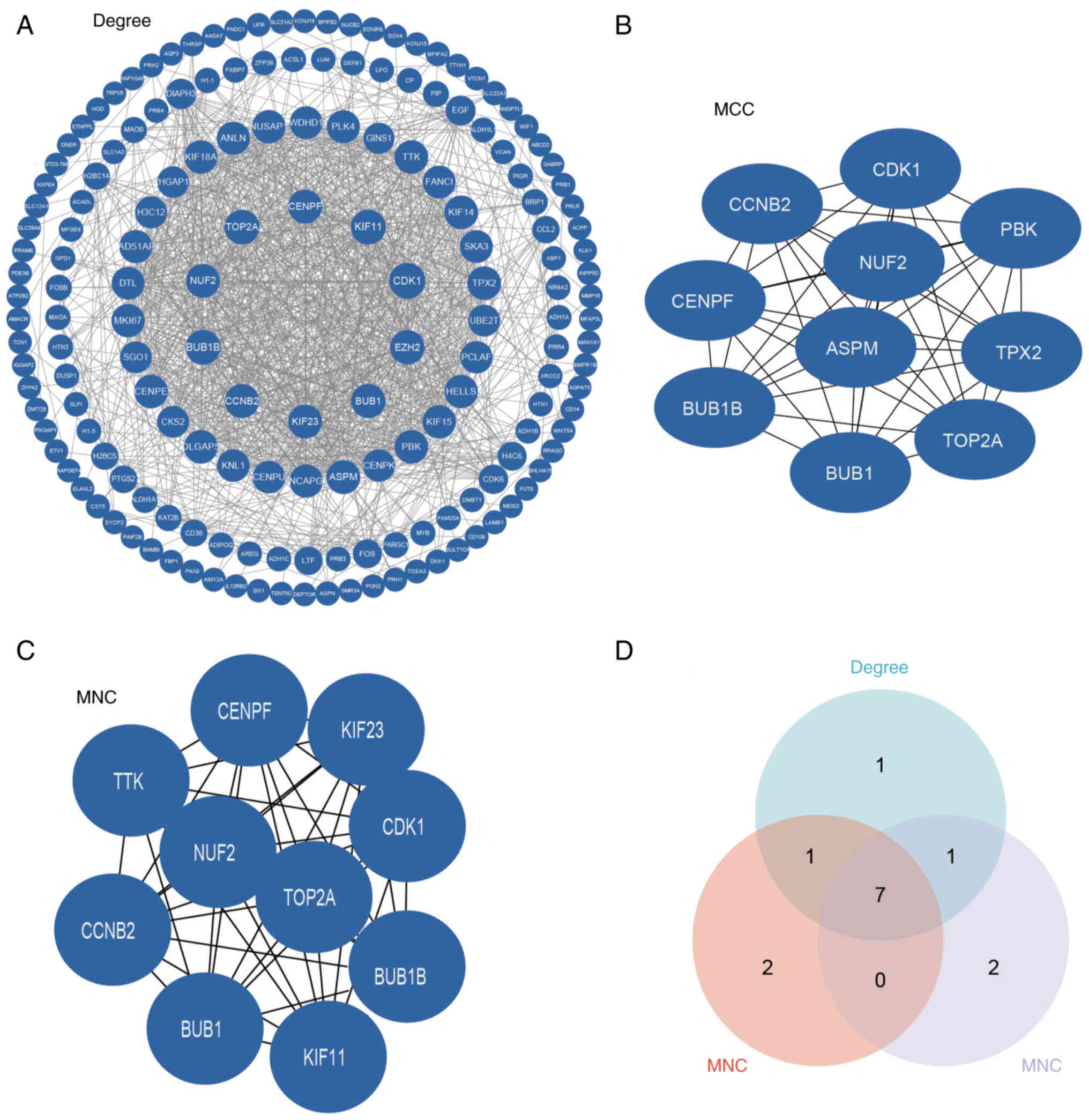
PPI network and module analysis
To identify functionally pivotal interactions, an
established DEG-associated PPI network was constructed using the
STRING database and analysed using Cytoscape software. Based on
network degree centrality analysis, the present study identified
the top 10 hub genes with the highest connectivity for further
investigation (Fig. 2A).
Subsequently, the present study implemented the MCC and MNC
algorithms to enhance the robustness of hub gene identification.
The top 10 highest-scoring genes from each method were obtained
(Fig. 2B and C). To identify key
genes, the present study first selected the top 10 candidates from
each method (degree, MCC and MNC) and then performed intersection
analysis to identify seven high-confidence hub genes (CDK1, budding
uninhibited by benzimidazoles 1 mitotic checkpoint serine/threonine
kinase B, DNA topoisomerase II α, cyclin B2, NUF2, budding
uninhibited by benzimidazoles 1 and centromere protein F; Fig. 2D). The role of NUF2 in ACC remains
unexplored; a PubMed (https://pubmed.ncbi.nlm.nih.gov/) search using the
keywords ‘NUF2’ and ‘adenoid cystic carcinoma’ yielded no relevant
publications.
Expression and verification of
NUF2
Analysis of the TIMER database using the ‘Gene_DE’
module demonstrated that NUF2 was significantly highly upregulated
in a variety of tumours compared with normal tissues, such as liver
hepatocellular carcinoma, oesophageal carcinoma, stomach
adenocarcinoma, glioblastoma multiforme, breast invasive carcinoma
and colon adenocarcinoma (P<0.001; Fig. 3A). In the combined and standardized
datasets (GSE88804 and GSE153002), the expression levels of NUF2
were also significantly higher in tumour tissue samples compared
with normal tissue samples (P<0.001; Fig. 3B). Similarly, NUF2 expression was
significantly upregulated in ACC within the GSE36820 validation set
(P<0.01; Fig. 3C). These results
were also supported by western blotting analysis, which revealed a
marked increase in NUF2 protein levels in ACC tissues (Fig. 3D). IHC analysis of a preliminary set
of paired samples (n=2) showed a trend of higher NUF2 expression in
ACC tissues (IHC scores, 6 and 6) compared with their matched
adjacent tissues (IHC scores, 3 and 4) (Fig. 3E).
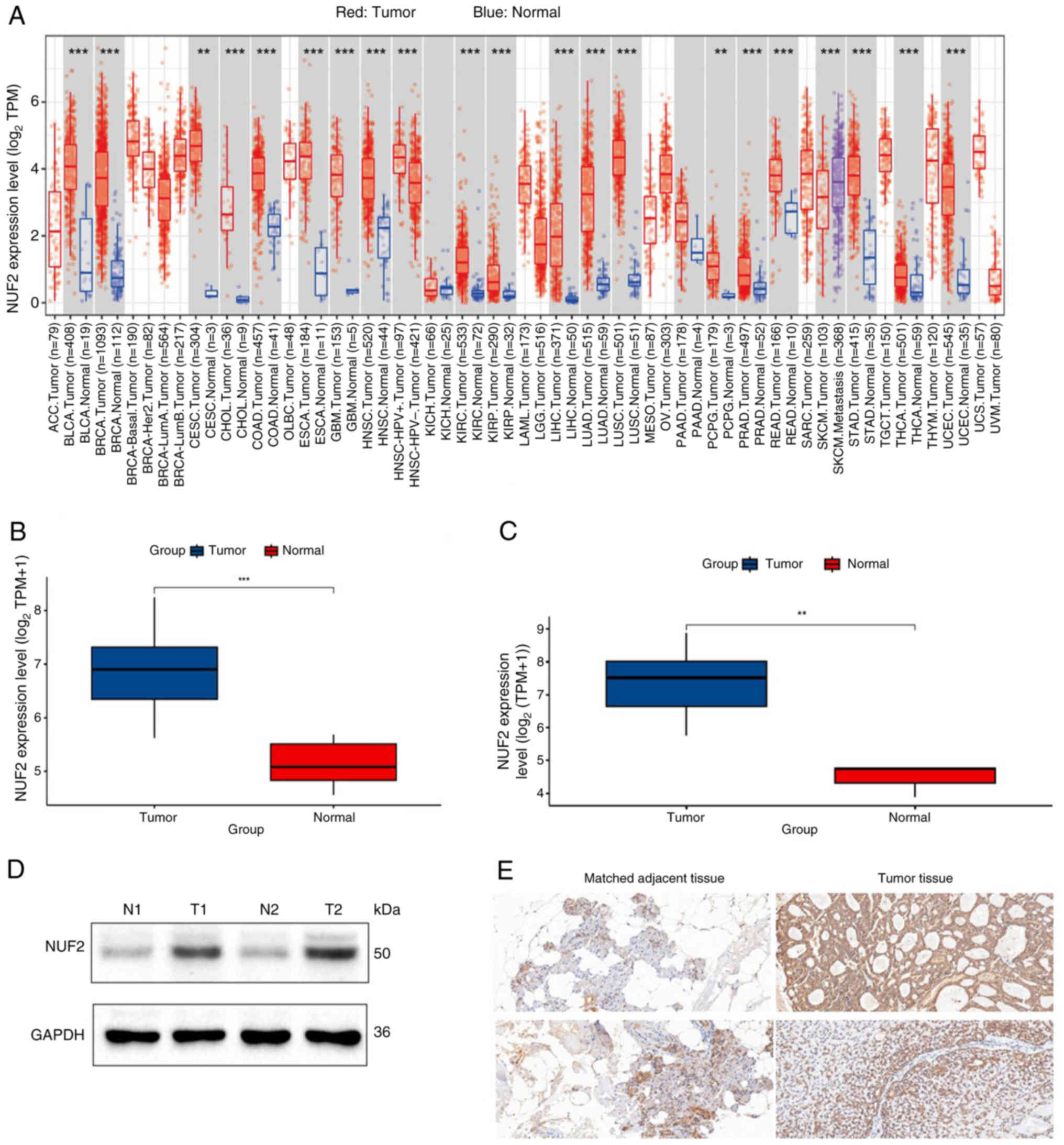 | Figure 3.Expression level of NUF2 and
immunohistochemistry. (A) Expression levels of NUF2 were analysed
with the TIMER database. (B) Expression levels of NUF2 were
verified with a merged dataset (GSE88804 and GSE153002). (C)
Expression levels of NUF2 were verified by the GSE36820 dataset.
(D) Expression levels of NUF2 in two ACC tissues and adjacent
normal tissues assessed using western blotting. (E)
Immunohistochemical staining results demonstrated that both tumour
tissue samples received a final score of 6, indicating strong NUF2
expression. By contrast, the matched adjacent tissues demonstrated
lower expression levels, with the upper adjacent tissue scoring 3
and the lower adjacent tissue scoring 4. Magnification, ×200.
**P<0.01 and ***P<0.001. siRNA, small interfering RNA; NUF2,
NDC80 kinetochore complex component; TIMER, Tumour Immune
Estimation Resource; GSE, Gene Set Enrichment; ACC, adenoid cystic
carcinoma; TPM, transcripts per million; N, normal; T, tumour. |
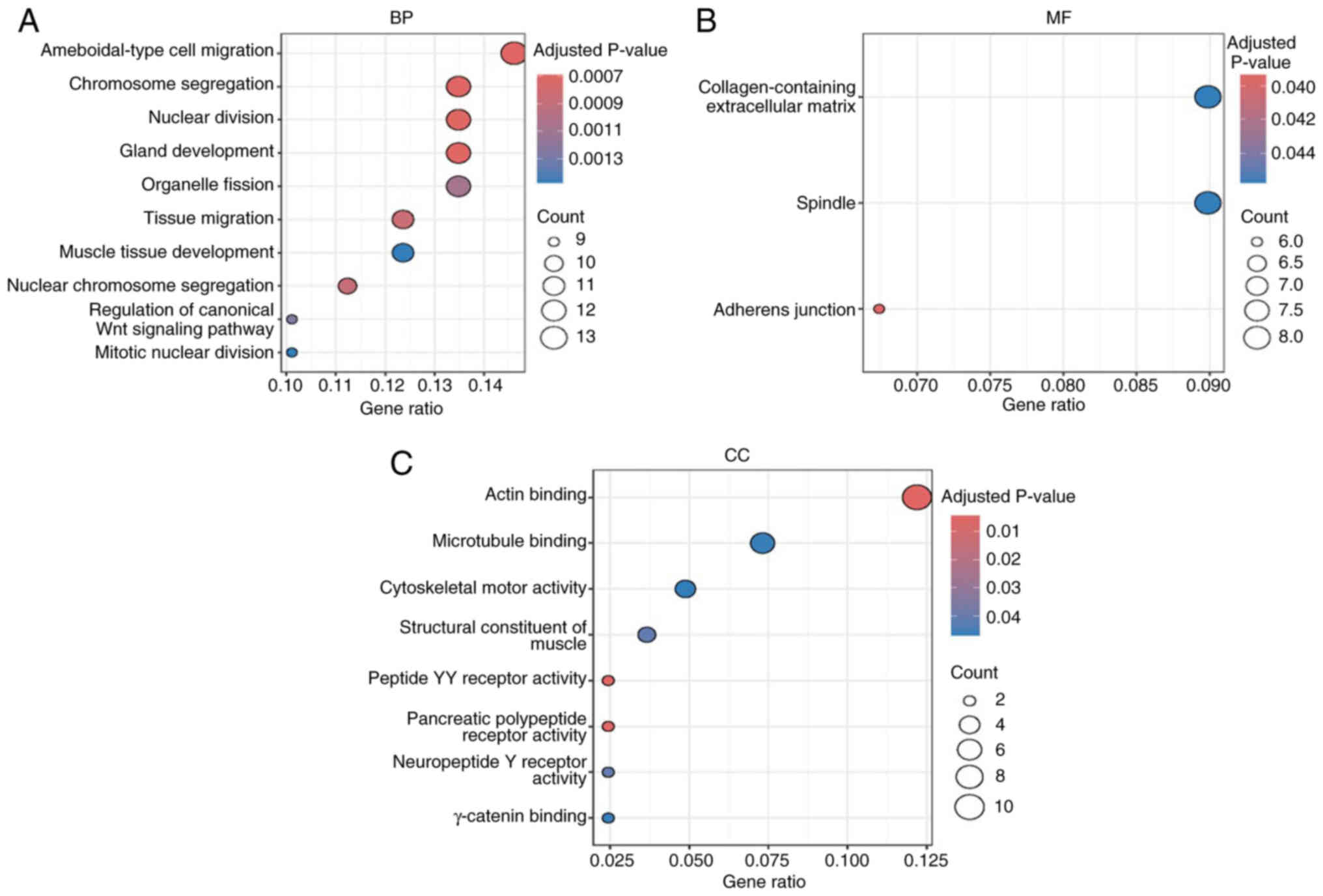
GO enrichment analysis of DEGs
In the present study, NUF2-related genes were
significantly enriched in ‘ameboidal-type cell migration’,
‘chromosome segregation’, ‘nuclear division’ (P<0.01) in
biological processes and ‘collagen-containing extracellular matrix’
(P<0.05) in molecular functions and ‘actin binding’ (P<0.05)
in cellular components (Fig. 4A-C),
suggesting its potential role in ACC cell metastasis and
proliferation.
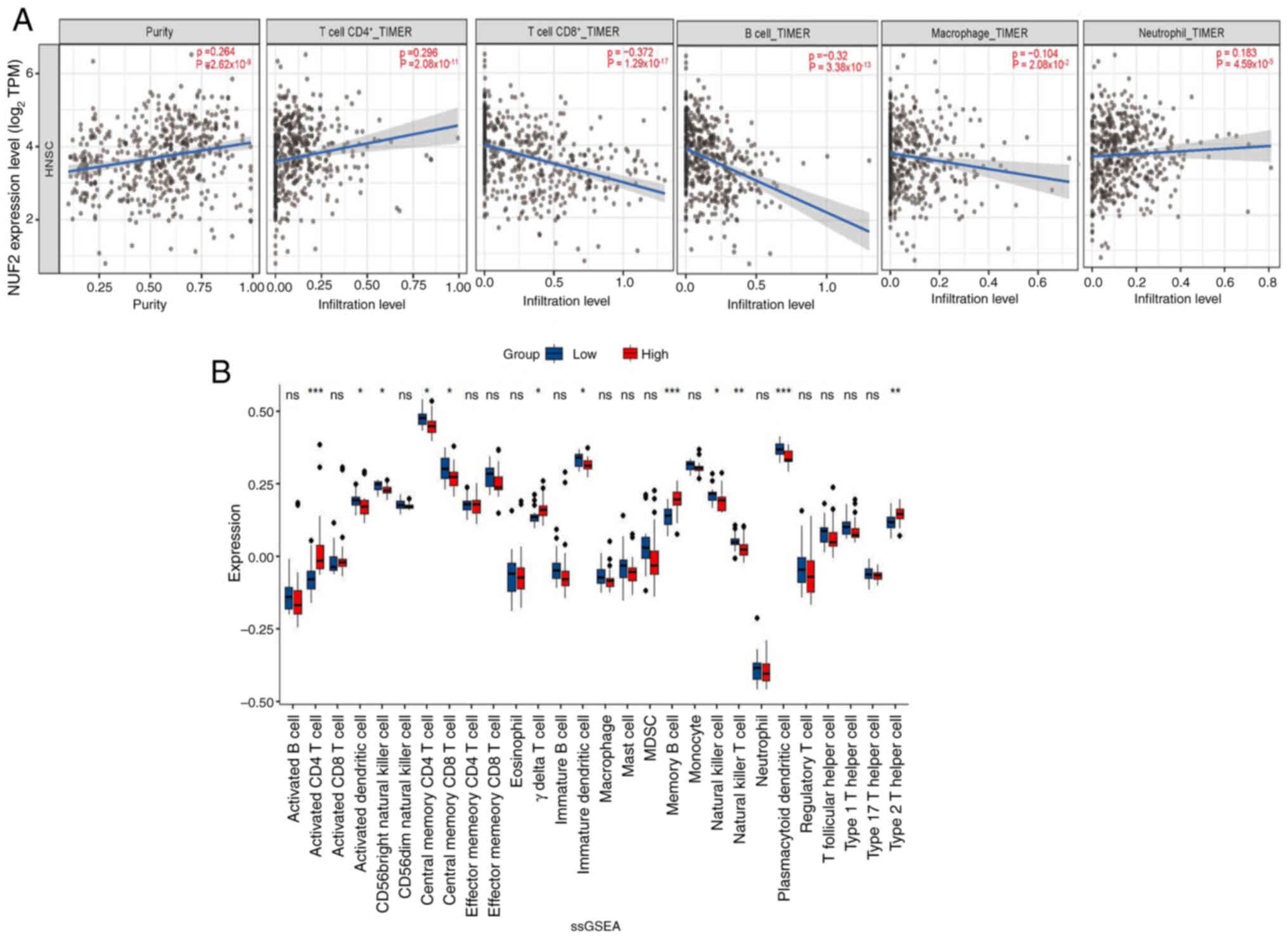
Correlations between immune cells and
the expression levels of NUF2 in ACC
To analyse the relationship between NUF2 expression
and the state of tumour-infiltrating immune cells, the TIMER
database was used to assess the association in HNSC tissues. The
present study identified that NUF2 expression showed a significant
but weak positive association with the infiltration levels of
CD4+ T cells (ρ=0.296; P=2.08×10−11),
neutrophils (ρ=0.183; P=4.59×10−05). By contrast, NUF2
expression was negatively correlated with the infiltration of
CD8+ T cells (ρ=−0.372; P=1.29×10−17) and B
cells (ρ=−0.32; P=3.38×10−13), with the correlation to
CD8+ T cells being moderate. Only a very weak negative association
was observed between NUF2 expression and macrophage infiltration
(ρ=−0.104; P=2.08×10−02; Fig. 5A).
Subsequently, ssGSEA was used to evaluate the
infiltration of 28 immune cells in each patient with ACC. The
results demonstrated that in the group with higher NUF2, activated
CD4+ T cells, γΔT cells, memory B cells and type 2 T
helper cells were significantly more abundant in infiltration
(P<0.05). The group with lower NUF2 expression demonstrated
significantly increased infiltration (P<0.05) of activated
dendritic cells, CD56 bright natural killer cells, central memory
CD4+ T cells, central memory CD8+ T cells,
immature dendritic cells, natural killer cells, natural killer T
cells and plasmacytoid dendritic cells (Fig. 5B).
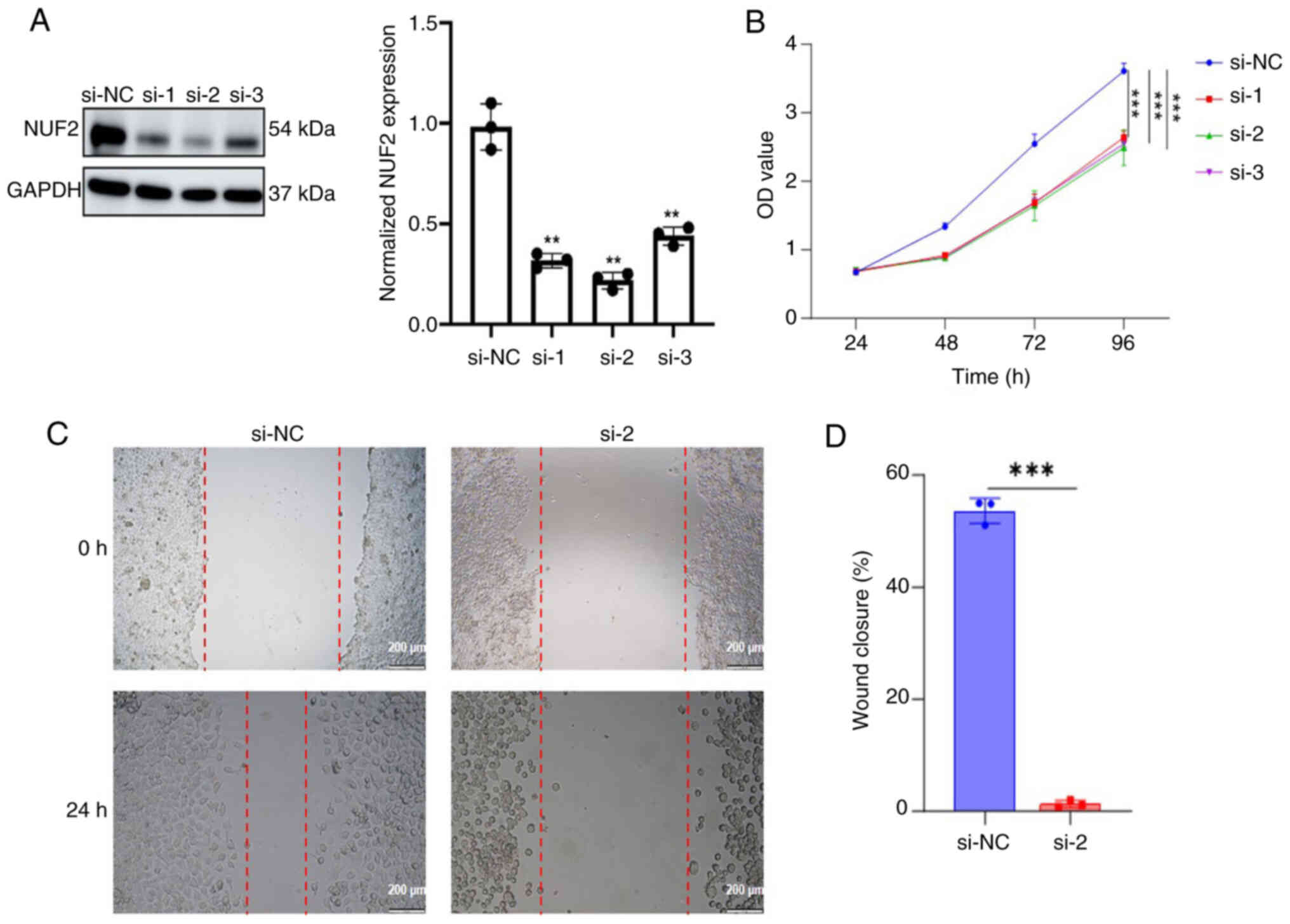
NUF2 promotes ACC cell proliferation
and migration in vitro
To assess the biological effect of NUF2 in ACC
cells, SACC-83 cells were transfected with siRNAs to silence NUF2
compared with the si-NC group. Western blotting was subsequently
used to evaluate the transfection efficiency. The siRNA with the
highest silencing efficiency, si-2, was selected for subsequent
functional studies (Fig. 6A).
In the present study, a CCK-8 assay was performed in
ACC cells. The downregulation of NUF2 expression significantly
inhibited (P<0.0001) the proliferation of ACC cells (Fig. 6B). Subsequently, the present study
explored the role of NUF2 in the migration of ACC cells using a
wound healing assay. The results revealed that knockdown of NUF2
significantly diminished (P=0.0004) the migration capacity of ACC
cells (Fig. 6C and D).
Discussion
As a result of the high risk of local recurrence and
delayed distant metastases, the treatment outcomes for ACC remain
suboptimal (29). Further research
on the molecular mechanisms of ACC may offer potentially effective
therapeutic strategies or promising biomarkers in the future.
However, the sensitivity and specificity of these methods are
limited. Increasing evidence suggests that NUF2 serves key roles in
the development and progression of several tumours, such as ovarian
cancer, multiple myeloma and hepatocellular carcinoma (20,30,31).
However, the role of NUF2 in ACC remains to be elucidated and
further research is warranted.
In the present study, the GEO database, western
blotting and IHC analysis were used for gene expression analysis.
The high expression level of NUF2 in tumour tissues and its low
expression in normal tissues are consistent with previous findings
(14,15,30).
Furthermore, several studies have reported that NUF2 upregulation
is associated with clinical stage and poor prognosis in
adrenocortical cancer, kidney renal clear cell carcinoma, kidney
renal papillary cell carcinoma and multiple myeloma (30,31).
Fundamental research and clinical trials are needed to further
validate NUF2 as a therapeutic target in ACC.
Several studies have reported that targeted
regulation of NUF2 can inhibit cancer cell proliferation and
migration. Liu et al (32)
reported that silencing NUF2 expression could slow cell
proliferation in human hepatocellular carcinomas. Furthermore,
downregulation of NUF2 expression reduced the migratory ability of
lung adenocarcinoma cells (18),
which is consistent with the present study finding. In the present
study, the GO analysis results revealed that NUF2 is potentially
involved in regulating both proliferation and migration processes
in ACC.
Malignant tumours are composed of tumour cells,
immune cells and non-immune cells. Immune and non-immune cells
constitute the tumour microenvironment (TME), which includes
components such as fibroblasts, nerves, immune cells and various
cytokines and vascular systems (33). Increasing research has highlighted
the key role of the TME in the genesis and progression of several
malignancies such as laryngeal squamous cell carcinoma, brain
metastasis (33–36) and liver cancer (37). Based on the status and distribution
of immune cells, tumours are categorized into three immune
phenotypes, including immune-desert (‘cold’ tumours),
immune-exclusive and immune inflammatory (‘hot’ tumours) (38). ‘Hot’ tumours are characterized by
abundant T-cell infiltration and exhibit good response to immune
checkpoint inhibitors (39).
Immunotherapy for numerous types of cancer has made notable
progress. Immunotherapy is now a main therapeutic choice for
advanced melanoma, renal cell carcinoma and renal cell carcinoma
and is regarded as a promising method for cancer therapy (40–42).
However, the expression levels of programmed death-ligand 1,
cytotoxic T-cell antigen 4 and programmed death receptor 1 are low
in the environment of ACC (43).
Furthermore, several studies on the use of immunotherapy in ACC
have reported unsatisfactory results (44,45).
Li et al (46) identified
that NUF2 is associated with the immune infiltration of certain
immune cells, such as CD8+ T Cells, B cells, natural
killer cells and neutrophils. and has a major function in the
regulation of cancer immunology. The present study results
regarding the immune microenvironment suggested that elevated NUF2
expression in ACC is correlated with the infiltration of activated
CD4+ T cells, memory B cells and type 2 T helper cells.
Further characterization indicated that high NUF2 expression was
associated with increased infiltration of activated CD4+
T cells, memory B cells and type 2 T helper cells, but decreased
infiltration of plasmacytoid dendritic cells, activated dendritic
cells and natural killer T cells. These findings suggest that the
expression levels of NUF2 may influence the recruitment or activity
of immune cells within the TME.
The present study had certain limitations. First, a
notable limitation to consider is that HNSC and salivary gland
cancer are distinct pathological entities. Consequently,
extrapolating immune-related findings from this HNSCC cohort to ACC
should be interpreted with caution. Second, the observed
correlation between NUF2 expression and immune cell infiltration
was inferred solely using in silico analysis using the
ssGSEA algorithm. Immune correlations derived from bulk RNA-seq
data can be susceptible to confounding factors such as tumour
purity and stromal contamination (47). Additionally, the correlation
analyses between NUF2 and immune cells derived from the TIMER
database, particularly those involving CD4+ T cells,
neutrophils and macrophages, demonstrated weak associations, and
their biological relevance remains to be determined. Third, the
present study had a limited sample size. Therefore, to overcome
these constraints, the collection of additional ACC specimens and
the use of multiplex IHC coupled with flow cytometry in future
studies may be used to validate the present study findings.
In summary, to the best of our knowledge, the
present study provides the first analysis of the expression pattern
of NUF2 in ACC. The present study results demonstrated that NUF2 is
upregulated in ACC compared with that of adjacent tissues and NUF2
expression is correlated with immune cell infiltration. These
findings may be potentially translated into effective clinical
interventions in the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XY and LL conceived the present study and provided
financial support. SH, YL, JZ, JH and HC performed the experiments,
collected the data and prepared the figures. XY and LL confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The collection of tumour samples and patient
information was approved by the Ethics Committee of Suzhou
Hospital, Affiliated Hospital of Medical School, Nanjing University
(approval no. IRB2020094; Suzhou, China). All patients signed
informed consent for the retention and analysis of their tissues
for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Coca-Pelaz A, Rodrigo JP, Bradley PJ,
Vander Poorten V, Triantafyllou A, Hunt JL, Strojan P, Rinaldo A,
Haigentz M Jr, Takes RP, et al: Adenoid cystic carcinoma of the
head and neck-An update. Oral Oncol. 51:652–661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duarte AF, Alpuim Costa D, Cacador N,
Boavida AM, Afonso AM, Vilares M and Devoto M: Adenoid cystic
carcinoma of the palpebral lobe of the lacrimal gland-case report
and literature review. Orbit. 41:605–610. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gou WB, Yang YQ, Song BW and He P: Solid
basal adenoid cystic carcinoma of the breast: A case report and
literature review. Medicine (Baltimore). 103:e370102024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee TH, Kim K, Oh D, Yang K, Jeong HS,
Chung MK and Ahn YC: Clinical outcomes in adenoid cystic carcinoma
of the nasal cavity and paranasal sinus: A comparative analysis of
treatment modalities. Cancers (Basel). 16:12352024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gural Z, Yucel S and Agaoglu F:
Bartholin's gland adenoid cystic carcinoma: Report of three cases
and the review of literature. Indian J Cancer. 61:346–349. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schembri-Wismayer D, Gupta S and Erickson
LA: Cutaneous adenoid cystic carcinoma. Mayo Clin Proc.
99:1017–1018. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baker GM, Selim MA and Hoang MP: Vulvar
adnexal lesions: A 32-year, single-institution review from
Massachusetts General Hospital. Arch Pathol Lab Med. 137:1237–1246.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Atallah S, Casiraghi O, Fakhry N, Wassef
M, Uro-Coste E, Espitalier F, Sudaka A, Kaminsky MC, Dakpe S, Digue
L, et al: A prospective multicentre REFCOR study of 470 cases of
head and neck Adenoid cystic carcinoma: Epidemiology and prognostic
factors. Eur J Cancer. 130:241–249. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Morais EF, da Silva LP, Moreira DGL,
Mafra RP, Rolim LSA, de Moura Santos E, de Souza LB and de Almeida
Freitas R: Prognostic factors and survival in adenoid cystic
carcinoma of the head and neck: A retrospective clinical and
histopathological analysis of patients seen at a cancer center.
Head Neck Pathol. 15:416–424. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dodd RL and Slevin NJ: Salivary gland
adenoid cystic carcinoma: A review of chemotherapy and molecular
therapies. Oral Oncol. 42:759–769. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ellington CL, Goodman M, Kono SA, Grist W,
Wadsworth T, Chen AY, Owonikoko T, Ramalingam S, Shin DM, Khuri FR,
et al: Adenoid cystic carcinoma of the head and neck: Incidence and
survival trends based on 1973–2007 surveillance, epidemiology, and
end results data. Cancer. 118:4444–4451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nabetani A, Koujin T, Tsutsumi C,
Haraguchi T and Hiraoka Y: A conserved protein, Nuf2, is implicated
in connecting the centromere to the spindle during chromosome
segregation: A link between the kinetochore function and the
spindle checkpoint. Chromosoma. 110:322–334. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie X, Jiang S and Li X: Nuf2 is a
prognostic-related biomarker and correlated with immune infiltrates
in hepatocellular carcinoma. Front Oncol. 11:6213732021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Wang Y, Wang J, Jiang W, Chen Y,
Shan J, Li X and Wu X: NUF2 regulated the progression of
hepatocellular carcinoma through modulating the PI3K/AKT pathway
via stabilizing ERBB3. Transl Oncol. 44:1019332024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng B, Wang S, Yuan X, Zhang J, Shen Z
and Ge C: NUF2 is correlated with a poor prognosis and immune
infiltration in clear cell renal cell carcinoma. BMC Urol.
23:822023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu X, Zou Y, Wu T, Ni J, Tan Q, Wang Q
and Zhang M: ANP32E contributes to gastric cancer progression via
NUF2 upregulation. Mol Med Rep. 26:2752022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhai X, Yang Z, Liu X, Dong Z and Zhou D:
Identification of NUF2 and FAM83D as potential biomarkers in
triple-negative breast cancer. PeerJ. 8:e99752020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang F, Huang X, Yang X, Zhou H and Wang
Y: NUF2 expression promotes lung adenocarcinoma progression and is
associated with poor prognosis. Front Oncol. 12:7959712022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lv S, Xu W, Zhang Y, Zhang J and Dong X:
NUF2 as an anticancer therapeutic target and prognostic factor in
breast cancer. Int J Oncol. 57:1358–1367. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ren M, Zhao H, Gao Y, Chen Q, Zhao X and
Yue W: NUF2 promotes tumorigenesis by interacting with HNRNPA2B1
via PI3K/AKT/mTOR pathway in ovarian cancer. J Ovarian Res.
16:172023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuce Sari S, Yazici G, Elmali A, Bayatfard
P, Koc I, Kiratli H and Cengiz M: Radiotherapy for adenoid cystic
carcinoma of the lacrimal gland: Study on twelve patients. Cancer
Radiother. 29:1046442025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andersson MK, Afshari MK, Andren Y, Wick
MJ and Stenman G: Targeting the oncogenic transcriptional regulator
MYB in adenoid cystic carcinoma by inhibition of IGF1R/AKT
signaling. J Natl Cancer Inst. 109:djx0172017. View Article : Google Scholar
|
|
23
|
Persson M, Andersson MK, Sahlin PE, Mitani
Y, Brandwein-Weber MS, Frierson HF Jr, Moskaluk C, Fonseca I,
Ferrarotto R, Boecker W, et al: Comprehensive molecular
characterization of adenoid cystic carcinoma reveals tumor
suppressors as novel drivers and prognostic biomarkers. J Pathol.
261:256–268. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hochberg Y and Hochberg Y: Controlling the
false discovery rate a practical and powerful approach to multiple
testing. J Royal Stat Soc Series B. 57:289–300. 1995. View Article : Google Scholar
|
|
25
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hutter C and Zenklusen JC: The cancer
genome atlas: Creating lasting value beyond its data. Cell.
173:283–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin J, Chen X, Yu H, Min S, Chen Y, Li Z
and Xie X: NUF2 drives clear cell renal cell carcinoma by
activating HMGA2 transcription through KDM2A-mediated H3K36me2
demethylation. Int J Biol Sci. 18:3621–3635. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stawarz K, Durzynska M, Galazka A,
Gorzelnik A, Zwolinski J, Paszkowska M, Bieńkowska-Pluta K and
Misiak-Galazka M: Current landscape and future directions of
therapeutic approaches for adenoid cystic carcinoma of the salivary
glands (Review). Oncol Lett. 29:1532025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Y, Chen L, Luo Y, Shen J and Zhou S:
Predictive value of NUF2 for prognosis and immunotherapy responses
in pan-cancer. Nan Fang Yi Ke Da Xue Xue Bao. 45:137–149. 2025.(In
Chinese). PubMed/NCBI
|
|
31
|
Zhang S, Zhang L, Cai L, Chen H, Wang Y,
Yuan Y, Zhang H and Wei X: NUF2 overexpression predicts poor
outcomes in multiple myeloma. Genes Dis. 12:1012682025. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Q, Dai SJ, Li H, Dong L and Peng YP:
Silencing of NUF2 inhibits tumor growth and induces apoptosis in
human hepatocellular carcinomas. Asian Pac J Cancer Prev.
15:8623–8629. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Elhanani O, Ben-Uri R and Keren L: Spatial
profiling technologies illuminate the tumor microenvironment.
Cancer Cell. 41:404–420. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rodrigo JP, Sanchez-Canteli M, Lopez F,
Wolf GT, Hernández-Prera JC, Williams MD, Willems SM, Franchi A,
Coca-Pelaz A and Ferlito A: Tumor-infiltrating lymphocytes in the
tumor microenvironment of laryngeal squamous cell carcinoma:
Systematic review and meta-analysis. Biomedicines. 9:4862021.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
de Visser KE and Joyce JA: The evolving
tumor microenvironment: From cancer initiation to metastatic
outgrowth. Cancer Cell. 41:374–403. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Affo S, Yu LX and Schwabe RF: The role of
cancer-associated fibroblasts and fibrosis in liver cancer. Annu
Rev Pathol. 12:153–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen DS and Mellman I: Elements of cancer
immunity and the cancer-immune set point. Nature. 541:321–330.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu Q, Hu J, Wang Y and Wang Z: The role of
tumor types in immune-related adverse events. Clin Transl Oncol.
27:3247–3260. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garg P, Pareek S, Kulkarni P, Horne D,
Salgia R and Singhal SS: Next-Generation immunotherapy: Advancing
clinical applications in cancer treatment. J Clin Med. 13:65372024.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu B, Zhou H, Tan L, Siu KTH and Guan XY:
Exploring treatment options in cancer: Tumor treatment strategies.
Signal Transduct Target Ther. 9:1752024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Waldman AD, Fritz JM and Lenardo MJ: A
guide to cancer immunotherapy: From T cell basic science to
clinical practice. Nat Rev Immunol. 20:651–668. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wolkow N, Jakobiec FA, Afrogheh AH, Kidd
M, Eagle RC, Pai SI and Faquin WC: PD-L1 and PD-L2 expression
levels are low in primary and secondary adenoid cystic carcinomas
of the orbit: Therapeutic implications. Ophthalmic Plast Reconstr
Surg. 36:444–450. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mosconi C, de Arruda JAA, de Farias ACR,
Oliveira GAQ, de Paula HM, Fonseca FP, Mesquita RA, Silva TA,
Mendonça EF and Batista AC: Immune microenvironment and evasion
mechanisms in adenoid cystic carcinomas of salivary glands. Oral
Oncol. 88:95–101. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sato R, Yamaki H, Komatsuda H, Wakisaka R,
Inoue T, Kumai T and Takahara M: Exploring immunological effects
and novel immune adjuvants in immunotherapy for salivary gland
cancers. Cancers (Basel). 16:12052024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li X, Zhang L, Yi Z, Zhou J, Song W, Zhao
P, Wu J, Song J and Ni Q: NUF2 is a potential immunological and
prognostic marker for non-small-cell lung cancer. J Immunol Res.
2022:11619312022.PubMed/NCBI
|
|
47
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|