Introduction
Cholangiocarcinoma (CCA) is a malignancy of bile
duct epithelia and the most common liver cancer in Northeast
Thailand. CCA is responsible for 71% of liver cancer in Khon Kaen,
representing the highest incidence of CCA in the world (1). Epidemiological and animal studies
showed the association of CCA development with liver fluke
(Opisthorchis viverrini) infection and carcinogenic
nitrosamine (1–4). The survival rate of CCA patients is
low because most patients are diagnosed at late stage of the
disease. Prognosis of CCA is poor and the majority of patients die
within 6–12 months after diagnosis. The relative survival of liver
cancer in Khon Kaen is 23.2% for one year, 11.1% for three years,
and 8.4% for five years (5).
Although the molecular mechanisms for the development of liver
fluke-associated CCA have been widely studied in the past decade,
the clearer image of how the studied genes mediate carcinogenesis
and pathogenesis of this disease remains obscure (6,7).
The trefoil factor (TFF) is a cluster of gene
located on chromosome 21q22.3 encoding polypeptides TFF1, TFF2, and
TFF3. TFF proteins are well known for their potent protective and
healing effects termed ‘restitution’ after mucosal damage in the
gastrointestinal tract. To achieve their role in epithelial
restitution, TFFs protect cells from apoptotic death and
stimulating cell migration (8,9). The
deregulation of these functions contributes to TFF-mediated cancer
development and progression (10–12).
Our previous study showed that the expression of TFF2 mRNA
in CCA was significantly higher than in normal tissues (13). Moreover, TFF2 positive
immunostaining in CCA was markedly increased compared with normal
and precancerous tissues suggesting its important role in tumor
progression (13). In addition, we
also demonstrated that TFF2 activates proliferation of CCA cells
via epidermal growth factor receptor (EGFR) and mitogen-activated
protein kinase (MAPK) signaling (13). While TFF2 and its role in CCA have
been defined, we raised the question whether there are splice
variants of TFF2 existed and whether these isoforms have
adversely affected on patient prognosis and tumor progression since
some tumors sustain their malignant behavior by aberrant expression
of splice variants (14). Likewise,
the presence of splice variants of many genes i.e. p53 gene
family has been shown to be associated with poor patient outcomes
(15,16).
In this study, we investigated the occurrence of a
novel alternatively spliced variant of the TFF2 gene in CCA
and quantified the splicing isoform TFF2 in CCA and normal
adjacent tissues. The ratio of splicing isoform and wild-type
TFF2 was analyzed for associations with clinical parameters.
In addition, correlation between expression of alternatively
spliced variant of TFF2 mRNA and wild-type TFF2 protein
expression was also tested.
Materials and methods
Cholangiocarcinoma (CCA) samples
Six CAA cell lines including KKU-OCA17, KKU-100,
KKU-M055, KKU-M139, KKU-M156 and KKU-M213 kindly provided by the
Liver Fluke and Cholangiocarcinoma Research Center (LFCRC), Faculty
of Medicine, Khon Kaen University were cultured in HAM-F12
supplemented with 10% fetal bovine serum (FBS). The cells were
maintained at 37°C in a humidified atmosphere with 5%
CO2 and 80–100% confluence of cell lines were
subpassaged or harvested by trypsinization.
Seventy-eight samples were obtained from resected
tissues of CCA patients undergoing surgery at Srinagarind Hospital,
Faculty of Medicine, Khon Kaen University. Signed informed consents
were received from all patients participating in this research
project. This study was approved by the Khon Kaen University Ethics
Committee for Human Research (HE522133). All resected tissues were
stored at −70°C until used. Histology was routinely examined and
the clinical data of the patients were collected.
RNA isolation and complementary DNA
(cDNA) preparation
Total RNA was extracted from fresh frozen CCA
tissues and CCA cell lines with RNeasy kit (Qiagen, Valencia, CA,
USA) according to the manufacturer’s instructions. RNA was
quantified using GE NanoVue spectrophotometer (GE Healthcare,
Buckinghamshire, UK). For reverse transcription, oligo-dT primed
first strand cDNA was synthesized from 1 μg template RNA using
Improm-II™ reverse transcription system kit (Promega, Madison, WI,
USA) according to the manufacturer’s instructions.
Reverse transcription-polymerase chain
reaction (RT-PCR)
To screen the expression pattern of TFF2
isoform mRNA, RT-PCR was performed with cDNA derived from CCA cell
lines and tumor tissues. The sequences of full length primers are
shown in Table I. The expected
amplified product size is 434 base pairs for wtTFF2 and 284
base pairs for splicing isoform of TFF2. PCR products were
selected for nucleotide sequencing to verify type of splicing
isoform of TFF2 and to construct the plasmid clones used as
a standard in quantitative reverse transcription-PCR (QRT-PCR)
method.
 | Table IPrimer sequences used for detection of
TFF2 gene expression. |
Table I
Primer sequences used for detection of
TFF2 gene expression.
| Primer name | Forward (5′→3′) | Reverse (5′→3′) | Product size
(bp) |
|---|
| RT-PCR |
| FL-TFF2 |
TGCAGCTGAGCTAGACATGG |
CAGATGCATCCTCTGGAACC | 434 |
| QRT-PCR |
| wtTFF2 |
TTCCCTGGAATCACCAGTGACC |
ATGACGCACTGATCCGACTCTTG | 119 |
| ΔEX2TFF2 |
CGGGGAGTGAGAAACCCTAGT |
CCACAGACTTCGGGAAGAAGC | 162 |
Quantitative reverse transcription-PCR
(QRT-PCR)
QRT-PCR was performed to quantify the level of gene
expression of wtTFF2, splicing isoform TFF2
(ΔEX2TFF2) and GAPDH (reference gene), using a SYBR
Green I assay (Amresco, Solon, OH, USA). The sequences of these
primers are summarized in Table I.
The GAPDH primer sequences are given elsewhere (17). PCR was conducted on a Rotor-Gene 6
(Corbett Research, Australia) and a melting curve profile was
analyzed for specific product. Copy number of gene expression was
generated from the standard curve, using plasmid clones as DNA
standards; copy number ranging 30-3×106 for
wtTFF2 and ΔEX2TFF2, and
3×102-3×107 for GAPDH. Each sample was
run in triplicate and the coefficient of variation (CV) <15% was
acceptable.
The relative expression of mRNA was normalized with
GAPDH before the ratio of ΔEX2TFF2/wtTFF2 was
obtained. The ΔEX2TFF2/wtTFF2 ratio was categorized into low
and high expression based on the cut-off value derived from
corresponding normal controls (cut-off value = upper quartile value
[75th percentile] + 1.5 [interquartile range]). Association of
ΔEX2TFF2/wtTFF2 ratio was analyzed with clinicopathological
data.
Western blotting
Total protein was extracted from CCA samples using
TRIzol® reagent (Invitrogen, Carlsbad, CA) according to
the manufacturer’s protocol. Protein was fractionated on 15%
SDS-PAGE and transferred to a parablot Polyvinylidine Fluoride
(PVDF) membrane (MN, Germany). The membrane was non-specifically
blocked with 5% skim milk in Tris Buffered Saline (TBS) (150 mM
NaCl, 50 mM Tris, pH 7.5) for 1 h, then incubated with primary
antibody at dilution 1:3000, 4°C overnight. Sources of primary
antibodies: rabbit polyclonal anti-human TFF2 (A gift from
Professor Andrew Giraud, Murdoch Children’s Research Institute,
Royal Children’s Hospital, Parkville, Australia); rabbit polyclonal
to β-actin (Abcam, Cambridge, UK). After washing, a dilution 1:3000
of secondary antibody, goat polyclonal to rabbit IgG conjugated
with horseradish peroxidase (Abcam, Cambridge, UK), was applied and
the immune complexes were detected by an enhanced chemiluminescence
(ECL) system (GE Healthcare). The band intensity was analyzed using
Image J software distributed by the National Institutes of Health
(http://rsb.info.nih.gov/ij/index.html). The expression
of wtTFF2 was quantitated by comparison with standard 100 nanogram
of glycosylated-recombinant human TFF2 (rhTFF2) (A gift from
Professor Andrew Giraud).
The wtTFF2 protein expression was categorized into
low and high expression based on the cut-off value derived from
corresponding normal controls [cut-off value = upper quartile value
(75th percentile) + 1.5 (interquartile range)]. The expression of
wtTFF2 was analyzed for the association with clinicopathological
data.
Statistical analysis
The difference of splice variant of TFF2 mRNA
expression in normal and tumor tissues was analyzed using
Mann-Whitney test. Correlations between splice variant of
TFF2 mRNA and wtTFF2 protein expression were analyzed
using Spearman’s rho test. Associations of splice variant mRNA and
wtTFF2 protein expression with clinicopathological data of CCA
patients were evaluated using χ2 test. Survival analysis
was analyzed using Kaplan Meier and log rank test. P<0.05 was
considered as statistical significance. All statistical analyses
were performed using SPSS.
Results
Expression pattern of TFF2 mRNA isoforms
in CCA
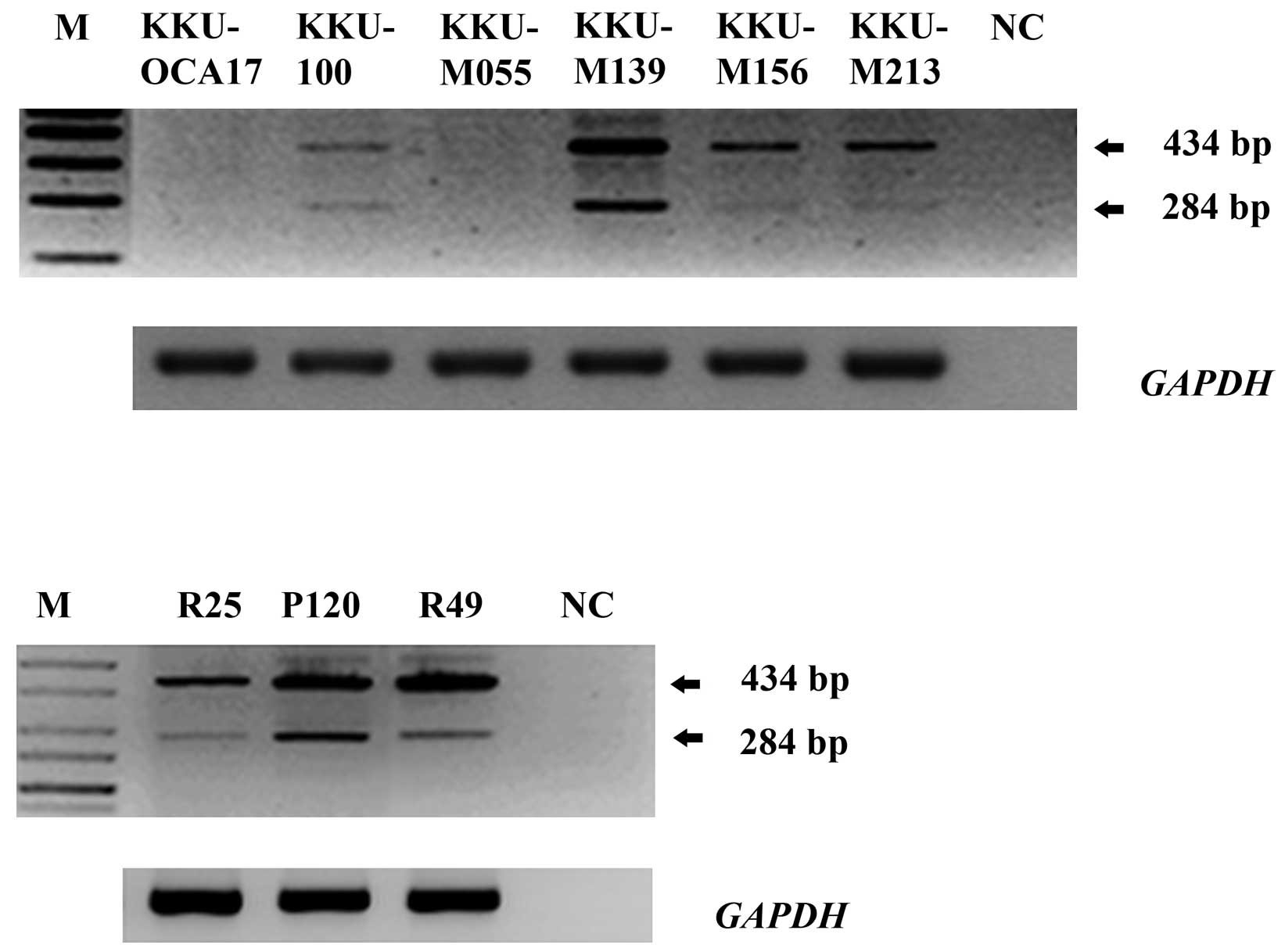
Six CCA cell lines including KKU-OCA17, KKU-M055,
KKU-100, KKU-M139, KKU-M156 and KKU-M213 were determined for
TFF2 mRNA expression by RT-PCR which could detect both full
length and splicing isoform. The PCR products revealed the
TFF2 full length (434 bp) and a smaller (284 bp) fragment.
Four out of six CCA cell lines, KKU-100, KKU-M139, KKU-M156 and
KKU-M213, exhibited both sizes, which were also detected in CCA
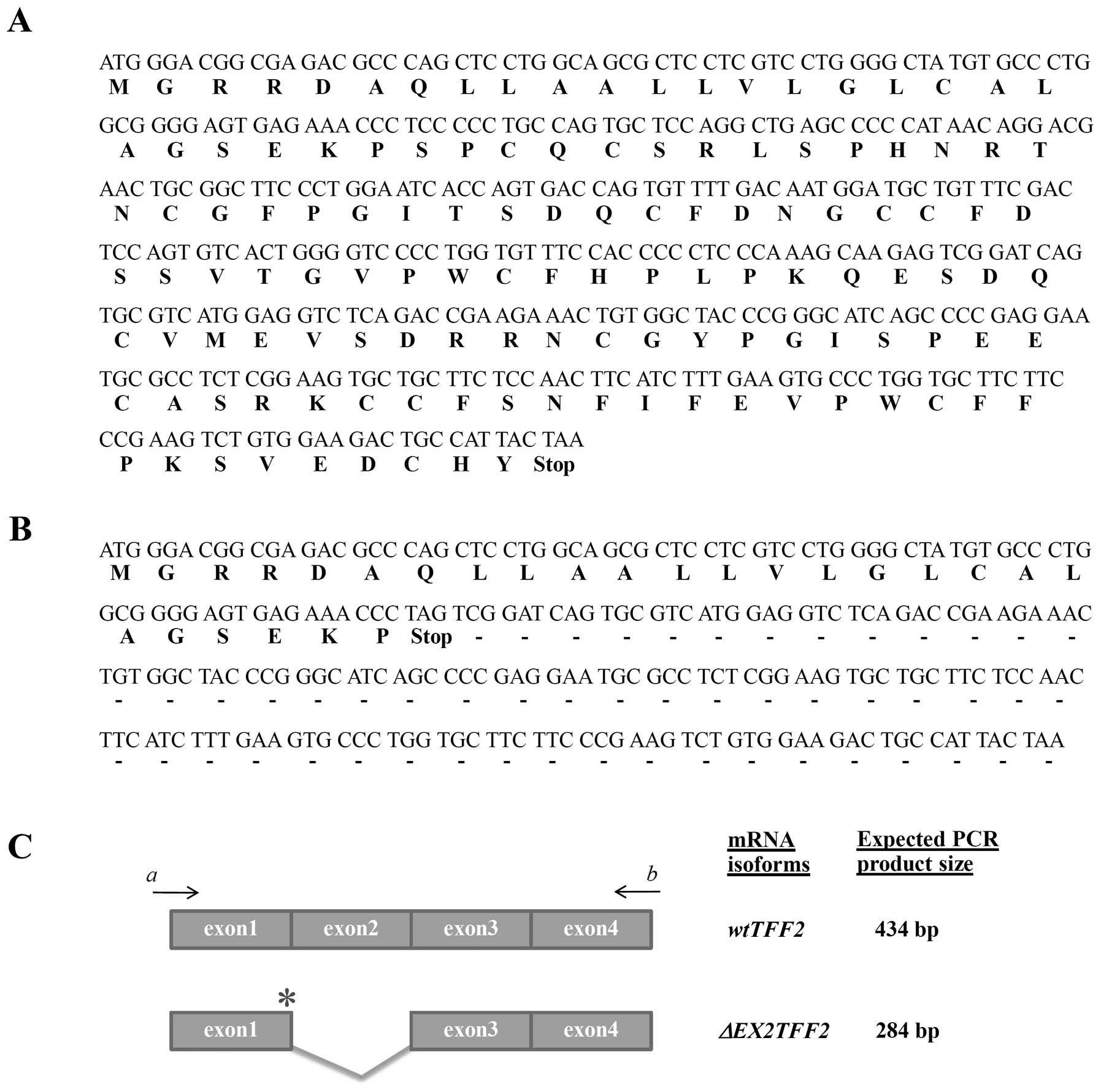
samples (Fig. 1). Nucleotide
sequencing was performed and showed exon 2 skipping
(ΔEX2TFF2) resulting in a premature termination codon (PTC)
within exon 1 in which only signal sequence was obtained (Fig. 2).
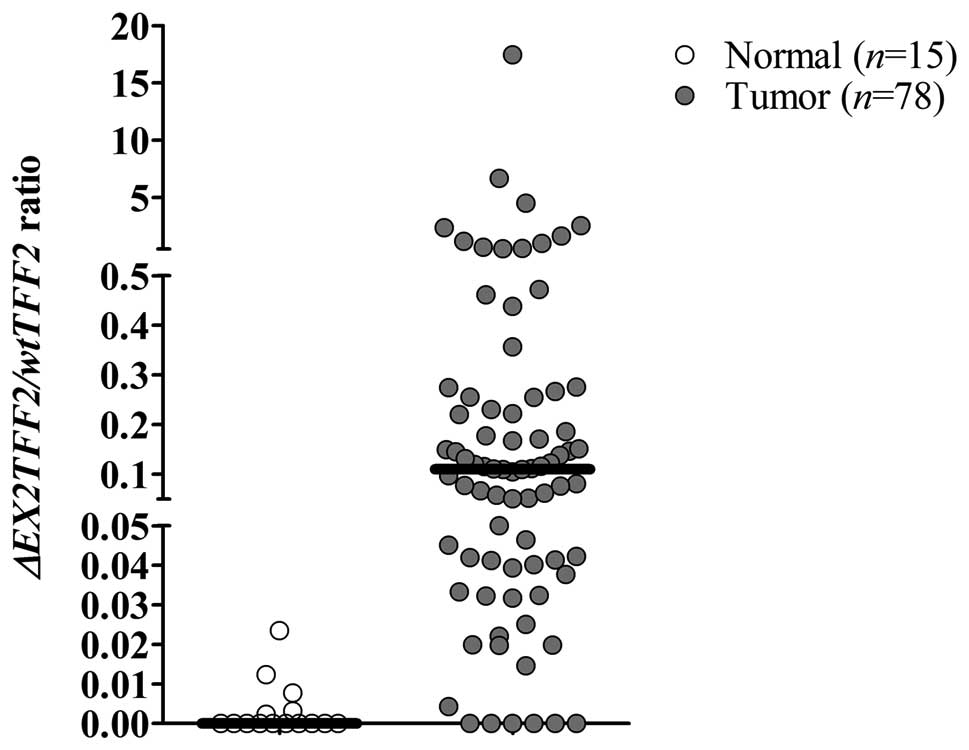
Expression of ΔEX2TFF2 in CCA samples.
ΔEX2TFF2
mRNA was quantified in 78 CCA and 15 normal adjacent
tissues using QRT-PCR. The median relative expression of
ΔEX2TFF2 mRNA in normal adjacent and CCA tissues were 0.00
(min: 0.00, max: 4.29×10−5) and 8.98×10−5
(min: 0.00, max: 1.80×10−2), respectively. The median of
ΔEX2TFF2/wtTFF2 ratio in normal adjacent and CCA tissues
were 0.00 (min: 0.00, max: 2.35×10−2) and 0.11 (min:
0.00, max: 17.47), respectively.
It has been shown that TFF2 is involved in
tumor progression in CCA (13).
Accordingly, alternative splicing may be one mechanism by which
cells employ to regulate the expression of TFF2 in term of
transcript and protein level. We then postulated that alternatively
spliced variant of TFF2 may regulate the expression of the
wild-type (wt) TFF2 in a negative feedback fashion and the
expression of ΔEX2TFF2 may be a good prognostic indicator in
CCA patients. We analyzed the median of ΔEX2TFF2/wtTFF2
ratio between CCA and normal adjacent tissues and found the
significant difference between these two groups (P<0.01,
Fig. 3). To analyze associations
between ΔEX2TFF2/wtTFF2 ratio and clinicopathological data
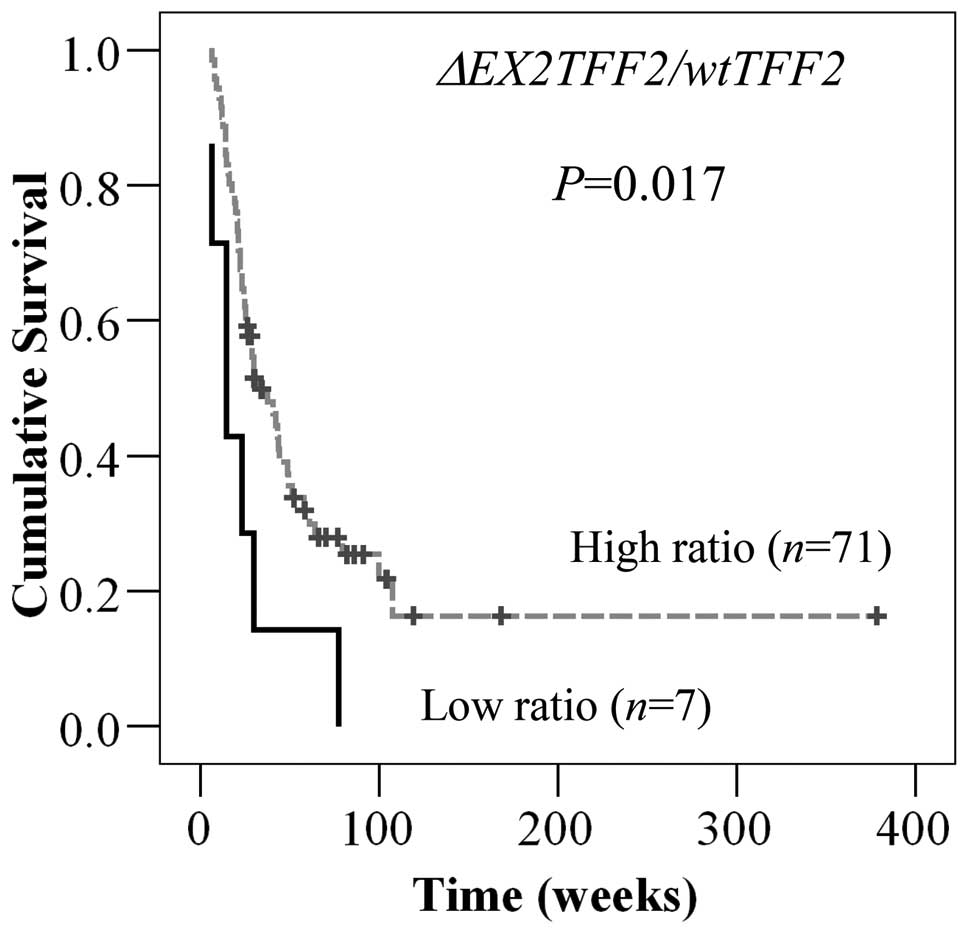
of CCA patients, we categorized ΔEX2TFF2/wtTFF2 ratio into
low and high expression based on the cut-off value, as described
above (cut-off = 6.93×10−3). As expected, patients with
high ratio of ΔEX2TFF2/wtTFF2 had longer overall survival
than patients with low ratio (P=0.017, Fig. 4). However, there were no
associations of ΔEX2TFF2/wtTFF2 ratio with age, gender and
histological grading.
Wild-type TFF2 protein expression in
CCA
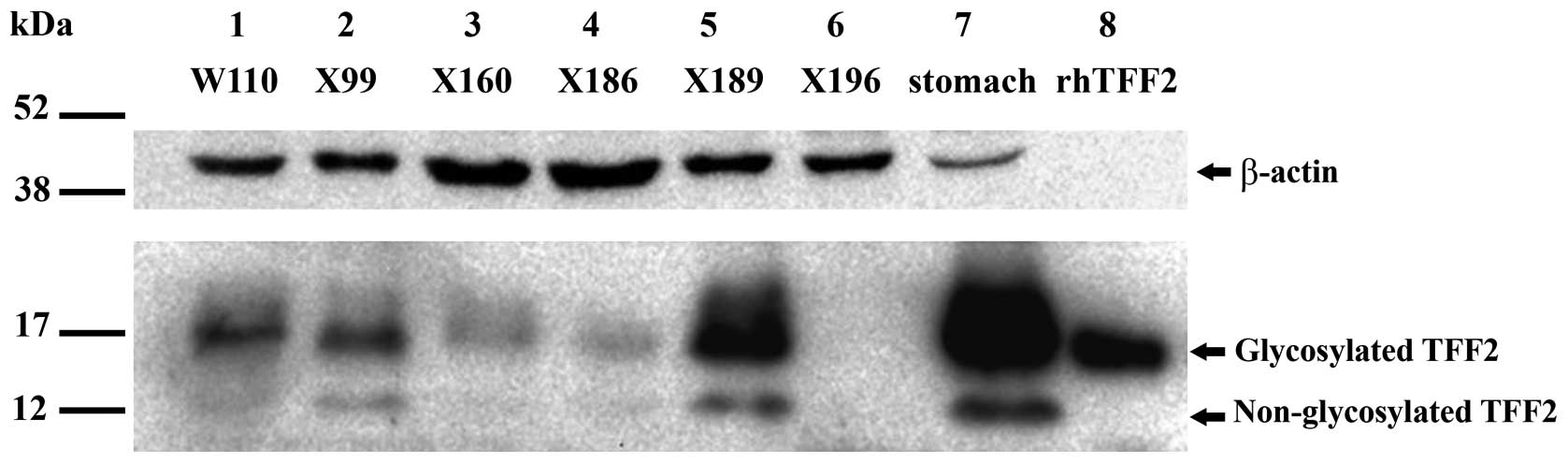
Expression of wtTFF2 protein was quantitated in 43
CCA tissues using western blotting. Recombinant human (rh) TFF2 and
protein derived from gastric tissue were used as a positive control
for TFF2 protein expression. The result showed that wtTFF2 protein,
glycosylated and non-glycosylated TFF2 (Fig. 5), was found in 9/43 CCA and 1/15
normal adjacent tissues. To analyze the association between wtTFF2
protein expression and clinocopathological data of CCA patients,
the expression of wtTFF2 protein was categorized into low and high
expression based on a cut-off value, as described above. The
cut-off value was 0.00. Thus, we categorized it into two groups,
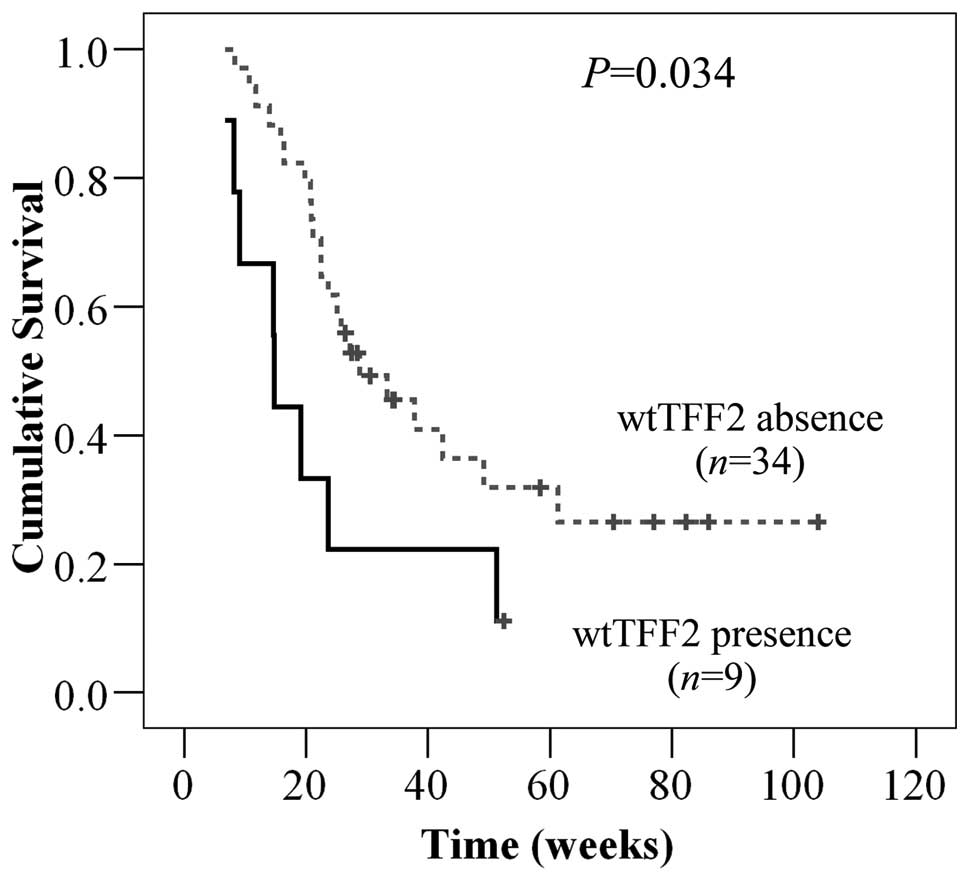
the presence and absence of wtTFF2. The presence of wtTFF2 was
significantly associated with the decreased overall survival time
in CCA patients (P=0.034, Fig. 6)
suggesting its important role in tumor progression. However, the
presence of wtTFF2 was not associated with age, gender and
histological grading. As postulated previously regarding the effect
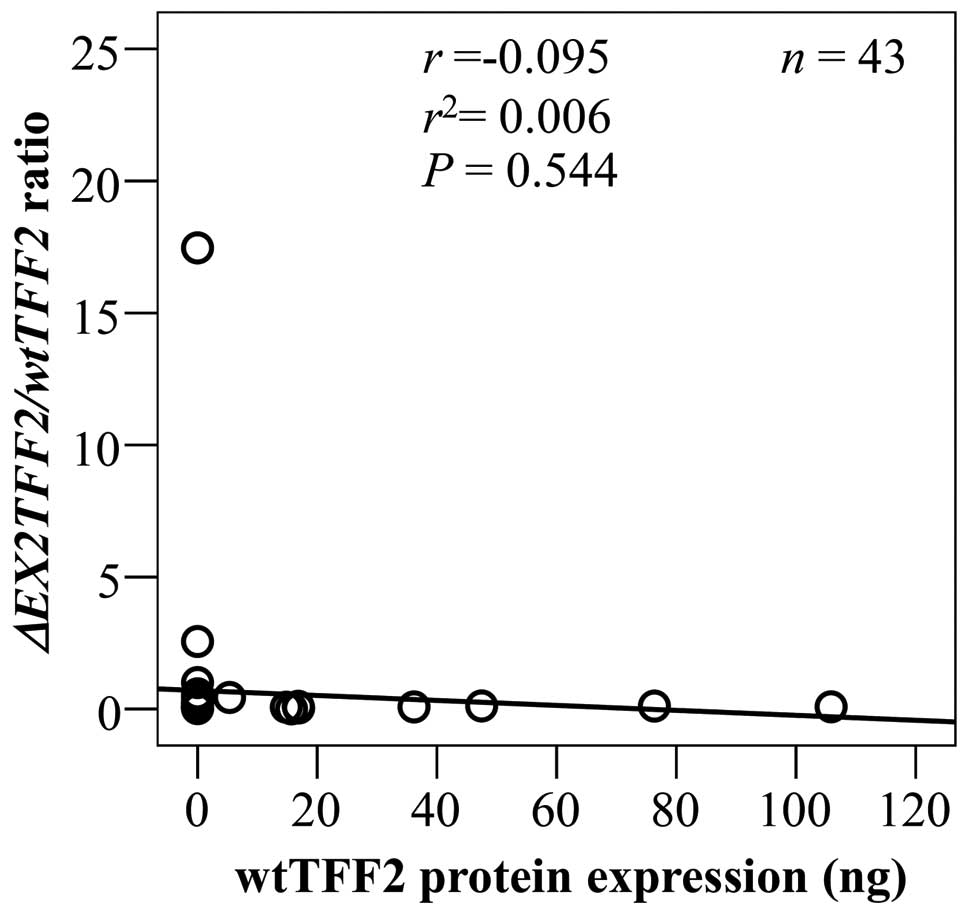
of splice variant on TFF2 protein expression, we tested the
correlation between ΔEX2TFF2/wtTFF2 ratio and wtTFF2 protein
expression. The results showed that ΔEX2TFF2/wtTFF2 ratio
was not significantly correlated with wtTFF2 protein level but it
had a trend negative correlation (r= −0.095, P=0.544, Fig. 7).
Discussion
We are the first to show the presence of an
alternatively spliced variant, exon 2 skipping, in CCA. This type
of splice variant, ΔEX2TFF2, resulted in a premature
termination codon (PTC) in the exon 1 in which only signal sequence
was encoded. The non-functional TFF2 cannot mediate cell
proliferation like its canonical TFF2 preventing tumor cell
progression. Our findings supported this postulation in which good
prognosis was found in patients with high ΔEX2TFF2/wtTFF2
ratio. Thus, ΔEX2TFF2/wtTFF2 ratio may serve as a prognostic
factor of good outcome in CCA. Moreover, the mRNA with PTC
subjected to be degraded by the nonsense mediated decay (NMD)
pathway (18,19) has been shown to have a very short
half-life. The presence of splice variant of TFF2 in CCA
patients suggested that alternative splicing may be a process which
regulates the expression of TFF2. Therefore, it is
conceivable that the overexpression of splice variant may play a
pivotal role as a dominant inhibitor of wild-type function. In this
study, we could not find a significant inverse correlation between
the ΔEX2TFF2/wtTFF2 ratio and wtTFF2 protein (r= −0.095)
suggesting that there are other mechanisms rather than alternative
splicing which control the expression of TFF2. Further study is
needed to address whether ΔEX2TFF2 plays a role in
translational regulation of wtTFF2 protein.
Our recent study reported that TFF2 positive
immunostaining was markedly increased in CCA compared with those in
normal bile ducts and dysplasia suggesting the role of TFF2 in
tumor progression (13). In this
study, we demonstrated that the expression of wtTFF2 protein was
associated with poor prognosis of CCA patients. This is consistent
with the previous studies which demonstrated that high expression
of TFF2 was an independent indicator of poor prognosis in CCA
(20) and gastric cancer patients
with TFF2-expressing tumors had a significantly worse disease-free
survival (11,21). Thus, wtTFF2 expression may serve as
a prognostic marker of poor outcome in CCA.
In conclusion, we identified a novel TFF2
splice variant, ΔEX2TFF2, and its significance in term of
ΔEX2TFF2/wtTFF2 ratio, which was upregulated in CCA. The
splice variant ΔEX2TFF2 might act as a negative regulator of
wtTFF2 expression. Clinically, the high ratio of
ΔEX2TFF2/wtTFF2 serves as a potential prolonged survival
marker of CCA patients. In contrast, the presence of wtTFF2 protein
is strongly associated with shorter overall survival time. This
study suggests the use of wtTFF2 inhibitor as a targeted therapy
for effective treatment of CCA.
Acknowledgements
This work was supported by the Higher Education
Research Promotion and National Research University Project of
Thailand, Office of the Higher Education Commission, through the
Center of Excellence in Specific Health Problems in Greater Mekong
Sub-region cluster (SHeP-GMS), Khon Kaen University; and Khon Kaen
University Research Affairs; and grants from the NHMRC, Australia,
and the Victorian Government’s Operational Infrastructure Support
Program. Kamlua S. was granted by the Centre for Research and
Development in Medical Diagnostic Laboratories (CMDL), Faculty of
Associated Medical Sciences; and Graduate School, Khon Kaen
University. We thank the Liver Fluke and Cholangiocarcinoma
Research Center (LFCRC), Faculty of Medicine, Khon Kaen University
for providing CCA samples, CCA cell lines and patients’ clinical
data.
References
|
1
|
Uttaravichien T, Bhudhisawasdi V,
Pairojkul C and Pugkhem A: Intrahepatic cholangiocarcinoma in
Thailand. J Hepatobiliary Pancreat Surg. 6:128–135. 1999.
View Article : Google Scholar
|
|
2
|
Sripa B, Kaewkes S, Sithithaworn P, et al:
Liver fluke induces cholangiocarcinoma. PLoS Med. 4:e2012007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Watanapa P and Watanapa WB: Liver
fluke-associated cholangiocarcinoma. Br J Surg. 89:962–970. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhamarapravati N, Thammavit W and
Vajrasthira S: Liver changes in hamsters infected with a liver
fluke of man, Opisthorchis viverrini. Am J Trop Med Hyg.
27:787–794. 1978.PubMed/NCBI
|
|
5
|
Vatanasapt V, Sriamporn S, Kamsa-ard S, et
al: Cancer survival in Khon Kaen, Thailand. IARC Sci Publ.
145:123–134. 1998.PubMed/NCBI
|
|
6
|
Sawanyawisuth K: Genes and
cholangiocarcinoma. Southeast Asian J Trop Med Public Health.
40:701–712. 2009.
|
|
7
|
Loilome W, Yongvanit P, Wongkham C, et al:
Altered gene expression in Opisthorchis viverrini-associated
cholangiocarcinoma in hamster model. Mol Carcinog. 45:279–287.
2006.
|
|
8
|
Hoffmann W: Trefoil factors TFF (trefoil
factor family) peptide-triggered signals promoting mucosal
restitution. Cell Mol Life Sci. 62:2932–2938. 2005.PubMed/NCBI
|
|
9
|
Thim L and May FE: Structure of mammalian
trefoil factors and functional insights. Cell Mol Life Sci.
62:2956–2973. 2005.PubMed/NCBI
|
|
10
|
Tu SP, Chi AL, Ai W, et al: p53 inhibition
of AP1-dependent TFF2 expression induces apoptosis and inhibits
cell migration in gastric cancer cells. Am J Physiol Gastrointest
Liver Physiol. 297:G385–G396. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dhar DK, Wang TC, Maruyama R, et al:
Expression of cytoplasmic TFF2 is a marker of tumor metastasis and
negative prognostic factor in gastric cancer. Lab Invest.
83:1343–1352. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
May FE, Semple JI, Prest SJ and Westley
BR: Expression and motogenic activity of TFF2 in human breast
cancer cells. Peptides. 25:865–872. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kosriwong K, Menheniott TR, Giraud AS,
Jearanaikoon P, Sripa B and Limpaiboon T: Trefoil factors: tumor
progression markers and mitogens via EGFR/MAPK activation in
cholangiocarcinoma. World J Gastroenterol. 17:1631–1641. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feltes CM, Kudo A, Blaschuk O and Byers
SW: An alternatively spliced cadherin-11 enhances human breast
cancer cell invasion. Cancer Res. 62:6688–6697. 2002.PubMed/NCBI
|
|
15
|
Hofstetter G, Berger A, Fiegl H, et al:
Alternative splicing of p53 and p73: the novel p53 splice variant
p53delta is an independent prognostic marker in ovarian cancer.
Oncogene. 29:1997–2004. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dominguez G, Garcia JM, Pena C, et al:
DeltaTAp73 upregulation correlates with poor prognosis in human
tumors: putative in vivo network involving p73 isoforms, p53, and
E2F-1. J Clin Oncol. 24:805–815. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Namwat N, Amimanan P, Loilome W, et al:
Characterization of 5-fluorouracil-resistant cholangiocarcinoma
cell lines. Chemotherapy. 54:343–351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gonzalez CI, Bhattacharya A, Wang W and
Peltz SW: Nonsense-mediated mRNA decay in Saccharomyces
cerevisiae. Gene. 274:15–25. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cuccurese M, Russo G, Russo A and
Pietropaolo C: Alternative splicing and nonsense-mediated mRNA
decay regulate mammalian ribosomal gene expression. Nucleic Acids
Res. 33:5965–5977. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thuwajit P, Chawengrattanachot W, Thuwajit
C, Sripa B, Paupairoj A and Chau-In S: Enhanced expression of mucin
6 glycoprotein in cholangiocarcinoma tissue from patients in
Thailand as a prognostic marker for survival. J Gastroenterol
Hepatol. 23:771–778. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dhar DK, Wang TC, Tabara H, et al:
Expression of trefoil factor family members correlates with patient
prognosis and neoangiogenesis. Clin Cancer Res. 11:6472–6478. 2005.
View Article : Google Scholar : PubMed/NCBI
|