Introduction
Ovarian carcinoma is the leading cause of death from
gynecological malignancies. The majority of ovarian cancer patient
deaths are due to the irreversible physiological effects of
metastases on normal organ function rather than from the primary
tumor (1). Despite recently
improved chemotherapeutic agents and an increased 5-year survival
rate, ovarian cancer mortality remains unchanged. A better
understanding of the metastatic mechanisms of ovarian cancer is
therefore needed to determine effective therapeutic interventions
to either eradicate or slow metastatic outgrowth.
Many factors are involved in regulating metastasis
through diverse mechanisms. Among metastasis suppressors, breast
cancer metastasis suppressor 1 (BRMS1) was originally shown to
functionally suppress the metastatic capacities of breast cancer
cells (2). Further studies showed
that BRMS1 is not only a metastasis suppressor gene in breast
cancer models but also in various other cancers, such as melanoma
and ovarian cancer (3,4). Zhang et al (4) demonstrated that low levels of BRMS1
expression correlated with poor prognosis in ovarian cancer
patients. They further showed that transfection of BRMS1
complementary DNA (cDNA) into the highly malignant ovarian
carcinoma cell line HO-8910PM significantly reduced cell adhesion,
motility and invasion in vitro and also decreased the
incidence of lung metastasis without affecting tumor growth. BRMS1
is thought to regulate metastasis through multiple mechanisms,
including restoration of gap junctions, reduction of
phosphoinositide signaling, interaction with the histone
deacetylase complex and regulation of the nuclear factor-κB (NF-κB)
pathway (5–7). In particular, several
metastasis-related genes were reported to be downregulated by BRMS1
through modulating the activity of NF-κB, including osteopontin
(OPN), urokinase-type plasminogen activator (uPA), microRNA-146,
interleukin-6 (IL-6) and chemokine receptor 4 (CXCR4) (8–12).
Chemokines are small cytokines that are
characterized by their capacity to induce directional cellular
migration towards a gradient of chemokines by binding to chemokine
receptors. One of the most extensively studied chemokine receptors
is CXCR4, which selectively binds the chemokine stromal
cell-derived factor-1 (SDF-1) also known as CXCL12 (13). Recent evidence suggests that the
SDF-1/CXCR4 pathway is involved in local invasion and metastasis of
many cancers, including breast cancer, gastric cancer and ovarian
cancer (14–16). Not only that, CXCR4 has been
observed to promote angiogenesis by stimulating the secretion of
several angiogenic factors, such as vascular endothelial growth
factor and IL-6 (17,18). Interestingly, a recent study by Yang
et al demonstrated that BRMS1 reduces CXCR4 expression in
lung cancer cells via abrogation of NF-κB activation (12); however, the functional implications
of BRMS1 and its relationship to the CXCR4 signaling pathway in
ovarian neoplasms are not clear.
Therefore, we investigated the potential mechanisms
of BRMS1-mediated metastasis suppression in ovarian cancer. In this
study, recombinant plasmid containing short-hairpin RNA (shRNA)
sequences targeting BRMS1 mRNA transcription regions was
constructed and transfected into ovarian cancer cells. Their
influences on cell adhesion, migration, invasion and angiogenesis
were observed, and the expression of CXCR4 was detected. Finally,
we employed an electrophoretic mobility shift assay (EMSA) to
explore whether BRMS1 regulates CXCR4 expression through the NF-κB
pathway. Our data indicate that BRMS1 negatively regulates
metastatic potential at least in part through the suppression of
NF-κB-dependent CXCR4 expression.
Materials and methods
Cell lines and cell culture
The human ovarian cancer cell line OVCAR3 (ATCC,
USA) was grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco,
Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS)
(Gibco, Invitrogen) and antibiotics (100 U/ml penicillin and 100
μg/ml streptomycin). Human umbilical venous endothelial cells
(HUVECs) were obtained from the Institute of Biochemistry and Cell
Biology of the Chinese Academy of Science (Shanghai) and cultured
in Kaighn’s modified Ham’s F-12K medium (Mediatech, Manassas, VA,
USA) supplemented with endothelial cell growth supplement (BD
Biosciences, Canada) and 10% FBS. Cultures were tested and shown
free of mycoplasma contamination. All cells were maintained in 5%
CO2 atmosphere at 37°C. For all functional and
biological assays, cells with >95% viability were used at 70–90%
confluence.
Plasmids construction
Based on the preliminary results of screening out
effective silencing siRNA sequences, the following double-stranded
RNA oligonucleotides specific for the BRMS1 coding region were
used: 5′-CACCGTTCGTACTT ATTCCTGATCACATCCTTCAAGAGAGGATGTGATCAG
GAATAAGTACGAATTTTTTG-3′ (sense), 5′-GATCCAA
AAAATTCGTACTTATTCCTGATCACATCCTCTCTTG
AAGGATGTGATCAGGAATAAGTACGAAC-3′ (antisense); Negative control
sequences with no significant homology to the BRMS1 gene and which
had the sequence not present in the human, mouse or rat genome
databases were: 5′-CACCGT TCTCCGAACGTGTCACGTCAAGAGATTACGTGACACG
TTCGGAGAATTTTTTG-3′ (sense), 5′-GATCCAAAAAAT
TCTCCGAACGTGTCACGTAATCTCTTGACGTGACACG TTCGGAGAAC-3′ (antisense).
All DNA chains were synthesized by GenePharma Co. (Shanghai,
China). The plasmids were extracted and the accuracy of the
constructs was confirmed by DNA sequencing.
Cell transfection
According to the manufacturer’s protocol for
Lipofectamine 2000 (Invitrogen), pGPU6/GFP/Neo-BRMS1 or
pGPU6/GFP/Neo-NC were transfected into OVCAR3 cells. After 6 h, the
cultures were replaced with 2 ml fresh medium supplemented with 10%
FBS and antibiotics. Then the cells were visualized under
fluorescence microscopy. After 48 h, 600 μg/ml G418 (Sigma, USA)
was added to the medium for selecting stable transfectants, and
individual clones were isolated and maintained in a medium
containing 300 μg/ml G418. Real-time PCR and Western blotting were
applied to analyze BRMS1 mRNA and protein levels, respectively. The
stably transfected OVCAR3 cells were named BRMS1-shRNA (transfected
with pGPU6/GFP/Neo-BRM-S1) and NC-shRNA, respectively.
Real-time reverse transcription
polymerase chain reaction (real-time PCR)
Total RNA from cells was extracted with TRIzol
Reagent (Invitrogen) following the manufacturer’s instruction. cDNA
was synthesized from total RNA using the PrimeScript RT reagent kit
(Takara, Japan). The cDNA specimens were amplified using the SYBR
Premix Ex Taq™ (Takara). GAPDH gene was used as an internal control
for standardization in triplicate. Cycle conditions were: 95°C for
30 sec, followed by 40 cycles of 95°C for 5 sec, 60°C for 34 sec
and finally 95°C for 15 sec, 60°C for 1 min. PCR amplification was
performed on the ABI 7500 Sequence Detection System (PE Applied
Biosystems, Foster City, CA, USA). The comparative Ct (ΔΔCT) method
was used to determine the expression fold change. The sequences of
the primers used were as follows: BRMS1 forward:
5′-ATGCCTGTCCAGCCTCC AAG-3′ and reverse
5′-GCGTCGCTCATAGTCCTCATCA-3′; CXCR4 forward:
5′-GGTGGTCTATGTTGGCGTCT-3′ and reverse 5′-CTCAGTGGAAACAGATGAAT-3′;
GAPDH forward: 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse 5′-TGGT
GAAGACGCCAGTGGA-3′.
Western blot analysis
Total cellular proteins from the cells were obtained
using RIPA lysis buffer (Santa Cruz Biotechnology, USA) containing
a cocktail of proteinase inhibitors and phosphatase inhibitors.
Protein concentrations were measured using the BCA protein assay
(Sigma). The proteins were subjected to 10% SDS denatured
polyacrylamide gel and transferred onto PVDF membranes. Membranes
were blocked in 5% non-fat milk for 1 h at 4°C and blotted with
rabbit anti-human antibody at the recommended dilution BRMS1
(1:500, BioWorld, USA), CXCR4 (1:100, Epitomics, USA) and β-actin
(1:500, BioWorld), and subsequently incubated with the appropriate
secondary antibody. After washing with TBST, visualization of the
second antibody was performed using a chemiluminescence detection
procedure according to the manufacturer’s protocol (Amersham
Biosciences, Japan). The LabWorks™ Image Acquisition and Analysis
Software (UVP, USA) was used to quantify band intensities. β-actin
was used as a loading control.
Cell adhesion assay
For this assay, 96-well plates were incubated with
50 μl (30 μg/ml) BD Matrigel™ Matrix (BD Biosciences, Germany) at
4°C overnight, then washed with PBS twice and blocked with 1% BSA
for 1 h at 37°C. Cells were trypsinized and seeded at
1×105/ml to each coated well and incubated for 2 h in 5%
CO2 atmosphere at 37°C, then rinsed three times with PBS
to remove non-adherent cells. Each well with 100 μl medium was
added 20 μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT), further incubated for 4 h, then the MTT was removed
and 150 μl dimethylsulfoxide (DMSO) was pipetted into each well.
The optical density (OD) was measured at 570 nm with a microplate
reader. The OD values were propotional to the number of cells with
adhesion; five duplicate wells were set up for each group.
Migration assay
Cell migration was assessed by adding
5×104 cells into the upper chamber of an 8-μm pore size
Transwell insert (Corning, USA) in serum-free media. These inserts
were placed in wells with serum-containing media. After seeding for
24 h, non-migrating cells were removed from the upper surface of
the filter with a cotton-tipped swab. The cells on the lower
surface of the filter were fixed in 4% paraformaldehyde and stained
using crystal violet staining solution. Five random fields were
counted at ×100 magnification. All the data presented are from at
least three independent experiments performed in duplicate.
Matrigel invasion assay
Transwell inserts (8-μm pore size) coated with 30 μl
Matrigel were placed in wells as previously described. In the top
chamber, 5×104 cells were plated in serum-free media and
incubated with serum-containing media as a chemoattractant in the
bottom chamber. Cells were then incubated at 37°C and allowed to
invade through the Matrigel barrier for 24 h. After incubation,
non-invading cells were removed using a cotton swab. Filters were
fixed and stained with crystal violet staining solution, and five
random fields were counted at ×100 magnification. All the data
presented are from at least three independent experiments performed
in duplicate.
In vitro tube formation assay
OVCAR3 cells were cultured in 6-well plates with
fresh complete medium for 24 h and 1 ml conditioned medium was
collected. For tube formation assay, the 48-well plates were coated
with Matrigel (100 μl per well) and kept in 5% CO2
atmosphere at 37°C for 30 min. Then, 5×104 HUVECs were
suspended in 500 μl conditioned medium and applied to the
pre-coated 48-well plates. After incubation at 37°C for another 24
h, images were captured under a microscope and the tubular
structures formed in the Matrigel were counted at ×100
magnification in five random fields.
Electrophoretic mobility shift assay
(EMSA)
The assay is based on that DNA-protein complexes
migrate slower than unbound DNA double-stranded oligonucleotides on
a native polyacrylamide gel, resulting in a ‘shift’ in the
migration of the labeled DNA band. The detection of bands was
performed by the LightShift™ Chemiluminescent EMSA kit (Pierce,
USA) that used a non-isotopic method to detect DNA-protein
interactions. Nuclear extracts were prepared from OVCAR3 cells
knockdown of BRMS1 and the control sample. Nuclear proteins were
incubated at room temperature for 10 min with oligonucleotide probe
bearing an NF-κB binding sequence on the CXCR4 promoter
(5′-TCCCCTGGGCTTCCCAAGCC-3′). The probe was labeled with a biotin
at its 5′-end. Another oligonucleotide with the same sequence but
without labeling was used as a competitive sequence at 500-fold
concentration. After the reaction the DNA-protein complexes were
subjected to a 6.5% native polyacrylamide gel electrophoresis and
transferred to a nylon membrane. Then the membrane was immediately
cross-linked for 15 min on a UV transilluminator equipped with 312
nm bulbs. Finally, a chemiluminescent detection method utilizing a
luminal/enhancer solution and a stable peroxide solution was used
as described by the manufacturer and membranes were exposed to
X-ray films for 2–5 min before developing.
Statistical analysis
All experiments were performed at least in
triplicate and data were compiled from three separate experiments.
The results were calculated as means ± SD. All statistical analyses
were determined by one-way ANOVA using the SPSS16.0 software. A
P-value <0.05 was considered significant.
Results
Specific inhibition of BRMS1 expression
by BRMS1-shRNA
Plamid vectors expressing either BRMS1-shRNA or
non-specific sequence control (NC) shRNA were constructed and
transfected into OVCAR3 cells. After 24 h, high transfection
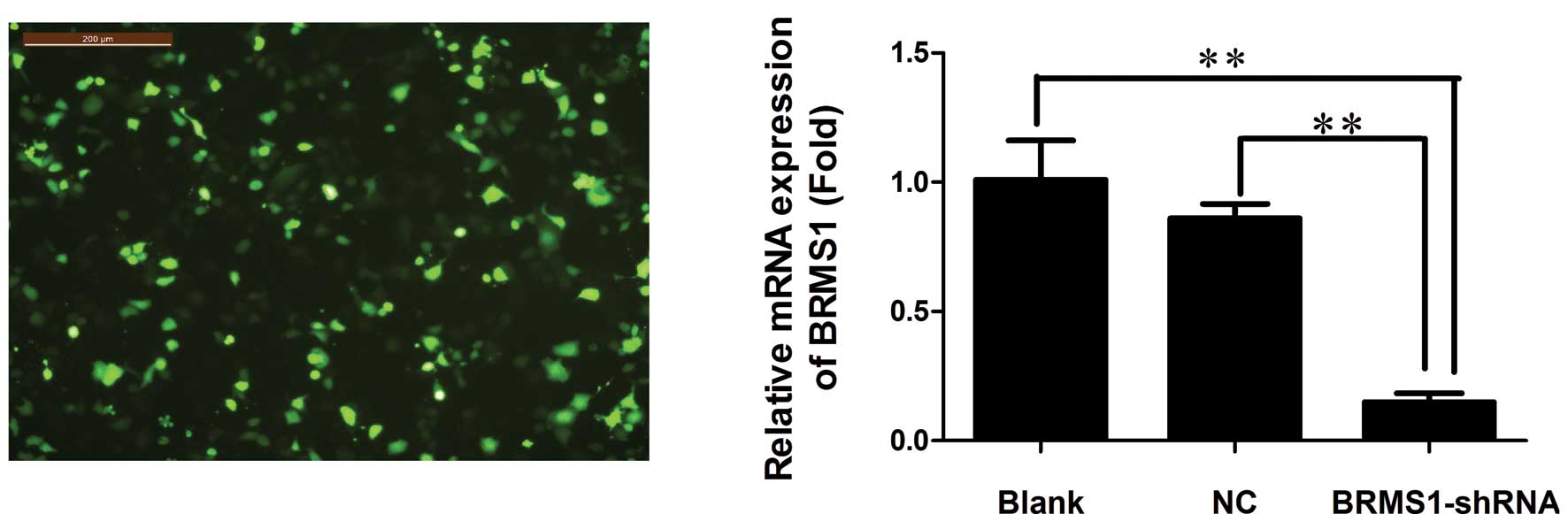
efficiency was observed by fluorescence microscopy (Fig. 1A). To determine silencing
efficiency, the expression levels of BRMS1 mRNA and protein were
measured by real-time PCR and Western blot analysis, respectively.
The BRMS1 mRNA level declined significantly in the BRMS1-shRNA
transfected cells, with an average inhibition of 85.15% compared to
the blank control group (P<0.01, Fig. 1B). BRMS1 protein expression was also
decreased, with an average inhibition of 46.67% in the BRMS1-shRNA
group (P<0.01, Fig. 1C). These
results suggested that pGPU6/GFP/Neo-BRMS1 could effectively
suppress BRMS1 expression at both the mRNA and protein levels in
OVCAR3 cells.
Effect of BRMS1-shRNA on adhesion
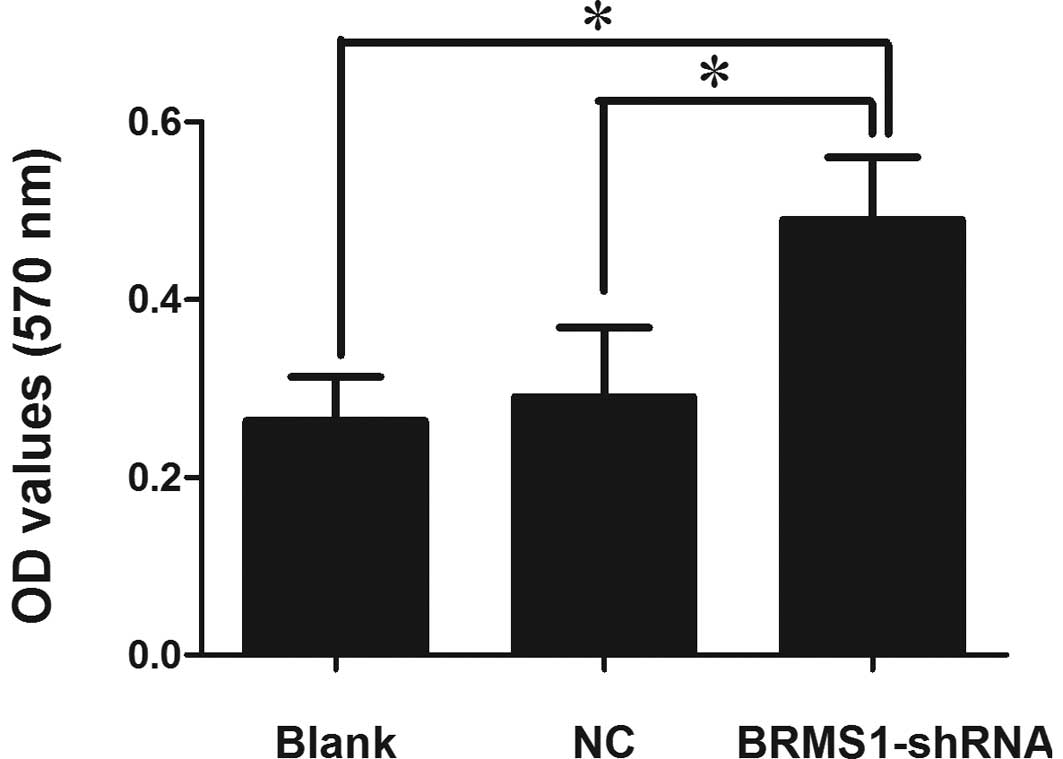
Cancer cell adhesion to the subendothelial
extracellular matrix is an important step in metastasis formation.
To assess the potential involvement of downregulation of BRMS1
expression on adhesion, a cell adhesion assay was employed. Our
results showed that BRMS1-shRNA cells markedly enhanced cell
adhesion to the Matrigel matrix. The OD values at 570 nm were
proportionate to the number of attached cells. Cell adhesion of
BRMS1-shRNA transfected OVCAR3 cells was increased by 1.88-fold
compared to the blank control cells (Fig. 2).
Effect of BRMS1-shRNA on migration and
invasion
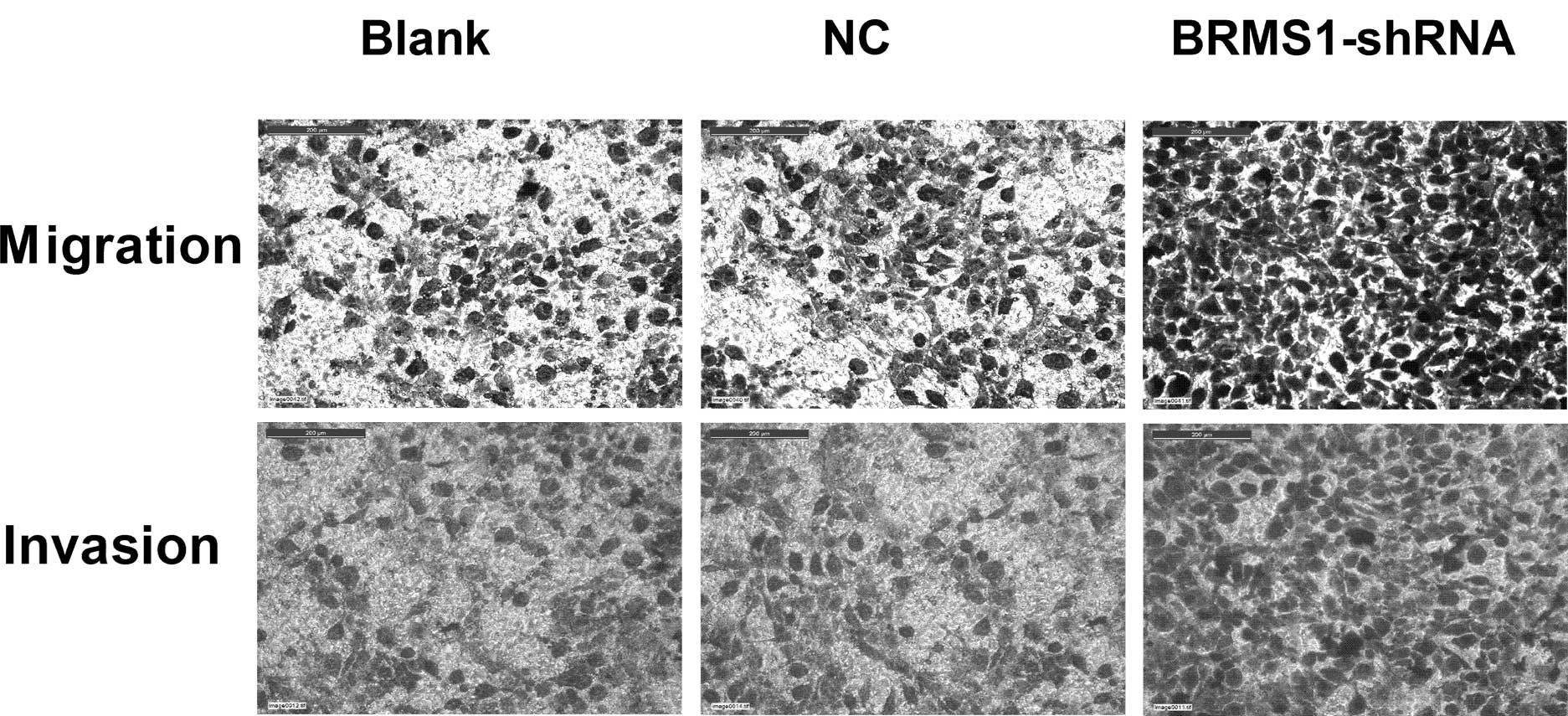
Migration and Matrigel invasion assay were performed
to examine the impact of BRMS1-shRNA on cell migration and
invasion, respectively. As shown in Fig. 3, BRMS1 knockdown increased OVCAR3
cell migration by 1.7-fold compared to untreated cells. The
Matrigel invasion assay results demonstrated that the invasiveness
of cells treated with BRMS1-shRNA increased by 1.81-fold compared
to the control group. Taken together, these data indicated that
BRMS1-shRNA promoted motility and invasion of OVCAR3 cells.
Effect of BRMS1-shRNA on
angiogenesis
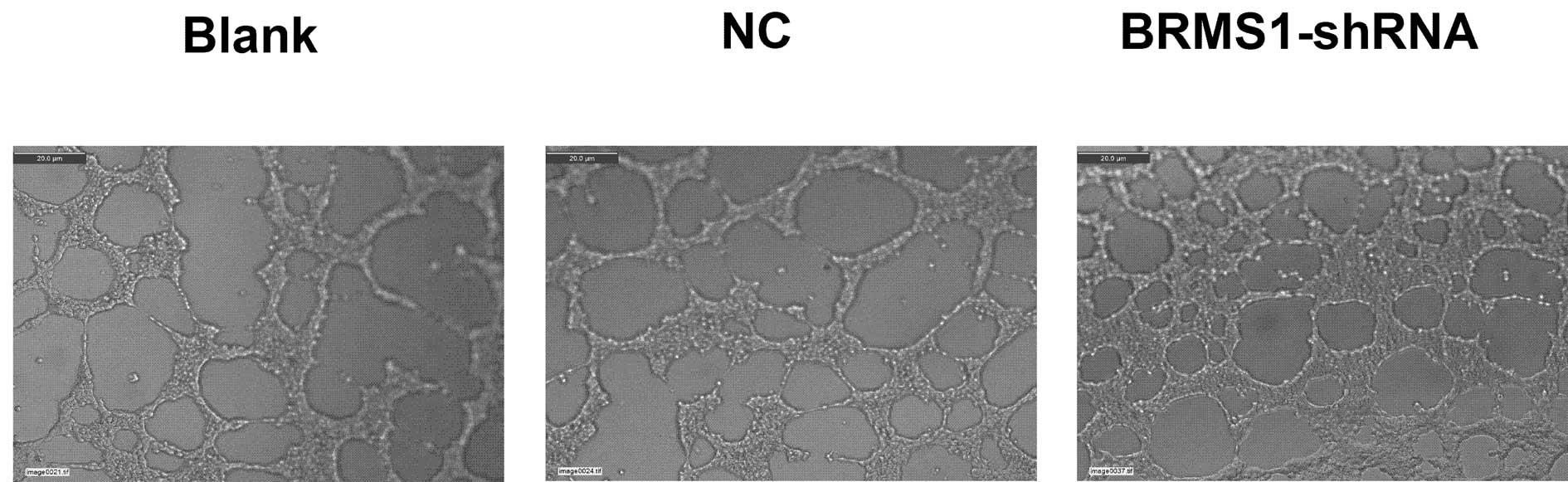
Angiogenesis plays a critical role in the growth and
metastatic potential of all solid tumors. To determine the effects
of BRMS1 silencing on ovarian cancer cell angiogenesis, we utilized
the tube formation assay. Compared to the corresponding control,
the average number of complete tubular structures formed by HUVECs
was increased by 1.97-fold in conditioned medium in OVCAR3 cells
transfected with BRMS1-shRNA. These data indicated that inhibition
of BRMS1 significantly enhanced the angiogenic capacity of the
ovarian cancer cells (Fig. 4).
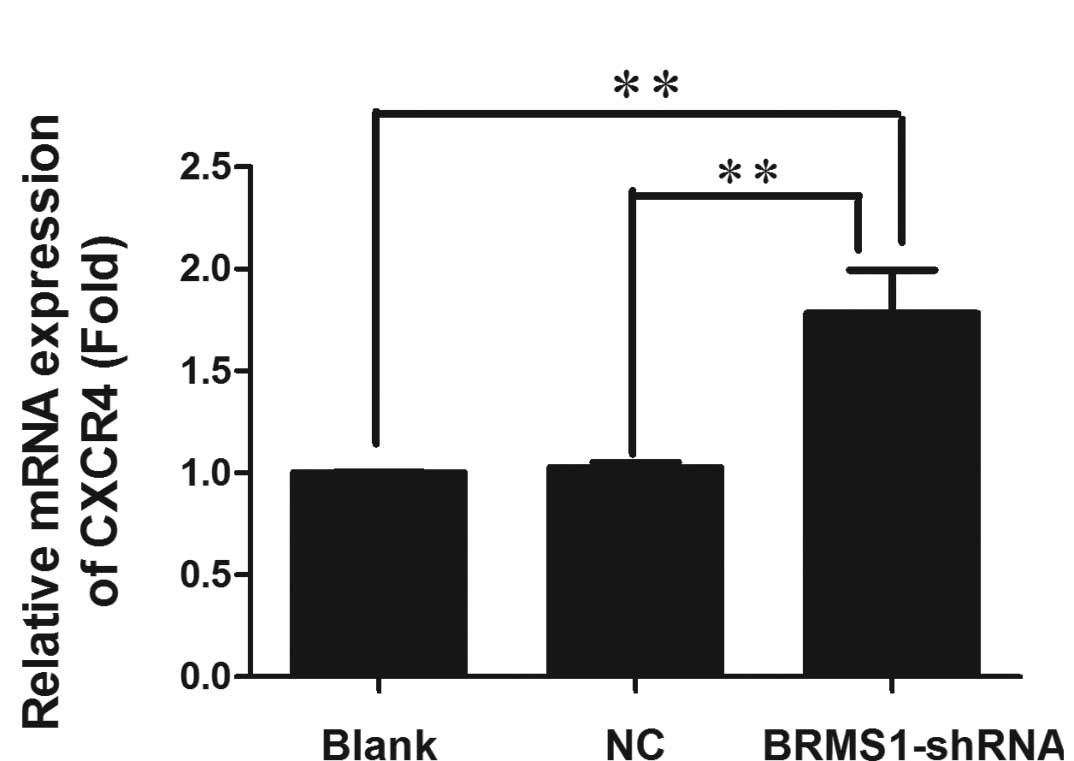
Upregulation of CXCR4 by BRMS1-shRNA
Recently, BRMS1 was shown to regulate metastatic
potential through the down-regulation of CXCR4 (12). To determine whether loss of BRMS1
regulated CXCR4 expression in ovarian cancer, CXCR4 mRNA and
protein levels of OVCAR3 cells transfected with either BRMS1-shRNA
or a negative control were measured by real-time RT-PCR and Western
blot analysis, respectively. As shown in Fig. 5, CXCR4 mRNA was 1.78-fold higher in
BRMS1-shRNA transfected cells compared to the blank control group.
Furthermore, Western blot analysis revealed that BRMS1 silencing in
OVCAR3 cells elevated CXCR4 protein levels by 1.26-fold. These
results elucidated that knockdown of BRMS1 upregulated CXCR4 at
both the transcriptional and translational levels. Taken together,
our data indicate that loss of BRMS1 expression induces adhesion,
migration, invasion and angiogenesis of OVCAR3 cells, which may be
due to upregulation of CXCR4.
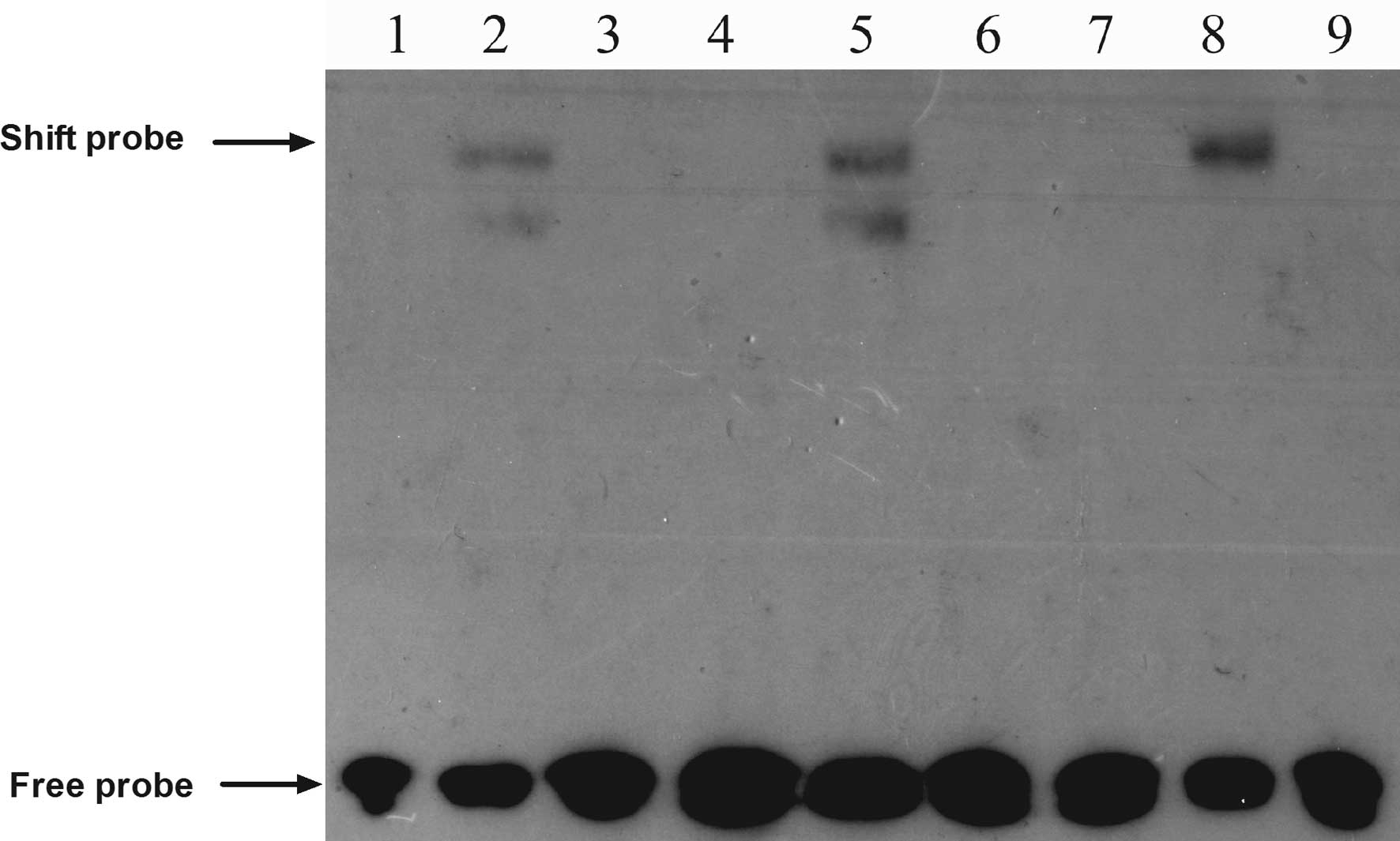
BRMS1 knockdown promotes CXCR4 expression
through activation of the NF-κB signaling pathway
A prior study revealed that NF-κB promoted breast
cancer migration and invasion by directly upregulating CXCR4
expression (19). Therefore, we
chose to perform an EMSA to explore whether BRMS1 regulates CXCR4
expression through the NF-κB pathway in ovarian cancer cells. The
EMSA results suggested that BRMS1 knockdown increased the DNA
binding activity of NF-κB to the CXCR4 promoter, compared to the
control groups, whereas the unlabeled competitive sequence markedly
inhibited this binding (Fig.
6).
Discussion
Metastasis is a multistep process involving
dissociation of malignant cells in the primary tumor, local
invasion, angiogenesis, intravasation, survival in the circulation,
extravasation and proliferation at a secondary site (20). These processes are modulated by many
factors among which the metastasis suppressors are of particular
importance for elucidating the underlying genetic and molecular
biological mechanisms of metastasis.
The metastasis suppressor gene BRMS1 was discovered
by Seraj et al (2) while
studying the non-random amplifications and deletions in chromosome
11 using differential display. It is located on chromosome 11q
13.1–13.2 and consists of 10 exons and 9 introns spanning
approximately 7 kb. Previous research showed that introducing BRMS1
into the highly metastatic ovarian cancer cell line HO-8910PM
significantly suppressed adhesion, motility and local invasion
without affecting tumor growth in vitro (4). Moreover, a recent report suggested
that BRMS1 was associated with tumor angiogenesis. Loss of BRMS1
resulted in deficient suppression of vasculogenesis and contributed
to melanoma metastasis (11). BRMS1
has also been shown to reduce the capacity of multiple human cancer
cell lines to metastasize to the lymph nodes, lungs and/or bone in
experimental models (2,21,22).
In addition, BRMS1 has clinical relevance for some tumor types.
BRMS1 mRNA expression was downregulated in breast tumor tissues
(7) and in breast cancer brain
metastases (23). Hicks et
al (24) claimed that
attenuation of BRMS1 expression in breast carcinomas was associated
with reduced disease-free survival in the context of hormone
receptor-negativity or HER2 overexpression. Furthermore, Zhao and
Wang (25) observed that BRMS1
expression in ovarian serous adenocarcinoma was significantly lower
than in both normal ovarian tissue and benign ovarian tumor tissue.
BRMS1 was correlated with surgical stage, lymph node metastasis and
tumor size. Another study determined that both BRMS1 mRNA and
protein levels were diminished in non-small cell lung cancer
(NSCLC) compared to the adjacent non-cancerous lung. Preservation
of BRMS1 expression was accordingly associated with improved
survival of NSCLC patients (26).
Recently, an increasing number of studies have demonstrated the
potential of using BRMS1 as a prognostic marker and therapeutic
target for breast cancer (27),
ovarian cancer (4), melanoma
(11) and NSCLC (26). Together, the data provide compelling
evidence that BRMS1 is an effective metastasis suppressor in
tumors; however, the mechanistic basis for its
metastasis-suppressive function in human ovarian cancer is poorly
defined.
In this study, we employed RNA interference (RNAi)
technology to knock down endogenous BRMS1 expression and analyzed
the influence of BRMS1 on the metastatic behavior of ovarian cancer
cells. Due to the stability and long-term effectiveness of shRNA,
BRMS1-shRNA was constructed and transfected into the human ovarian
cancer cell line OVCAR3. Our data revealed that the expression of
BRMS1 mRNA and protein was decreased in OVCAR3 cells following
BRMS1-shRNA transfection, with inhibition rates of 85.15% at the
mRNA level and 46.67% at the protein level. We then focused on cell
adhesion, migration, invasiveness and angiogenesis, all of which
are essential steps for the establishment of metastasis. We found
that BRMS1 silencing increased adhesion, migration and invasion,
and induced vascularization of ovarian cancer cells. Consistent
with results reported in the literature, we determined that BRMS1
is indeed an effective metastasis suppressor in ovarian cancer.
Numerous studies have confirmed that many BRMS1
downstream targets are involved in regulating tumor progression and
metastatic behaviors. It has also been reported that these
processes are associated with NF-κB signaling pathways. Cicek et
al (7) demonstrated that BRMS1
expression led to the inhibition of IκBα phosphorylation and
degradation and subsequently to a reduction in NF-κB nuclear
translocation. Expression analysis has indicated that the OPN is
decreased when BRMS1 is overexpressed in MDA-MB-435 cells;
interestingly, a mechanism by which BRMS1 reduces OPN expression
levels is via abrogation of NF-κB activation (8). Another study revealed that BRMS1
expression stimulated p65 dissociation from the NF-κB binding site
of the uPA promoter, which resulted in reduced transactivation of
uPA expression (9). Moreover, BRMS1
has been shown to negatively regulate melanoma angiogenesis by
suppressing NF-κB activity and IL-6 expression (11). Perhaps most interestingly, BRMS1 was
shown to reduce CXCR4 expression via abrogation of NF-κB signaling,
which led to metastasis suppression in lung cancer cells (12). CXCR4 is a seven-domain transmembrane
chemokine receptor that is predominantly expressed on lymphocytes
where it activates chemotaxis. SDF-1 is the only physiological
ligand for CXCR4. The SDF-1/CXCR4 axis has been recently shown to
be involved in stimulating multiple metastatic processes in many
different neoplasms, including migration, invasion, angiogenesis
and proliferation (13–16). Chu et al (17) also demonstrated that CXCR4
overexpression increased vascularity, which may help promote human
basal cell carcinoma metastasis. Conversely, both knockdown of
CXCR4 and use of a neutralizing antibody against CXCR4 in ovarian
carcinoma decreased invasion (28).
Moreover, a prior report showed that NF-κB could promote migration
and organ-specific homing of cancer cells through the induction of
CXCR4. The NF-κB binding site has also been identified in the
proximal region of the CXCR4 promoter and is postulated to play a
role in CXCR4 expression in human breast cancer cells (19,28,29).
Because both BRMS1 and CXCR4 are involved in regulating the NF-κB
signaling pathway, we hypothesized that BRMS1 might modulate
metastasis of ovarian cancer cells in part by regulating CXCR4
expression. Our data suggested that inhibiting BRMS1 in OVCAR3
cells could lead to the upregulation of CXCR4. We further
investigated whether the increase in CXCR4 expression resulting
from BRMS1 silencing was due to activation of the NF-κB pathway. To
address this question we used an EMSA targeting NF-κB binding in
the CXCR4 promoter. We determined that blocking BRMS1 obviously
increased NF-κB binding to the CXCR4 promoter compared to the
parental cells, whereas an unlabeled competitive sequence markedly
inhibited this binding. Taken together, these data provided
mechanistic support for our hypothesis that BRMS1 regulates CXCR4
expression through the NF-κB pathway.
In summary, we report that knockdown of BRMS1 in
ovarian cancer cells is associated with upregulation of CXCR4
mediated by NF-κB activation, which then increases the metastatic
potential. Our results contribute to the better understanding of
the tumor-suppressive functions of BRMS1 in ovarian cancer and
suggest that BRMS1 restoration may be a promising approach for
anti-metastasis therapy for human ovarian cancer.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of Guangdong Province (no. 2009B060700080) and
the Science and Information Technology of Guangzhou (no.
2010GN-E00221).
References
|
1
|
Moss C and Kaye SB: Ovarian cancer:
progress and continuing controversies in management. Eur J Cancer.
38:1701–1707. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seraj MJ, Samant RS, Verderame MF and
Welch DR: Functional evidence for a novel human breast carcinoma
metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer
Res. 60:2764–2769. 2000.PubMed/NCBI
|
|
3
|
Shevde LA, Samant RS, Goldberg SF,
Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT and Welch DR:
Suppression of human melanoma metastasis by the metastasis
suppressor gene, BRMS1. Exp Cell Res. 273:229–239. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang S, Lin QD and Di W: Suppression of
human ovarian carcinoma metastasis by the metastasis-suppressor
gene, BRMS1. Int J Gynecol Cancer. 16:522–531. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen X, Xu Z and Wang Y: Recent advances
in breast cancer metastasis suppressor 1. Int J Biol Markers.
26:1–8. 2011. View Article : Google Scholar
|
|
6
|
DeWald DB, Torabinejad J, Samant RS,
Johnston D, Erin N, Shope JC, Xie Y and Welch DR: Metastasis
suppression by breast cancer metastasis suppressor 1 involves
reduction of phosphoinositide signaling in MDA-MB-435 breast
carcinoma cells. Cancer Res. 65:713–717. 2005.
|
|
7
|
Cicek M, Fukuyama R, Welch DR, Sizemore N
and Casey G: Breast cancer metastasis suppressor 1 inhibits gene
expression by targeting nuclear factor-kappaB activity. Cancer Res.
65:3586–3595. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Samant RS, Clark DW, Fillmore RA, Cicek M,
Metge BJ, Chandramouli KH, Chambers AF, Casey G, Welch DR and
Shevde LA: Breast cancer metastasis suppressor 1 (BRMS1) inhibits
osteopontin transcription by abrogating NF-κB activation. Mol
Cancer. 6:62007.PubMed/NCBI
|
|
9
|
Cicek M, Fukuyama R, Cicek MS, Sizemore S,
Welch DR, Sizemore N and Casey G: BRMS1 contributes to the negative
regulation of uPA gene expression through recruitment of HDAC1 to
the NF-κB binding site of the uPA promoter. Clin Exp Metastasis.
26:229–237. 2009.PubMed/NCBI
|
|
10
|
Hurst DR, Edmonds MD, Scott GK, Benz CC,
Vaidya KS and Welch DR: Breast cancer metastasis suppressor 1
up-regulates miR-146, which suppresses breast cancer metastasis.
Cancer Res. 69:1279–1283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li J, Cheng Y, Tai D, Martinka M, Welch DR
and Li G: Prognostic significance of BRMS1 expression in human
melanoma and its role in tumor angiogenesis. Oncogene. 30:896–906.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Zhang B, Lin Y, Yang Y, Liu X and
Lu F: Breast cancer metastasis suppressor 1 inhibits SDF-1α-induced
migration of non-small cell lung cancer by decreasing CXCR4
expression. Cancer Lett. 269:46–56. 2008.PubMed/NCBI
|
|
13
|
Kruizinga RC, Bestebroer J, Berghuis P, de
Haas CJ, Links TP, de Vries EG and Walenkamp AM: Role of chemokines
and their receptors in cancer. Curr Pharm Des. 15:3396–3416. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Manu KA, Shanmugam MK, Rajendran P, Li F,
Ramachandran L, Hay HS, Kannaiyan R, Swamy SN, Vail S, Kapoor S,
Ramesh B, Bist P, Koay ES, Lim LH, Ahn KS, Kumar AP and Sethi G:
Plumbagin inhibits invasion and migration of breast and gastric
cancer cells by downregulating the expression of chemokine receptor
CXCR4. Mol Cancer. 10:1072011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao BC, Wang ZJ, Mao WZ, Ma HC, Han JG,
Zhao B and Xu HM: CXCR4/SDF-1 axis is involved in lymph node
metastasis of gastric carcinoma. World J Gastroenterol.
17:2389–2396. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kajiyama H, Shibata K, Terauchi M, Ino K,
Nawa A and Kikkawa F: Involvement of SDF-1α/CXCR4 axis in the
enhanced peritoneal metastasis of epithelial ovarian carcinoma. Int
J Cancer. 122:91–99. 2008.
|
|
17
|
Chu CY, Cha ST, Lin WC, Lu PH, Tan CT,
Chang CC, Lin BR, Jee SH and Kuo ML: Stromal cell-derived factor-1
α (SDF-1 α/CXCL12)-enhanced angiogenesis of human basal cell
carcinoma cells involves ERK1/2-NF-κB/interleukin-6 pathway.
Carcinogenesis. 30:205–213. 2009.
|
|
18
|
Ping YF, Yao XH, Jiang JY, Zhao LT, Yu SC,
Jiang T, Lin MC, Chen JH, Wang B, Zhang R, Cui YH, Qian C, Wang J
and Bian XW: The chemokine CXCL12 and its receptor CXCR4 promote
glioma stem cell-mediated VEGF production and tumour angiogenesis
via PI3K/AKT signaling. J Pathol. 224:344–354. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Helbig G, Christopherson KW II,
Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE and
Nakshatri H: NF-κB promotes breast cancer cell migration and
metastasis by inducing the expression of the chemokine receptor
CXCR4. J Bilo Chem. 278:21631–21638. 2003.
|
|
20
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hedley BD, Vaidya KS, Phadke P, Mackenzie
L, Dales DW, Postenka CO, MacDonald IC and Chambers AF: BRMS1
suppresses breast cancer metastasis in multiple experimental models
of metastasis by reducing solitary cell survival and inhibiting
growth initiation. Clin Exp Metastasis. 25:727–740. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Phadke PA, Vaidya KS, Nash KT, Hurst DR
and Welch DR: BRMS1 suppresses breast cancer experimental
metastasis to multiple organs by inhibiting several steps of the
metastatic process. Am J Pathol. 172:809–817. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stark AM, Tongers K, Maass N, Mehdorn HM
and Held-Feindt J: Reduced metastasis-suppressor gene
mRNA-expression in breast cancer brain metastases. J Cancer Res
Clin Oncol. 131:191–198. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hicks DG, Yoder BJ, Short S, Tarr S,
Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M,
Choueiri TK, Tubbs RR, Gaile D, Nowak N, Accavitti-Loper MA, Frost
AR, Welch DR and Casey G: Loss of breast cancer metastasis
suppressor 1 protein expression predicts reduced disease-free
survival in subsets of breast cancer patients. Clin Cancer Res.
12:6702–6708. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao XL and Wang P: Expression of SATB1
and BRMS1 in ovarian serous adenocarcinoma and its relationship
with clinicopathological features. Sichuan Da Xue Xue Bao Yi Xue
Ban. 42:82–85. 1052011.(In Chinese).
|
|
26
|
Smith PW, Liu Y, Siefert SA, Moskaluk CA,
Petroni GR and Jones DR: Breast cancer metastasis suppressor 1
(BRMS1) suppresses metastasis and correlates with improved patient
survival in non-small cell lung cancer. Cancer Lett. 276:196–203.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Z, Yamashita H, Toyama T, Yamamoto
Y, Kawasoe T and Iwase H: Reduced expression of the breast cancer
metastasis suppressor 1 mRNA is correlated with poor progress in
breast cancer. Clin Cancer Res. 12:6410–6414. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miyanishi N, Suzuki Y, Simizu S, Kuwabara
Y, Banno K and Umezawa K: Involvement of autocrine CXCL12/CXCR4
system in the regulation of ovarian carcinoma cell invasion.
Biochem Biophys Res Commun. 403:154–159. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chua AW, Hay HS, Rajendran P, Shanmugam
MK, Li F, Bist P, Koay ES, Lim LH, Kumar AP and Sethi G: Butein
downregulates chemokine receptor CXCR4 expression and function
through suppression of NF-κB activation in breast and pancreatic
tumor cells. Biochem Pharmacol. 80:1553–1562. 2010.PubMed/NCBI
|