Introduction
Head and neck squamous cell carcinoma (HNSCC) is a
common neoplasm associated with exposure to tobacco and alcohol. It
accounts for up to 5% of all newly diagnosed malignancies worldwide
and is the sixth most common cancer in the world (1). The incidence of this type of tumor is
expected to rise as a result of the increasing number of female and
adolescent smokers. Despite considerable advances in diagnosis and
treatment of HNSCC, the overall survival rate has remained constant
at 60% over the past 30 years in the United States (2). The lack of progress in head and neck
oncology emphasizes the need for basic studies on the molecular
biology of HNSCC.
In HNSCC genetic analyses demonstrated the frequent
loss of genomic material at several chromosomal loci suggesting the
involvement of diverse tumor suppressor genes (TSGs) in the genesis
of HNSCC (3,4). More recent, in addition to genetic
alterations, promoter hypermethylation has been recognized as
another mechanism of TSG inactivation. Accordingly, several studies
have shown that methylation of CpG islands located within the
promoter regions of tumor suppressor genes is a frequent event in
the development of HNSCC and other human malignancies (5–7).
Among multiple TSGs that have been found to be
hypermethylated in a variety of malignancies are the RASSF1A
and the MLH1 at chromosome arm 3p (8,9) and
the MGMT TSG at 10q (10).
RASSF1A is a member of a new group of RAS
effectors which are involved in cell cycle control, microtubule
stabilization, cellular adhesion and motility as well as apoptosis.
RASSF1A acts as a TSG by controlling mitotic function and
decreasing the risk of aneuploidy leading to increased genomic
stability (11). MLH1 is a
mismatch repair gene, functions to correct replicate mismatches
that escape DNA polymerase proofreading, and hence plays an
important role in the maintenance of genetic stability (12). The MGMT TSG is a DNA-repair
gene, which prevents the alkylation of guanine (13).
To date, reports have revealed varying frequencies
of TSG silencing by hypermethylation in HNSCC. The usefulness of
the analysis of the promoter methylation status for prognostic
purposes has been shown (14,15) as
well as normalization of hypermethylation by drugs in cancer
patients. e.g., the efficiency of 5-azacytidine (5-Aza,
Vidaza®) and decitabine (Dacogen®) are
established for the therapy of acute myeloid leukemia and
myelodysplastic syndromes (16,17).
Considering these findings, we examined the
methylation status and the expression of MGMT, MLH1
and RASSF1A in 23 HNSCC biopsy samples and in one HNSCC cell
line to establish a potential role of the hypermethylated TSGs in
HNSCC development. Furthermore, we investigated the possibility of
restoring the methylation status of the TSGs by treatment with
5-Aza and the functional impact of 5-Aza treatment on proliferation
of the tumor cells.
Materials and methods
Patients and specimens
A total of 23 patients (19 males, 4 females) with
histological confirmed squamous cell carcinoma and one HNSCC cell
line were included in this study (for patient and tumor
characteristics see Table I). The
specimens obtained in the operation room were fixed in formalin for
24 h, paraffin-embedded and used for later analysis. Clinical
information was obtained from the patients charts. Patients ranged
in age from 45 to 83 (mean age at operation 62). As controls, three
samples of healthy gingiva were analyzed. This study was approved
by the Institutional Review Board and performed in accordance to
the actual version of the declaration of Helsinki. Informed consent
was obtained. All patients were operated between March 2005 and
April 2006 at the Department of Otorhinolaryngology, Head and Neck
Surgery, University Medical Center of the Johannes Gutenberg
University Mainz.
 | Table IPatient, specimen and cell line
characteristics. |
Table I
Patient, specimen and cell line
characteristics.
| Age | Gender | Site | TNM |
|---|
| Patient no. |
| 1 | 54 | Male | Larynx | T3N0M0 |
| 2 | 68 | Male | Oropharynx | T1N0M0 |
| 3 | 52 | Female | Hypopharynx | T2N2bM0 |
| 4 | 70 | Male | Floor of mouth | T1N0M0 |
| 5 | 61 | Male | Oropharynx | T2N0M0 |
| 6 | 65 | Male | Larynx | T3N0M0 |
| 7 | 65 | Male | Hypopharynx | T2N2bM0 |
| 8 | 58 | Female | Hypopharynx | T2N1M0 |
| 9 | 74 | Male | Oropharynx | T2N2cM0 |
| 10 | 77 | Male | Nasal sinus | T2N0M0 |
| 11 | 46 | Male | Oropharynx | T2N1M0 |
| 12 | 45 | Male | Oropharynx | T3N0M0 |
| 13 | 63 | Male | Hypopharynx | T2N0M0 |
| 14 | 53 | Male | Floor of mouth | T3N0M0 |
| 15 | 51 | Male | Oropharynx | T2N2bM0 |
| 16 | 64 | Male | Oropharynx | T4N2bM0 |
| 17 | 67 | Male | Tongue | T4N0M0 |
| 18 | 65 | Male | Larynx | T3N0M0 |
| 19 | 61 | Male | Oropharynx | T1N1M0 |
| 20 | 61 | Male | Larynx | T1N0M0 |
| 21 | 67 | Female | Larynx | T1N0M0 |
| 22 | 61 | Male | Hypopharynx | T3N2bM0 |
| 23 | 83 | Female | Oropharynx | T2N1M0 |
| Control no. |
| 24 | | NA | Healthy
gingiva | - |
| 25 | | NA | Healthy
gingiva | - |
| 26 | | NA | Healthy
gingiva | - |
| Cell line | | | | |
| UM-SCC 33 | | Female | Nasal sinus | T4N3aM0 |
Cell culture
For our experiments the cell line UM-SCC 33 derived
from squamous cell carcinoma of the head and neck (HNSCC) was used
(Table I) (18,19).
The cell line was maintained in DMEM/Ham's F12
(PAA), supplemented with 5% FCS (Greiner), and antibiotic solution
(penicillin 100 U/ml and streptomycin 100 μg/ml, (PAA) at 37°C in
5% CO2. The cell line was treated with 0.2 and 2 μM
5-Aza (Vidaza®) (Sigma-Aldrich) for 72 h.
DNA isolation and bisulfite
modification
Genomic DNA was extracted using the DNeasy tissue
kit (Qiagen) and 2 μg DNA each were subjected for bisulfite
treatment (20). Bisulfite
modification of genomic DNA converts unmethylated cytidine residues
to uridine residues that are then converted to thymidine during
subsequent PCR. We used the Epitect® Bisulfite kit
(Qiagen). By the use of conversion specific primers during
MSP-analysis the methylation status was then analyzed.
MSP (methylation specific PCR)
Methylation in the region near the start codon of
MGMT, MLH1 and RASSF1A was assessed using
bisulfite-treated DNA. To increase the sensitivity and specificity
we applied a two-step PCR approach. First, we amplified the TSG
promoter regions with the primers MGMT-outer-F (5′-TGG TA
ATT AAG GTA TAG AG-3′, upstream), MGMT-outer-R (5′-CCA ATC
CAC AAT CAC TCA-3′, downstream), MLH1-outer-F (5′-TTT TAG
GAG TGA AGG AGG-3′ upstream, MLH1-outer-R (5′-ATA AAA CCC
TAT ACC TAA TC-3′, downstream), RASSF1A-outer-F (5′-GAG GAG
GGG ATG AAG GAG G-3′, upstream) and RASSF1A-outer-R (5′-CTC
CAA CCA AAT ACA ACC CT-3′ downstream).
The PCR conditions were 95°C for 5 min; 40 cycles at
95°C for 30 sec, 53°C for 45 sec, and 72°C for 45 sec; and a final
extension at 72°C for 10 min. Ten microliters of each sample was
subjected to the second round of inner PCR amplified with the
followingMSP primers: for the MGMT TSG, MSP-F (5′-TTT CGA
CGT TCG TAG GTT TTC GC-3′ upstream) and MSP-R (5′-GCA CTC TTC CGA
AAA CGA AAC G-3′, downstream) and unmethylated DNA-specific primers
(UMSP), UMSP-F (5′-TTT GTG TTT TGA TGT TTG TAG GTT TTT GT-3′,
upstream) and UMSP-R (5′-AAC TCC ACA CTC TTC CAA AAA CAA AAC A-3′
downstream). For the MLH1 gene MSP analysis was performed
with the following primers: MSP-F (5′-ACG TAG ACG TTT TAT TAG GGT
CGT-3′, upstream), MSP-R (5′-CCT CAT CGT AAC TAC CCG CG-3′,
downstream), UMSP-F (5′-TTT TGA TGT AGA TGT TTT ATT AGG GTT GT-3′,
upstream) and UMSP-R (5′-ACC ACC TCA TCA TAA CTA CCC ACA-3′
downstream).
For the RASSF1A gene inner PCR was performed
with the following primers: MSP-F (5′-GGG TTT TGC GAG AGC GCG-3′,
upstream), MSP-R (5′-GCT AAC AAA GCG GAA CCG-3′ downstream), UMSP-F
(5′-GGT TTT GTG AGA GTG TGT TTA G-3′, upstream) and UMSP-R (5′-CAC
TAA CAA ACA CAA ACC AAA C-3′ downstream).
The PCR conditions were 94°C for 15 min; 40 cycles
at 94°C for 30 sec, 62°C for 30 sec, and 72°C for 30 sec; and a
final extension at 72°C for 10 min. Primers for MSP and UMSP were
as previously described (10,21,22).
The sequences of the primers were derived from sequences AL 355531
(MGMT), AC 002481 (RASSF1A) and AB 017806
(MLH1). The 81 bp and 93 bp PCR products of the MGMT
analysis as well as the 115 bp, 124 bp products of the MLH1
inner PCR and the 169 bp products of the RASSF1A analysis,
respectively, were separated by electrophoresis on a 2% agarose gel
and stained with ethidium bromide. Distilled water was used as
negative control. Bisulfite-treated lymphocyte DNA from healthy
volunteers served as a positive control for unmethylated DNA. This
DNA was methylated by the use of SssI methyltransferase
(NEB) and used after bisulfite modification as a positive control
for amplification of methylated DNA.
DNA sequencing
The order of nucleotides in the promoter region of
each gene were analyzed by DNA cycle-sequencing using the BigDye™
kit (ABI, Foster City, CA, USA). Briefly, extracted DNA samples
from the cell line before and after treatment with 5-Aza have been
modified with sodium bisulfite treatment. Amplification was
performed by the use of M13 extended outer primers of MSP-analysis.
Sequences were than determined using an ABI capillary sequencer
310.
Immunohistochemistry
Immunohistochemical analysis of tumor samples was
performed using standard procedures. In brief, formalin-fixed,
paraffin-embedded tissues were used. Heat-induced antigen retrieval
was performed using microwave treatment (3 times for 5 min each,
600 W in 10 mM citrate buffer, pH 6.0) of all slides after dewaxing
and rehydration followed by blocking of endogenous peroxidase with
3% H2O2/methanol. Pre-incubation with 10%
normal serum and 2% bovine albumin/PBS for 75 min to avoid
unspecific binding, was followed by the incubation with the
specific primary antibodies (MGMT, 1:20 BD Pharmingen, NY; MLH1,
1:20, BD Pharmingen; RASSF1A, 1:50, Genway, CA) for 1 h at room
temperature. The slides were consecutively incubated with
biotinylated secondary antibody (goat-anti-mouse, 1:250, Dako A/S,
Glostrup Denmark) for 45 min and then for 30 min with
streptavidin-peroxidase. The visualization of the immunoreaction
was performed with 3,3′-diaminobenzidine. All washing procedures
were performed in PBS; dilutions of antibodies were prepared in 2%
bovine albumin/PBS at room temperature. Negative controls were
performed as previously described, substituting the primary
antibody with PBS.
Quantification of the expression
For evaluation of the MGMT-expression in tumor
samples we measured the stained area and intensity of each section
in five fields by a computer-based image analysis method,
previously described in detail by us (23). In brief, stainings were quantified
by the multiplication of the stained area by the staining intensity
and expressed as arbitrary units (A.U.).
Alamar blue assay-proliferation
For determination of 5-Aza mediated functional
consequences we incubated the cell line UM-SCC 33 for 72 h with 0.2
or 2 μM 5-Aza. Briefly, media were changed and 10% v/v Alamar Blue
reagent (Biozol) was added to each well. The Alamar
Blue® assay is based on a redox indicator, changing its
color from blue (oxidized) to fluorescent red (reduced). After 4-h
fluorescence was measured. Color changes are a measure of cellular
metabolism, corresponding to the viability and proliferative
activity of the cells (24). Each
experiment was repeated three times.
Statistics
A one-sided t-test was applied to assess the
statistical significance. All calculations were performed using the
SAS software, version 6.12 (Statistical Analysis Systems, SAS
Institute Inc., Cary, NC, USA). p-values <0.05 are
indicated.
Results
To examine if the promoters of MGMT,
MLH1 and RASSF1A in HNSCC are methylated, we analyzed
these tumor suppressor genes for their methylation status in 23
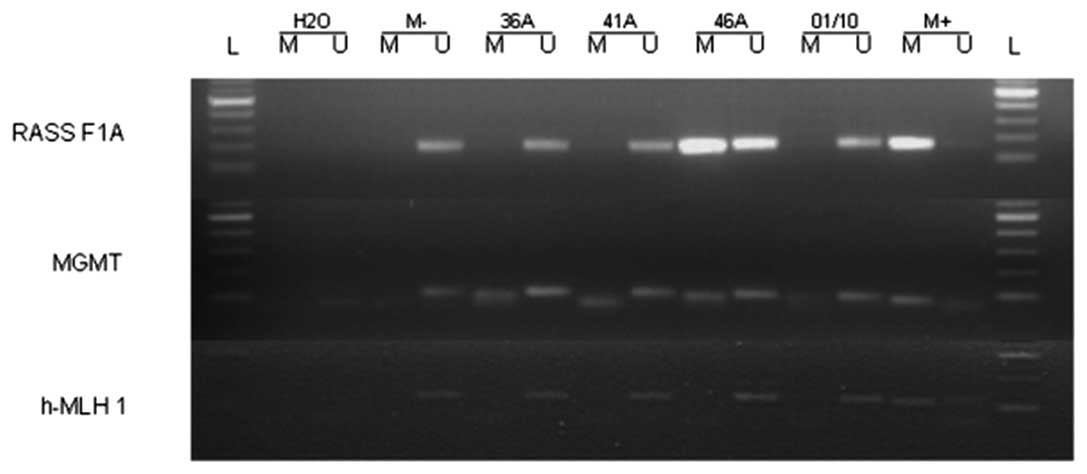
samples of primary HNSCC and one HNSCC cell line by MSP (Table II). We found that the MGMT
promoter was methylated in 13 out of the 23 (57%) analyzed primary
tumor samples and in the examined cell line. The MLH1
promoter was found to be methylated in one out of the 23 (4%) tumor
samples while the MLH1 promoter in the UM-SCC cell line was
unmethylated. MSP analysis of the RASSF1A promoter showed
promoter methylation in 3 out of 23 (13%) tumor samples and
promoter methylation in the cell line (Table II). Fig. 1 shows representative examples of the
MSP-analysis. To compare the methylation status, we additionally
analyzed three samples of healthy gingiva by MSP-PCR. We found that
in all three samples the promoter region of MLH1,
RASSF1A and MGMT was unmethylated. DNA sequencing
verified the results of the MSP analysis in the UM-SCC 33 cell
line. To analyze a possible association of hypermethylated promoter
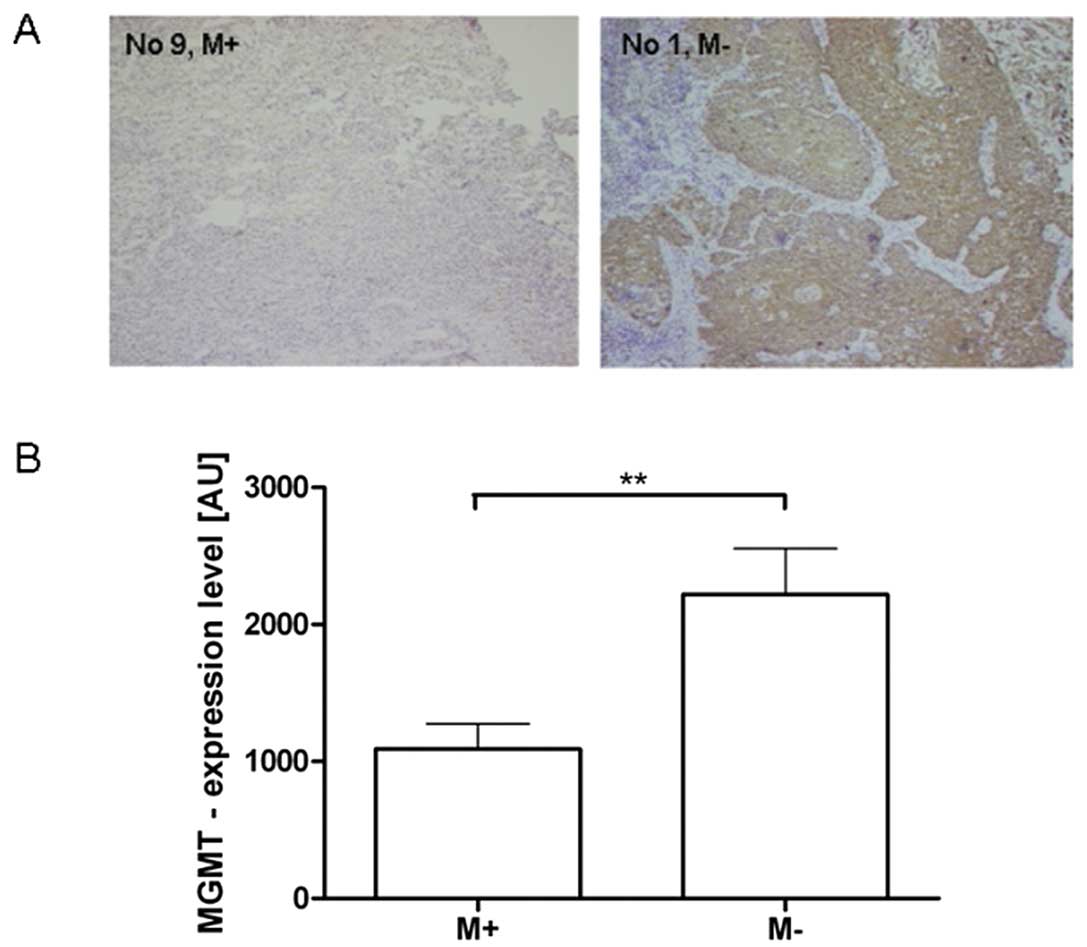
regions and decreased expression levels, we quantified the
MGMT-levels by semi-quantative immunohistochemical analysis.
 | Table IIAnalysis of RASSF1A,
MLH1 and MGMT promoter methylation by MSP in primary
HNSCC and cell lines and MGMT expression. |
Table II
Analysis of RASSF1A,
MLH1 and MGMT promoter methylation by MSP in primary
HNSCC and cell lines and MGMT expression.
| RASSF1A
methylation | MLH1
methylation | MGMT
methylation | MGMT expression
(A.U) |
|---|
| Patient no. |
| 1 | − | − | − | 3286 |
| 2 | − | − | − | 2596 |
| 3 | − | − | − | 2788 |
| 4 | + | − | − | NA |
| 5 | − | − | − | 1238 |
| 6 | − | − | − | 3485 |
| 7 | − | − | + | 2550 |
| 8 | − | − | + | 1318 |
| 9 | − | − | + | 532 |
| 10 | − | − | − | 2818 |
| 11 | − | − | + | 884 |
| 12 | − | − | + | 1491 |
| 13 | − | − | + | 709 |
| 14 | − | − | + | 865 |
| 15 | − | − | − | 1625 |
| 16 | − | − | + | 478 |
| 17 | − | − | − | 565 |
| 18 | + | − | + | NA |
| 19 | − | + | + | 952 |
| 20 | + | − | − | 1576 |
| 21 | − | − | + | 617 |
| 22 | − | − | + | NA |
| 23 | − | − | + | 1604 |
| Control no. |
| 24 | − | − | − | |
| 25 | − | − | − | |
| 26 | − | − | − | |
| Cell line |
| UM-SCC 33 | − | − | − | |
MGMT expression varied from 478 to 3485 A.U.
(mean 1.599±976 A.U.). MGMT expression statistically
significantly (p<0.01) decreased in the tumor samples with a
hypermethylated MGMT promoter region (Fig. 2). Due to limited tumor samples with
hypermethylated RASSF1A and MLH1 promoter regions, no
correlation analysis of hypermethylated promoter regions and
expression levels was performed.
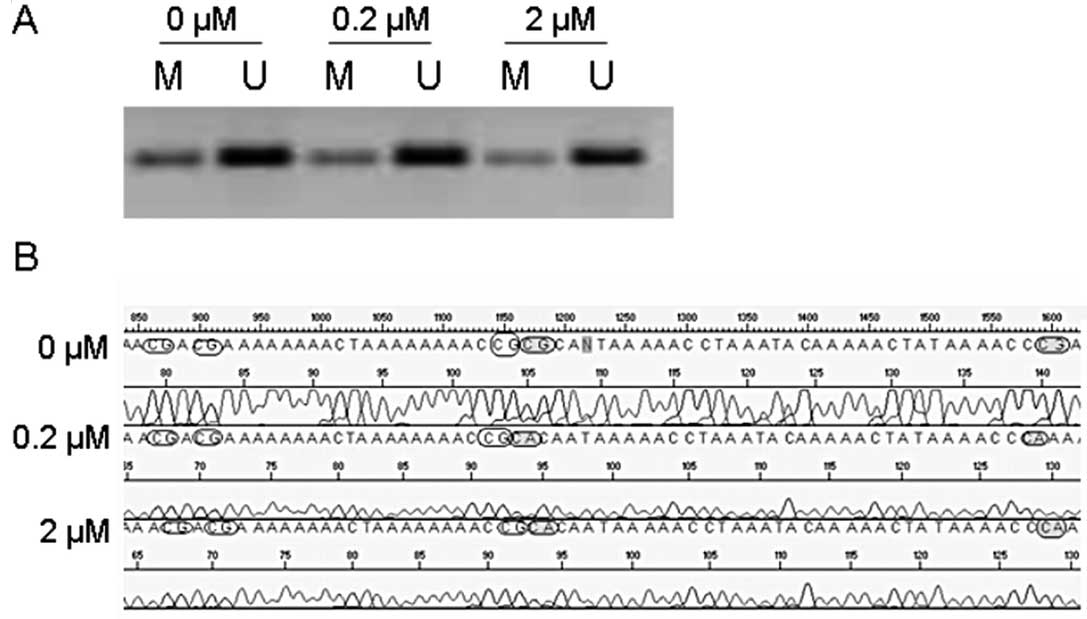
After treatment of the UM-SCC33 cell line with 5-Aza
for 72 h we observed no demethylating effect by MSP analysis
(representative result is shown in Fig.
3A). However, sequence analysis showed that 5-Aza treatment led
to an increased number of unmethylated CpG islands in the
methylated promoter region of RASSF1A in the UM-SCC33 cell
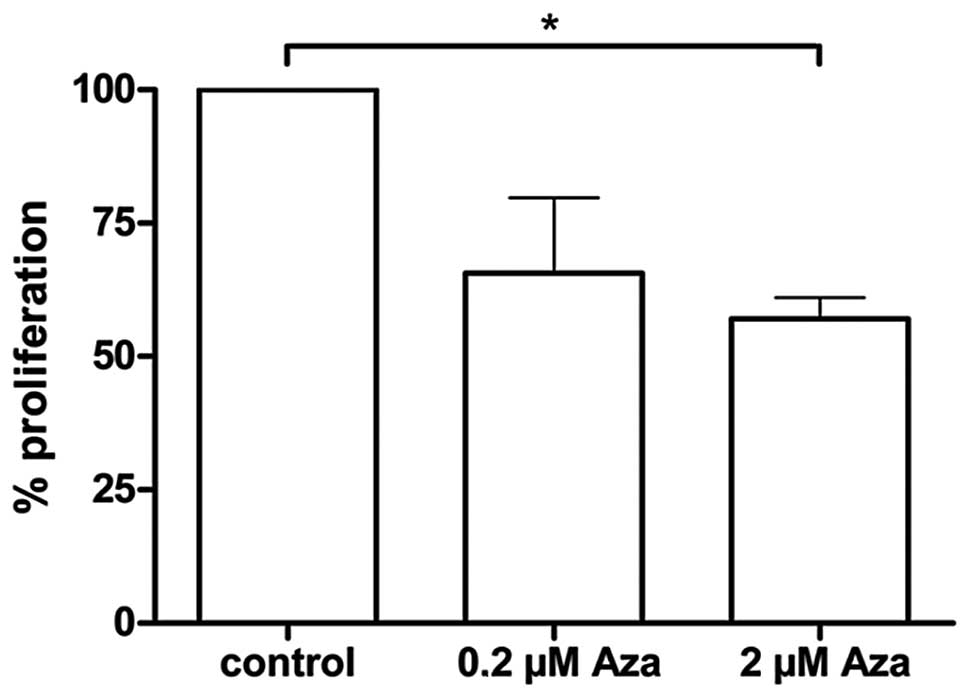
line (Fig. 3B). The MTT assay
revealed a tendency for reduced tumor cell proliferation 4 h after
treatment with 5-Aza for 72 h at a concentration of 0.2 μM (not
significant) and a statistically significant reduction (p<0.05)
of the proliferation at the same point of time with the 2 μM 5-Aza
concentration (Fig. 4).
Discussion
Aberrations of genomic material as well as
epigenetic modifications of genome-like promoter hypermethylations
may result in the deregulation of tumor suppressor genes and
finally in cancer (14). In HNSCC
recurrent chromosomal losses have been described (3,4,14)
suggesting the involvement of several tumor suppressor genes in the
genesis of HNSCC. In this study we analyzed the methylation status
of three potential TSGs. MLH1 is a mismatch repair gene,
located at 3p, a chromosomal arm showing high level of allelic
losses in many cases of malignancies (9,12).
Several studies reported deletion or hypermethylation of the gene
in hereditary colon cancer, gastric, endometrium, prostate but also
HNSC cancer (21). The TSG
RASSF1A, also located at 3p, plays a major role in the
regulation of mitosis and it has been reported to be methylated in
the vast majority of lung cancers and to a lesser extent in breast,
ovarian and HNSC cancer (8,11,22).
MGMT is a TSG, located at 10q, playing vital roles in
preventing induction of mutations and cancer related to alkylating
agents (10,13). Aberrations of MGMT have been
reported in lung, colon, brain, liver and HNSC cancer (10).
We analyzed the methylation status of these three
TSGs in tumor samples from 23 patients with HNSCC and in one cell
line by MSP PCR. We found that the MLH1 promoter was
methylated in one tumor (4%) while no methylation could be observed
in the cell line.
A direct association between hypermethylation of
MLH1 and cancer has been reported in colon cancer but
reports on the methylation status of the same gene in HNSCC are
inconclusive and have ranged from 0 to 88%. High percentage (88%)
of MLH1 methylation was reported by Liu et al but
only in samples previous found to have loss of expression of the
gene product (25). Steinmann et
al reported that 69% of the 54 HNSCC samples showed
hypermethylation of the MLH1 promoter (26), while two studies with 96 and 57
samples did not find methylation at all (27,28).
Thus, our results concerning the MLH1 promoter methylation
are in agreement with other reports. The widely divergent findings
reported in the literature may be attributed to different
sensitivities of the techniques as well as to insufficient DNA
quality because of tissue preservation or bisulfite treatment.
Furthermore, Wright and Stewart argued the importance of
histological grading for MLH1 expression, reporting that a
high sample number (70%) lacked MLH1 expression in poorly
differentiated colon adenocarcinoma. Thus, the reported MLH1
methylation status may also depend on the variety of the
histological grading of a study group in HNSCC (29). Yamamoto et al reported a high
risk of developing secondary carcinoma in the gastrointestinal
tract, in patients with defective protein expression of MLH1
(30), while in a further study a
significant correlation between the methylation of mismatch repair
genes and multiple oral malignancies was found (31). Since our patient cohort consisted
from patients with solitary tumors, that could be a further reason
for the very rare MLH1 methylation reported in our study.
Taken together, the above data suggest that aberrant MLH1
methylation-mediated transcriptional silencing might play a role in
the development of multiple synchronous or metachronous
malignancies, but it may be of minor importance for the development
of solitary HNSCC lesions.
The methylation of the RASSF1A promoter was
also rare in 3 out of 23 (13%) tumor samples while the
RASSF1A promoter was found methylated in the UM-SCC 33 cell
line. These results are in accordance to the study of Dong et
al who reported of RASSF1A methylation in 15% of primary
HNSCC and higher frequency in cell lines (32) and to Hogg et al who reported
of 17% RASSF1A methylation in HNSCC while poorly
differentiated HNSCC were more commonly methylated for
RASSF1A than moderately and well differentiated HNSCC
(8). Steinmann et al also
report of 18% RASSF1A methylation in an analysis of 54 HNSCC
tumor samples (26). Notably, Lo
et al have reported 14 of 21 primary nasopharyngeal
carcinomas to show RASSF1A promoter methylation (33). This difference in the prevalence of
the RASSF1A methylation may reflect the known differences in
the disease. Hence epigenetic inactivation of RASSF1A plays
an important role in the development of cancer but is apparently
less important in HNSCC where genes other than RASSF1A may
be of greater importance.
The MGMT TSG was found in 13 out of the 23
(57%) analyzed primary tumor samples as well as in the examined
cell line methylated. Accordingly, we found statistically
significantly decreased protein levels of MGMT in the
hypermethylated tumors. In the literature the incidence of
MGMT promoter hypermethylation in HNSCC ranged from 18% to
54% (26,34,35).
Frequent MGMT methylation can increase the sensitivity
towards the mutagenic effects of DNA alkylating agents like
cigarette smoke nitrosamines. On the other hand, it can facilitate
the cytotoxic effects of DNA alkylating chemotherapeutics in
patients with malignant astrocytomas, glioma and diffuse large
B-cell lymphoma (36,37).
In gastric cancer it has been previously reported
that a correlation between MGMT methylation and lymph node
metastasis exists (38). It can be
hypothesized that in HNSCC the epigenetic loss of MGMT
function may increase the mutation rates as a result of an impaired
repair of DNA damage induced by cigarette smoke nitrosamines. This
may facilitate the cell to acquire an enhanced migration potential
and invasiveness. In our patient cohort the MGMT
hypermethylated samples were equally divided between nodal positive
and nodal negative patients.
The frequent MGMT promoter hypermethylation
in our study is in line with the results previously described. Our
data suggest that although MGMT silencing by promoter
hypermethylation might not be important for the development of
nodal metastasis in HNSCC, it might play a role in the
tumorigenesis of HNSCC, by increasing the sensitivity towards the
mutagenic effects of DNA alkylating agents.
After treatment of the cell line with 5-Aza, no
demethylating effect could be shown by MSP. DNA-sequencing of the
promoter region of the three TSG in the UM-SCC 33 cell line, after
treatment with 5-Aza showed an increase in the number of
unmethylated CpG islands in the methylated promoter region of the
RASSF1A and MGMT TSG in the UM-SCC 33 cell line. This
can be explained through the higher sensitivity of DNA-sequencing
to reveal such minor changes. The ineffective demethylation results
are supported by other studies (39,40).
Costello et al reported that only around 40% of the
aberrantly methylated genes can be reactivated by 5-Aza in cultured
cancer cell lines (41). Karpf and
Jones (42) reported a similar
result in colon cell lines, and they suggested that 5-Aza-treated
tumor cell lines may no longer express activating factors required
for the transcription of these particular CpG-island carrying
genes.
This partial promoter demethylation was accompanied
in our study by a significant decrease of proliferative activity.
The observed demethylating effect of 5-Aza on the proliferative
activity of UM-SCC 33 tumor cell line cannot be attributed only to
demethylation of MGMT and RASSF1A promoter, since
further hypermethylated TSGs might also be involved. Nevertheless,
5-Aza treatment reduces the proliferative activity of tumor cells
with hypermethylated TSG promoter in vitro and shows that
5-Aza treatment may be feasible in the therapy of HNSCC patients
with TSG promoter hypermethylation.
The value of demethylating treatment with 5-Aza has
previously been shown for hematologic disorders (16,17).
In these diseases the reactivation of the TSG p15, an inhibitor of
cyclin-dependent kinases, is assumed to be the underlying mode of
action. Even though our data revealed only limited demethylating
effects through 5-Aza demethylating treatment in HNSCC patients
with hypermethylation of TSGs, further studies are warranted to
elucidate the potential use of demethylating treatment concepts in
the treatment of HNSCC.
Acknowledgements
We are greatly indebted to Bettina Mros for her
excellent technical assistance. This study was funded by a grant
provided by the Foundation Tumour Research Head and Neck,
Wiesbaden, Germany. The foundation is a non-profit organization.
The funders played no role in the experiment design, execution,
analysis or preparation of the study.
References
|
1
|
Vokes EE, Weichselbaum RR, Lippman SM and
Hong WK: Head and neck cancer. N Engl J Med. 328:184–194. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics. CA Cancer J Clin. 60:277–300. 2010.
|
|
3
|
Gollin SM: Chromosomal alterations in
squamous cell carcinomas of the head and neck: window to the
biology of disease. Head Neck. 23:238–253. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brieger J, Jacob R, Riazimand HS, Essig E,
Heinrich UR, Bittinger F and Mann WJ: Chromosomal aberrations in
premalignant and malignant squamous epithelium. Cancer Genet
Cytogenet. 144:148–155. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ha PK and Califano JA: Promoter
methylation and inactivation of tumour suppressor genes in oral
squamous cell carcinoma. Lancet Oncol. 7:77–82. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esteller M, Corn PG, Baylin SB and Herman
JG: A gene hypermethylation profile of human cancer. Cancer Res.
61:3225–3229. 2001.PubMed/NCBI
|
|
7
|
Brieger J, Pongsapich W, Mann SA, Hedrich
J, Fruth K, Pogozelski B and Mann WJ: Demethylation treatment
restores hic1 expression and impairs aggressiveness of head and
neck squamous cell carcinoma. Oral Oncol. 46:678–683. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hogg RP, Honorio S, Martinez A,
Agathanggelou A, Dallol A, Fullwood P, Weichselbaum R, Kuo MJ,
Maher ER and Latif F: Frequent 3p allele loss and epigenetic
inactivation of the RASSF1A tumour suppressor gene from region
3p21.3 in head and neck squamous cell carcinoma. Eur J Cancer.
38:1585–1592. 2002. View Article : Google Scholar
|
|
9
|
Viswanathan M, Tsuchida N and Shanmugam G:
Promoter hypermethylation profile of tumor-associated genes p16,
p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma.
Int J Cancer. 105:41–46. 2003. View Article : Google Scholar
|
|
10
|
Kato K, Hara A, Kuno T, Mori H, Yamashita
T, Toida M and Shibata T: Aberrant promoter hypermethylation of p16
and MGMT genes in oral squamous cell carcinomas and the surrounding
normal mucosa. J Cancer Res Clin Oncol. 132:735–743. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van der Weyden L, Tachibana KK, Gonzalez
MA, Adams DJ, Ng BL, Petty R, Venkitaraman AR, Arends MJ and
Bradley A: The RASSF1A isoform of RASSF1 promotes microtubule
stability and suppresses tumorigenesis. Mol Cell Biol.
25:8356–8367. 2005.PubMed/NCBI
|
|
12
|
Jiricny J and Nyström-Lahti M: Mismatch
repair defects in cancer. Curr Opin Genet Dev. 10:157–161. 2000.
View Article : Google Scholar
|
|
13
|
Pegg AE: Mammalian O6-alkylguanine-DNA
alkyltransferase: regulation and importance in response to
alkylating carcinogenic and therapeutic agents. Cancer Res.
50:6119–6129. 1990.PubMed/NCBI
|
|
14
|
Baylin SB, Esteller M, Rountree MR,
Bachman KE, Schuebel K and Herman JG: Aberrant patterns of DNA
methylation, chromatin formation and gene expression in cancer. Hum
Mol Genet. 10:687–692. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Belinsky SA, Liechty KC, Gentry FD, Wolf
HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, et
al: Promoter hypermethylation of multiple genes in sputum precedes
lung cancer incidence in a high-risk cohort. Cancer Res.
66:3338–3344. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Daskalakis M, Nguyen TT, Nguyen C,
Guldberg P, Kohler G, Wijermans P, Jones PA and Lubbert M:
Demethylation of a hypermethylated P15/INK4B gene in patients with
myelodysplastic syndrome by 5-aza-2′-deoxycytidine (decitabine)
treatment. Blood. 100:2957–2964. 2002.PubMed/NCBI
|
|
17
|
Ruter B, Wijermans PW and Lubbert M: DNA
methylation as a therapeutic target in hematologic disorders:
recent results in older patients with myelodysplasia and acute
myeloid leukaemia. Int J Hematol. 80:128–135. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Welkoborsky HJ, Jacob R, Riazimand SH,
Bernauer HS and Mann WJ: Molecular biologic characteristics of
seven new cell lines of squamous cell carcinomas of the head and
neck and comparison to fresh tumor tissue. Oncology. 65:60–71.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin CJ, Grandis JR, Carey TE, Gollin SM,
Whiteside TL, Koch WM, Ferris RL and Lai SY: Head and neck squamous
cell carcinoma cell lines: established models and rationale for
selection. Head Neck. 29:163–188. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nan HM, Song YJ, Yun HY, Park JS and Kim
H: Effects of dietary intake and genetic factors on
hypermethylation of the hMLH1 gene promoter in gastric cancer.
World J Gastroenterol. 11:3834–3841. 2005.PubMed/NCBI
|
|
22
|
Burbee DG, Forgacs E, Zöchbauer-Müller S,
Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S,
et al: Epigenetic inactivation of RASSF1A in lung and breast
cancers and malignant phenotype suppression. J Natl Cancer Inst.
93:691–699. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gosepath J, Brieger J, Lehr HA and Mann
WJ: Expression, localization and significance of vascular
permeability/vascular endothelial growth factor in nasal polyps. Am
J Rhinol. 19:7–13. 2005.PubMed/NCBI
|
|
24
|
Ahmed SA, Gogal RM and Walsh JE: A new
rapid and simple non-radioactive assay to monitor and determine the
proliferation of lymphocytes: an alternative to
[3H]thymidine incorporation assay. J Immunol Methods.
170:211–224. 1994.PubMed/NCBI
|
|
25
|
Liu K, Zuo C, Luo QK, Suen JY, Hanna E and
Fan CY: Promoter hypermethylation and inactivation of hMLH1, a DNA
mismatch repair gene, in head and neck squamous cell carcinoma.
Diagn Mol Pathol. 12:50–56. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Steinmann K, Sandner A, Schagdarsurengin U
and Dammann RH: Frequent promoter hypermethylation of tumor-related
genes in head and neck squamous cell carcinoma. Oncol Rep.
22:1519–1526. 2009.PubMed/NCBI
|
|
27
|
Ogi K, Toyota M, Ohe-Toyota M, Tanaka N,
Noguchi M, Sonoda T, Kohama G and Tokino T: Aberrant methylation of
multiple genes and clinicopathological features in oral squamous
cell carcinoma. Clin Cancer Res. 8:3164–3171. 2002.PubMed/NCBI
|
|
28
|
Wang Y, Irish J, MacMillan C, Brown D,
Xuan Y, Boyington C, Gullane P and Kamel-Reid S: High frequency of
microsatellite instability in young patients with head-and-neck
squamous-cell carcinoma: lack of involvement of the mismatch repair
genes hMLH1 and hMSH2. Int J Cancer. 93:353–360. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wright CL and Stewart ID: Histopathology
and mismatch repair status of 458 consecutive colorectal
carcinomas. Am J Surg Pathol. 27:1393–1406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamamoto M, Taguchi K, Baba H, Endo K,
Kohnoe S, Okamura T and Maehara Y: Loss of protein expression of
hMLH1 and hMSH2 with double primary carcinomas of the stomach and
colorectum. Oncol Rep. 16:41–47. 2006.PubMed/NCBI
|
|
31
|
Czerninski R, Krichevsky S, Ashhab Y,
Gazit D, Patel V and Ben-Yehuda D: Promoter hypermethylation of
mismatch repair genes, hMLH1 and hMSH2 in oral squamous cell
carcinoma. Oral Dis. 15:206–213. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dong SM, Sun DI, Benoit NE, Kuzmin I,
Lerman MI and Sidransky D: Epigenetic inactivation of RASSF1A in
head and neck cancer. Clin Cancer Res. 9:3635–3640. 2003.PubMed/NCBI
|
|
33
|
Lo KW, Kwong J, Hui AB, Chan SY, To KF,
Chan AS, Chow LS, Teo PM, Johnson PJ and Huang DP: High frequency
of promoter hypermethylation of RASSF1A in nasopharyngeal
carcinoma. Cancer Res. 61:3877–3881. 2001.PubMed/NCBI
|
|
34
|
Zuo C, Ai L, Ratliff P, Suen JY, Hanna E,
Brent TP and Fan CY: O6-methylguanine-DNA methyltransferase gene:
epigenetic silencing and prognostic value in head and neck squamous
cell carcinoma. Cancer Epidemiol Biomarkers Prev. 13:967–975.
2004.PubMed/NCBI
|
|
35
|
Paluszczak J, Misiak P, Wierzbicka M,
WoŸniak A and Baer-Dubowska W: Frequent hypermethylation of DAPK,
RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell
carcinomas and adjacent normal mucosa. Oral Oncol. 47:104–107.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Esteller M, Gaidano G, Goodman SN, Zagonel
V, Capello D, Botto B, Rossi D, Gloghini A, Vitolo U, Carbone A, et
al: Hypermethylation of the DNA repair gene O(6)-methylguanine DNA
methyltransferase and survival of patients with diffuse large
B-cell lymphoma. J Natl Cancer Inst. 94:26–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Esteller M, Garcia-Foncillas J, Andion E,
Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB and Herman JG:
Inactivation of the DNA-repair gene MGMT and the clinical response
of gliomas to alkylating agents. N Engl J Med. 343:1350–1354. 2000.
View Article : Google Scholar
|
|
38
|
Park TJ, Han SU, Cho YK, Paik WK, Kim YB
and Lim IK: Methylation of O(6)-methylguanine-DNA methyltransferase
gene is associated significantly with K-ras mutation, lymph node
invasion, tumor staging, and disease free survival in patients with
gastric carcinoma. Cancer. 92:2760–2768. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cameron EE, Bachman KE, Myöhänen S, Herman
JG and Baylin SB: Synergy of demethylation and histone deacetylase
inhibition in the re-expression of genes silenced in cancer. Nat
Genet. 21:103–107. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shen WJ, Dai DQ, Teng Y and Liu HB:
Regulation of demethylation and re-expression of RASSF1A gene in
gastric cancer cell lines by combined treatment of 5-Aza-CdR and
NaB. World J Gastroenterol. 14:595–600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Costello JF, Frühwald MC, Smiraglia DJ,
Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P,
Lang JC, et al: Aberrant CpG-island methylation has non-random and
tumour-type-specific patterns. Nat Genet. 24:132–138. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Karpf AR and Jones DA: Reactivating the
expression of methylation silenced genes in human cancer. Oncogene.
21:5496–5503. 2002. View Article : Google Scholar : PubMed/NCBI
|