Introduction
microRNA-21 negatively regulates several targets,
and thus impacts tumorigenesis. However, the exact mechanism of
action in human gastric carcinoma is poorly understood, and there
is currently no direct evidence that demonstrates a correlation
between microRNA-21 function and phenotype. In this study, we
investigate the function of microRNA-21 as a potent oncomir and
probe the relationship between microRNA-21, the targets of
microRNA-21, and the phenotypic alterations. microRNAs (miRNAs) are
small, non-coding RNAs of 18- to 23-nucleotides found in a diverse
array of organisms. They have a broad impact on gene expression
through translational repression or post-transcriptional
suppression (1). microRNAs play an
important role in numerous biological processes, such as
development, differentiation, and the cellular stress response
(2–4). Recent studies have linked deregulation
of miRNAs to various diseases including cancer (5,6).
Gastric cancer is the most common malignancy in China and the
fourth most common cancer world-wide; moreover, the prognosis for
gastric cancer is quite poor. Globally, it is the second and fourth
leading cause of cancer-related death in men and women,
respectively (7). Despite progress
in the development of new management strategies, gastric cancer
remains difficult to diagnose at an early stage. According to the
International Union Against Cancer (UICC) and American Joint
Committee on Cancer (AJCC) staging systems (8), nearly 65% of patients in the USA are
initially diagnosed with gastric cancer at an advanced stage
(T3/T4), and 85% of patients have lymph node metastasis (9). The mean survival time for these
patients is 24 months and the 5-year survival rate is only 20–40%
after surgery (10). Thus, the
discovery of new biomarkers is of critical importance to allow for
early diagnosis of gastric cancer.
Reports have indicated that dysregulation of miRNAs
is associated with the formation and progression of gastric cancer
(11,12). Therefore, miRNAs are potentially
useful biomarkers for clinical diagnosis. Dramatic upregulation of
microRNA-21 (miR-21) has been reported in numerous types of cancer
(13–15). Therefore, miR-21 is recognized as an
oncomir. Several targets of miR-21 have been experimentally
validated, including PDCD4 (16)
and RECK (17); however, ectopic
expression of these targets may exert differing functional effects
on tumorigenesis. Moreover, uncontrolled proliferation, lack of
apoptosis, and invasiveness, which are all modulated by miR-21,
have also been identified in tumorigenesis, leaving several
questions unanswered. For example, PTEN is reported to be a direct
target of miR-21 in HCC, but the phenotype alteration caused by
miR-21-mediated PTEN regulation remains unclear. In this study, the
mechanism of oncomir miR-21 in gastric cancer was studied with
respect to the regulation of PTEN. We discovered that miR-21
expression was markedly increased in gastric cancer tissues
compared to normal tissues. More importantly, we demonstrated that
miR-21 promoted gastric cancer cell proliferation by directly
targeting PTEN, thereby increasing gastric cancer cell invasiveness
by directly targeting PTEN.
Materials and methods
Cell lines, cell culture, and human
tissue samples
The human gastric cancer cell lines SGC-7901,
MKN-28, MKN-45, and AGS were purchased from Shanghai Institutes for
Biological Sciences, Chinese Academy of Sciences, and NCI-N87,
BGC-823, HTB-103, CRL-5974, and CRL-5971 were purchased from the
American Type Culture Collection (ATCC). The immortalized normal
gastric mucosal epithelial cell line GES-1 was a gift from
Professor Feng Bin (Sichuan University, Chengdu, China). The cells
were routinely cultured in RPMI-1640 supplemented with 10%
heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin and
100 μg/ml streptomycin in a humidified cell incubator with an
atmosphere of 5% CO2 at 37°C. Cells growing at an
exponential rate were used for the experiments.
Fresh frozen human tumor samples and human
non-neoplastic gastric tissues were obtained from 30 patients with
gastric cancer undergoing radical gastrectomy at the Department of
Surgery, Ruijin Hospital Shanghai Jiao Tong University School of
Medicine from May 2005 to August 2008. None of the patients had
received preoperative treatment, such as radiation therapy or
chemotherapy. Sections from each specimen were independently
examined by two pathologists, and histological typing was performed
using Lauren’s classification. TNM classification of malignant
tumors was assigned in accordance to the International Union
Against Cancer (1997).
RNA isolation and miRNA cloning
Total RNA was extracted and isolated from tissue
samples or cell lines using either the mirVana miRNA isolation kit
(Ambion, Austin, TX) or the TRIzol method. The quality and quantity
of the RNA samples were assessed by standard electrophoresis and
spectrophotometric methods. The expression level of mature miR-21
was measured by qRT-PCR according to the TaqMan®
MicroRNA Assay protocol (Applied Biosystems) and normalized using
U6 small nuclear RNA (RNU6B; Applied Biosystems) by the
2−ΔCt method. The relative expression ratio of miR-21 in
each paired tumour to non-tumour tissue sample was calculated using
the 2−ΔΔCt method. The miR-21 expression level was
defined as being up-regulated in tumour tissue with a relative
expression ratio >1, and was defined as downregulated in tumour
tissue with a relative expression ratio <1.
Transfection with antisense
oligonucleotides
The stability-enhanced miRNA precursor that mimicks
miR-21 and the control non-specific miRNA precursor (pre-miR
precursor, negative control) and the miR-21 inhibitor (miR-21
inhibitor, negative control) were purchased from Shanghai
GenePharma Co., Ltd. BGC-823 cells were trypsinised, counted, and
seeded onto 6-well plates the day prior to transfection to ensure
50% cell confluence on the day of transfection. Transfection of
miRNA precursors/inhibitors into BGC-823 cells was performed using
Lipofectamine 2000 (Invitrogen) in accordance with the
manufacturer’s advised procedure. The miRNA precursors/inhibitors
were used at a final concentration of 100 nM. At 48 h
post-transfection, qRT-PCR and western blot analysis were
performed. Transfection efficiency was monitored by the
transfection of Cy3-labeled pre-miR™ negative control #1
(Ambion).
Cell proliferation assay
Cell proliferation was monitored by the colorimetric
water-soluble tetrazolium salt (CCK8) assay using a Cell Counting
Kit-8 (Dojindo) according to the manufacturer’s instructions. At 24
h post-transfection with miR-21 pre-cursor/inhibitor or control
oligotides, BGC-823 cells were seeded onto 96-well plates
(2×103 cells/well), and cell proliferation was
documented every 24 h for 4 days. The number of viable cells was
assessed by measurement of the absorbance at 450 nm using a
Safire2 microplate reader (TECAN).
Cell cycle and apoptosis analysis
At 48 h post-transfection with the miR-21
precursor/inhibitor or control precursor (100 nM), BGC-823 cells
were collected by trypsinisation and washed with phosphate-buffered
saline (PBS). For cell cycle analysis, the cells were fixed with
75% ethanol and stored at 4°C overnight. The following day, fixed
cells were washed with PBS, treated with RNase A (50 μg/ml), and
stained with propidium iodide (PI) (50 μg/ml) for 30 min in the
dark. The stained cells were analyzed by flow cytometry
(FACSCalibur, Becton-Dickinson). The cell debris and fixation
artifacts were gated out and the cell populations at the G0/G1, S,
and G2/M phases were quantified using the Flowjo7.6.2 (Treestar).
At least 10,000 cells in each sample were analyzed to obtain a
measurable signal. For apoptosis analysis, an Annexin-V-FITC
Apoptosis Detection Kit I (BD Pharmingen) was used according to the
manufacturer’s instructions. In brief, cells were washed with PBS
and resuspended in 1X binding buffer at a concentration of
1×106 cells/ml; next, 5 μl of FITC Annexin-V and 5 μl PI
were added to 100 μl of the cell suspension and the samples were
incubated for 15 min in the dark, after incubation, 400 μl 1X
binding buffer was added. Apoptosis was analyzed by flow cytometry
(FACSCalibur, Becton-Dickinson) using the Cell-Quest software
(Becton-Dickinson). The cells undergoing apoptosis were
Annexin-V-FITC-positive and PI-negative.
Cell migration assay
Migration of BGC-823 cells was assessed using the
QCM™ 24-Well Colorimetric Cell Migration Assay Kit (Millipore)
according to the manufacturer’s instructions. Briefly, at 24 h
post-transfection with miR-21 inhibitor or control (100 nM),
2×104 BGC-823 cells in 300 μl serum-free medium were
added to the upper chamber. A volume of 0.5 ml of 10%
FBS-containing medium was then added to the lower chamber as a
chemoattractant. Cells were incubated for another 36 h at 37°C, and
non-migrating cells on the upper surface of the membrane were then
scraped off with cotton swabs. Cells that migrated to the bottom of
the membrane were stained for 30 min with the cell stain provided
in the assay kit. Stained cells were visualised under a microscope.
To minimize the bias, at least three randomly selected fields were
quantified using ×100 magnification, and the average number of
cells was taken. For scratch wound-healing motility assays, BGC-823
cells were seeded on 6-well plates and allowed to grow to
confluence. Confluent monolayers were scratched with a pipette tip
and maintained under standard conditions for 24–48 h. Plates were
washed once with fresh medium to remove non-adherent cells, and
then photographed. Cells were treated with precursor (100 nM) for
24 h before wounding as well as throughout the assay period.
Cloning of 3′UTR of PTEN into pMIR-REPORT
luciferase vector
Total-RNA from BGC-823 cells was initially reverse
transcribed into cDNA with Oligo(dT)16, which was used
as the template, and wild-type and mutant PTEN 3′-UTRs were
amplified by 5′-GGCACTAGTTATACTGGT
TCACATCCTACCCCTTTGCACTTGTGGCAACAGAT AAGTTTGCAGTTGGCTAAGAGAGGTT-3′,
and 5′-GAT AAGCTTCATTCCCCTAACCCGAATACATGCATTAG
AATGTAGCAAAACCCTTCGGAAACCTCTCTTAGCC AACTGC-3′;
5′-GGCACTAGTTATACTGGTTCAC ATCCTACCCCTTTGCACTTGTGGCAACAGCTGAAT
CTGCAGTTGGCTAAGAGAGGTT-3′, and 5′-GATAAG
CTTCATTCCCCTAACCCGAATACATGCATTAGAAT
GTAGCAAAACCCTTCGGAAACCTCTCTTAGCCAAC TGC-3′. The final pieces of
wild-type and mutant PTEN 3′-UTRs were cloned into the SpeI
and HindIII sites of the pMIR-REPORT luciferase vector
(Ambion) and named pMIR/PTEN and pMIR/PTEN/mut, respectively. Both
constructs were verified by sequencing.
Luciferase activity assay
BGC-823 cells were cultured in 6-well plates, and
each was transfected with 1 μg of either pMIR/PTEN vector or
pMIR/PTEN/mut vector containing Firefly luciferase along with 0.05
μg of the pRL-TK vector (Promega) containing Renilla luciferase and
30 nM miR-21 inhibitor or control oligonucleotide. Transfection was
performed using Lipofectamine 2000 (Invitrogen). At 24 h
post-transfection, relative luciferase activity was calculated by
normalising the Firefly luminescence to the Renilla luminescence
using the Dual-Luciferase Reporter Assay (Promega) according to the
manufacturer’s instructions.
Western blot analysis and
immunohistochemistry
Cultured cells were lysed using RIPA buffer (Pierce)
in the presence of Protease Inhibitor Cocktail (Pierce). Tissue
samples were lysed using the T-PER Tissue Protein Extraction
Reagent (Pierce) in the presence of Protease Inhibitor Cocktail
(Pierce). The protein concentration of the lysates was measured
using a BCA Protein Assay Kit (Pierce). Equivalent amounts of
protein were resolved and mixed with 5X Lane Marker Reducing Sample
Buffer (Pierce), electrophoresed in a 12.5% SDS-acrylamide gel, and
transferred to Immobilon-P Transfer Membrane (Millipore). The
membranes were blocked with 5% non-fat milk in Tris-buffered saline
and then incubated with mouse anti-human PTEN monoclonal antibody
(Abcam) followed by horseradish peroxidase-conjugated secondary
antibody (Abcam). Signals were detected with Immobilon Western
chemiluminescent HRP Substrate (Millipore). GAPDH (Abcam) served as
the loading control.
Tissues were fixed in 10% neutralised formalin and
embedded in paraffin blocks. Sections (4 μm) were then prepared for
immunohistochemical examination. After deparaffinisation and
rehydration, antigen retrieval was performed by boiling in 10
mmol/l of citrate buffer (pH 6.0) for 10 min. After inhibition of
endogenous peroxidase activity for 30 min with methanol containing
0.3% H2O2, the sections were blocked with 2%
bovine serum albumin in PBS for 30 min and incubated with mouse
anti-human PTEN monoclonal antibody (Abcam, dilution 1:500). The
immune complex was visualised with the Dako REAL™EnVision™
Detection System, Perox-idase/DAB, Rabbit/Mouse (Dako), according
to the manufacturer’s procedure. The cytoplasm was counterstained
with hematoxylin.
Statistical analysis
The relationships between the miR-21 expression
level and clinicopathological parameters were analysed using the
Pearson χ2 test. For comparisons between two different
groups, statistical significance was determined using the Student’s
t-test. All statistical analyses were performed using the SAS 6.12
software package. A two-tailed value of P<0.05 was considered
statistically significant.
Results
The expression of miR-21 is upregulated
in gastric cancer and correlates with clinicopathological
parameters
To explore the role of miRNAs in gastric cancer, the
expression level of miRNAs was a primary consideration. We used
miRNA microarray to analyse the miRNA expression profile of nine
gastric cancer cell lines and six normal gastric mucosa, and we
identified 146 under-expressed and 17 over-expressed miRNAs in all
of the gastric cancer cell lines. It has been previously reported
that some dysregulated miRNAs, such as miR-106a, miR-141, miR-143,
miR-145, miR-218, miR-31, Let-7a, miR-17-5p, miR-221, miR-93, and
miR-136 are altered in gastric cancer (18–24),
which is in concordance with our results. Interestingly, we found
significant upregulation of miR-21 in gastric cancer cell lines,
which has not been previously described. Furthermore, qRT-PCR was
performed to detect the miR-21 expression level in the nine gastric
cancer cell lines in order to validate the high-expression trend of
miR-21 in the gastric cancer cell lines obtained from the miRNA
microarray analysis; the results were then compared to one
immortalized normal gastric mucosal epithelial cell line (GES-1).
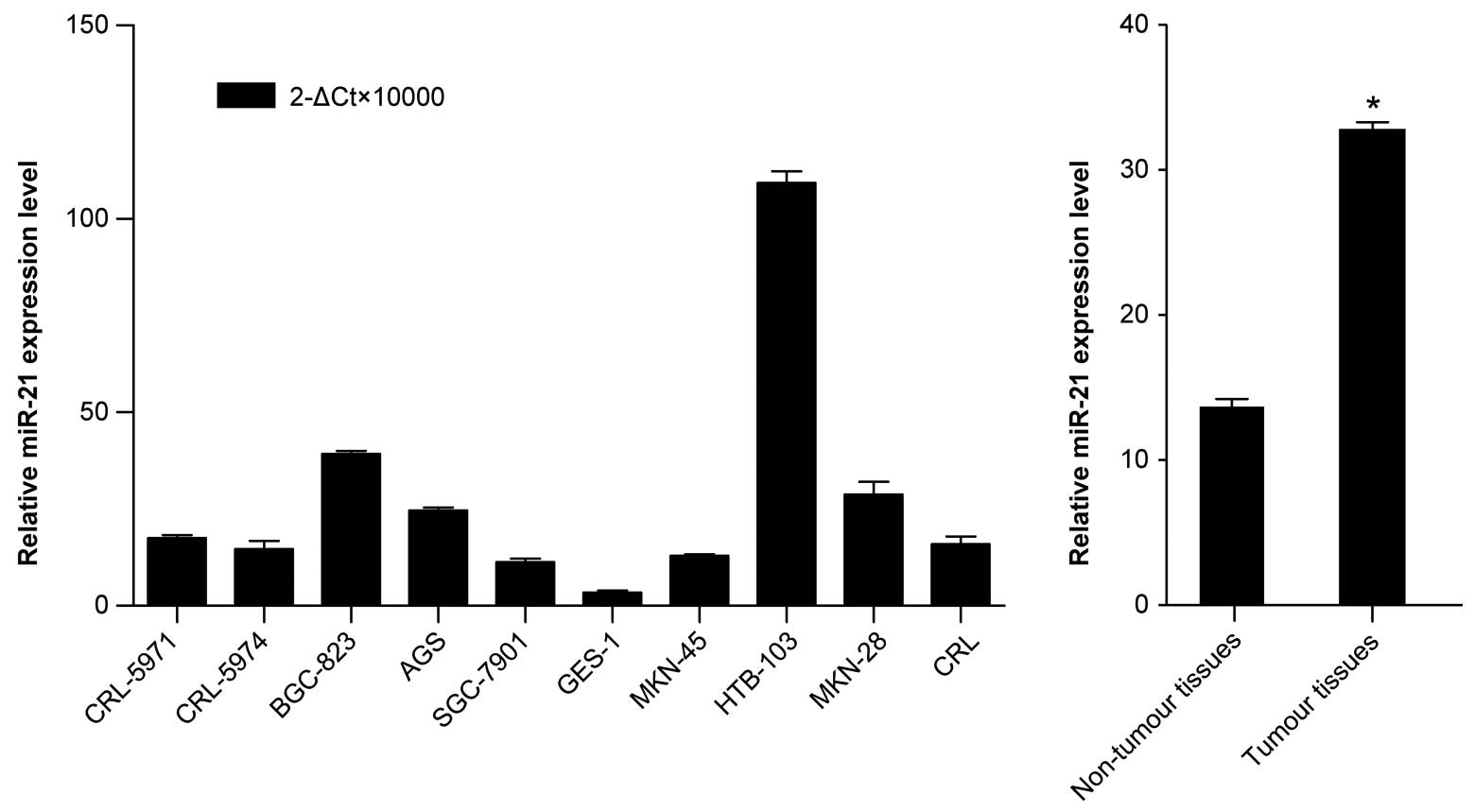
As shown in Fig. 1A, miR-21 was
significantly upregulated in most gastric cancer cell lines
compared with GES-1, thus validating the miRNA microarray results.
Similarly, by analyzing the clinical gastric cancer tissues, we
found that the average expression level of miR-21 was also
significantly upregulated in tumour tissues compared with its
matched non-tumour tissues (Fig.
1B). Collectively, these results provided strong evidence that
miR-21 is prominently upregulated in gastric cancer. Extensive
analysis indicated that among the 30 gastric cancer tissues, 80%
(24/30) of the tumour tissues exhibited upregulation of miR-21
compared with the matched non-tumour samples (relative expression
ratio >1.0). Furthermore, 55% (16/30) of the tumour tissues
exhibited greater significant upregulation of miR-21 (relative
expression ratio >2.0). Based upon a relative expression ratio
of >2.0, the miR-21 high-expression group demonstrated a trend
toward differentiation. However, miR-21 expression demonstrated no
relationship with age, gender, tumour site, or TNM stage (Table I).
 | Table IRelationship between miR-21 expression
level and clinicopathological parameters. |
Table I
Relationship between miR-21 expression
level and clinicopathological parameters.
| Clinicopathological
parameters | n | mean ± SD | P-value |
|---|
| Age (years) |
| ≤59 | 14 | 9.00±9.81 | 0.675 |
| >59 | 16 | 7.60±8.27 | |
| Gender |
| Male | 17 | 10.±10.36 | 0.104 |
| Female | 13 | 5.42±5.71 | |
| Bormann type |
| I, II | 6 | 7.23±8.03 | 0.758 |
| III, IV | 24 | 8.51±9.23 | |
| Location |
| Middle proximal | 10 | 9.25±9.76 | 0.672 |
| Distal | 20 | 7.75±8.64 | |
| Diameter (cm) |
| ≤5 | 18 | 10.58±10.24 | 0.138 |
| >5 | 12 | 5.53±5.67 | |
| Histological
type |
| Intestinal | 10 | 9.17±9.25 | 0.562 |
| Diffuse | 20 | 7.54±8.32 | |
| Depth of
invasion |
| T1, T2 | 8 | 2.23±4.16 | 0.042 |
| T3, T4 | 22 | 9.8±10.36 | |
| Lymph node
metastasis |
| No | 9 | 3.13±2.17 | 0.028 |
| Yes | 21 | 10.3±9.67 | |
| Differentiation |
| High/middle | 9 | 3.24±1.47 | 0.004 |
| Moderate/low | 21 | 10.4±9.88 | |
| TNM stage |
| I, II | 7 | 5.22±7.76 | 0.312 |
| III, IV | 23 | 9.17±9.16 | |
Ectopic expression of miR-21
promotes/inhibits the growth of gastric cancer cells
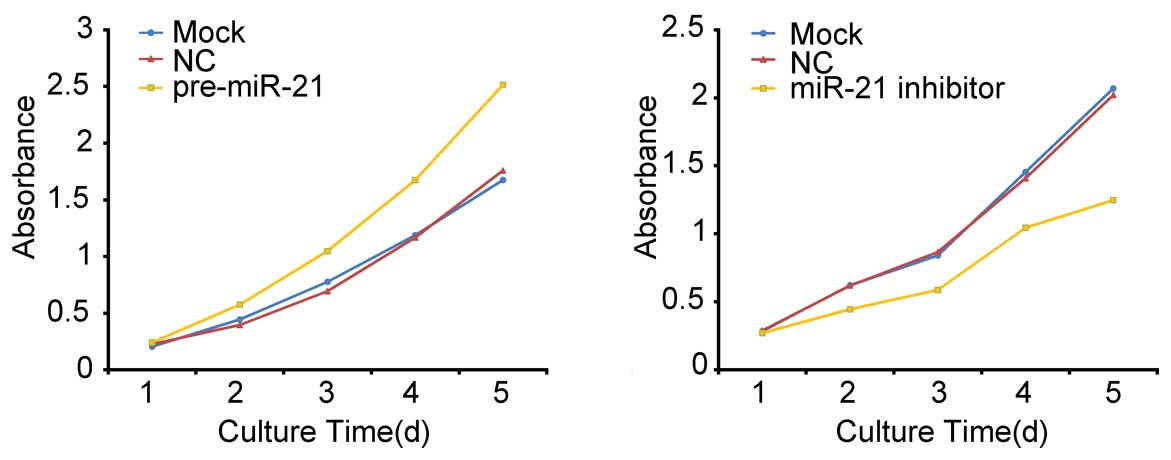
Because miR-21 is markedly upregulated in gastric
cancer, it may thus function as a tumour promoter. Therefore, we
tested whether over-expression/low-expression of miR-21 in BGC-823
cells affects cell growth. In a CCK8 assay, cells transfected with
miR-21 precursor/inhibitor grew more rapidly/slowly than the
control group (Fig. 2). The
dramatic contrast in proliferative activity indicates that
over-expression/low-expression of miR-21 promotes/inhibits the
gastric cancer BGC-823 cell growth activity. These results suggest
that over-expression/low-expression of miR-21 promotes/inhibits
cell growth in vitro. The transfection efficiency was
monitored using a Cy3-labeled pre-miR™ negative control.
Low-expression of miR-21 induces cell
cycle arrest in G1/S phase and affects gastric cancer cell
apoptosis
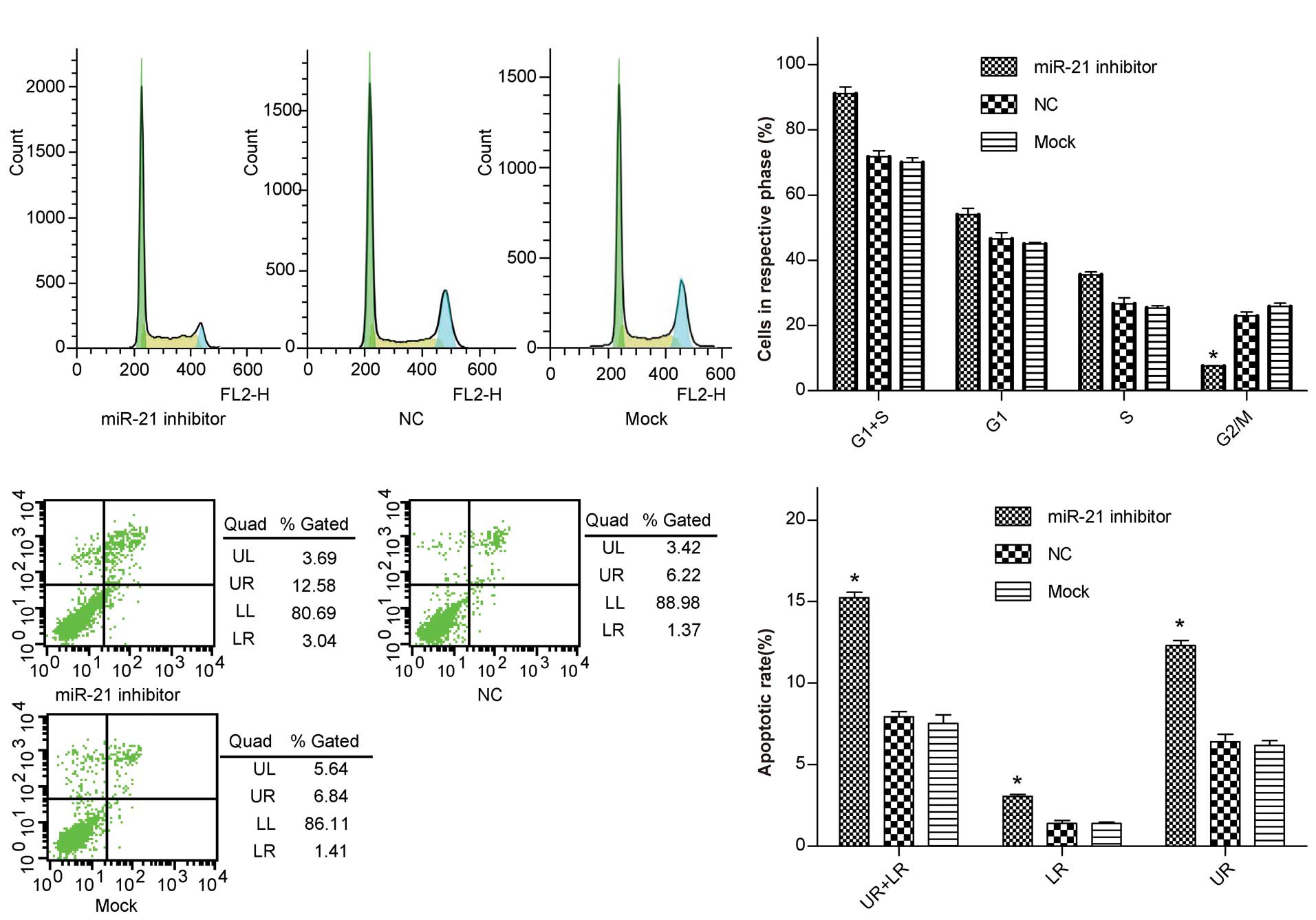
To elucidate the mechanism of miR-21-mediated cell
growth promotion in gastric cancer cells, cell cycle analysis was
performed. The results demonstrated that, when compared with the
control group, the percentage of miR-21 inhibitor-transfected
BGC-823 cells in G1/S phase increased from 70% to 91% (P<0.05),
whereas the percentage of cells in G2/M phase decreased from 22.9
to 7.6% (P<0.05) (Fig. 3A and
B), and there was a significant difference in the apoptotic
rate between the differently treated groups (12.3 vs. 6.4%,
P<0.05) (Fig. 3C and D). These
results indicate that low-expression of miR-21 induces G1/S phase
arrest in BGC-823 cells, which in turn contributes to the
stimulating growth properties of miR-21.
Ectopic expression of miR-21
promotes/inhibits migration of gastric cancer cells in vitro
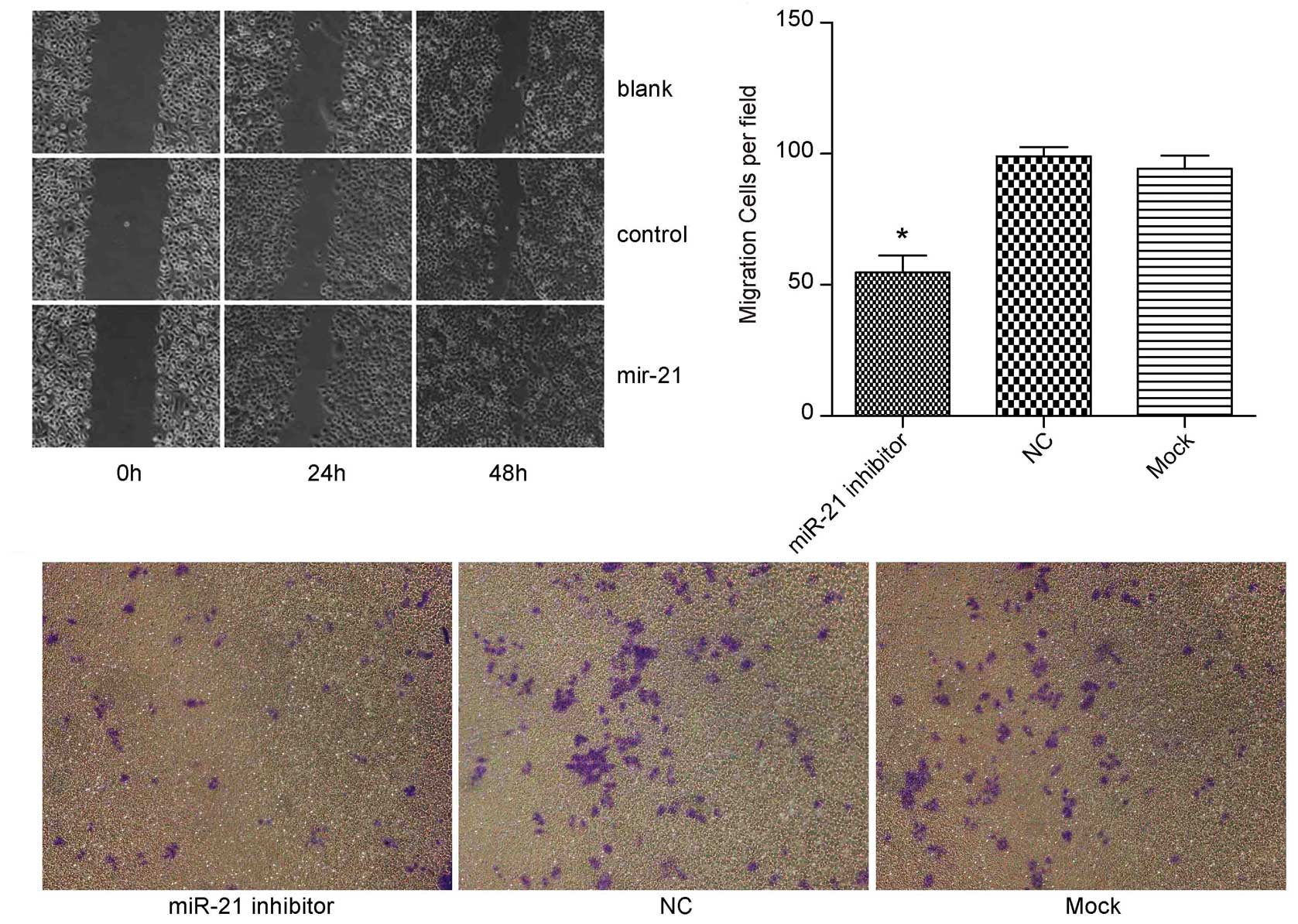
We further assessed the effects of miR-21 on cell
migration, a key determinant of malignant progression and
metastasis. As shown in Fig. 4A,
miR-21 precursor group scratch wound-healing motility was faster
compared to the control group; (Fig. 4B
and C) ectopic expression of miR-21 led to significantly
decreased migration (miR-21 inhibitor group, 50±18 cells per field;
control inhibitor group, 100±28 cells per field, P<0.05) of
BGC-823 cells. These results propose a functional role for miR-21
in mediating cell migration in gastric cancer and suggest a
mechanism by which upregulation of miR-21 potentially contributes
to tumour metastasis in gastric cancer.
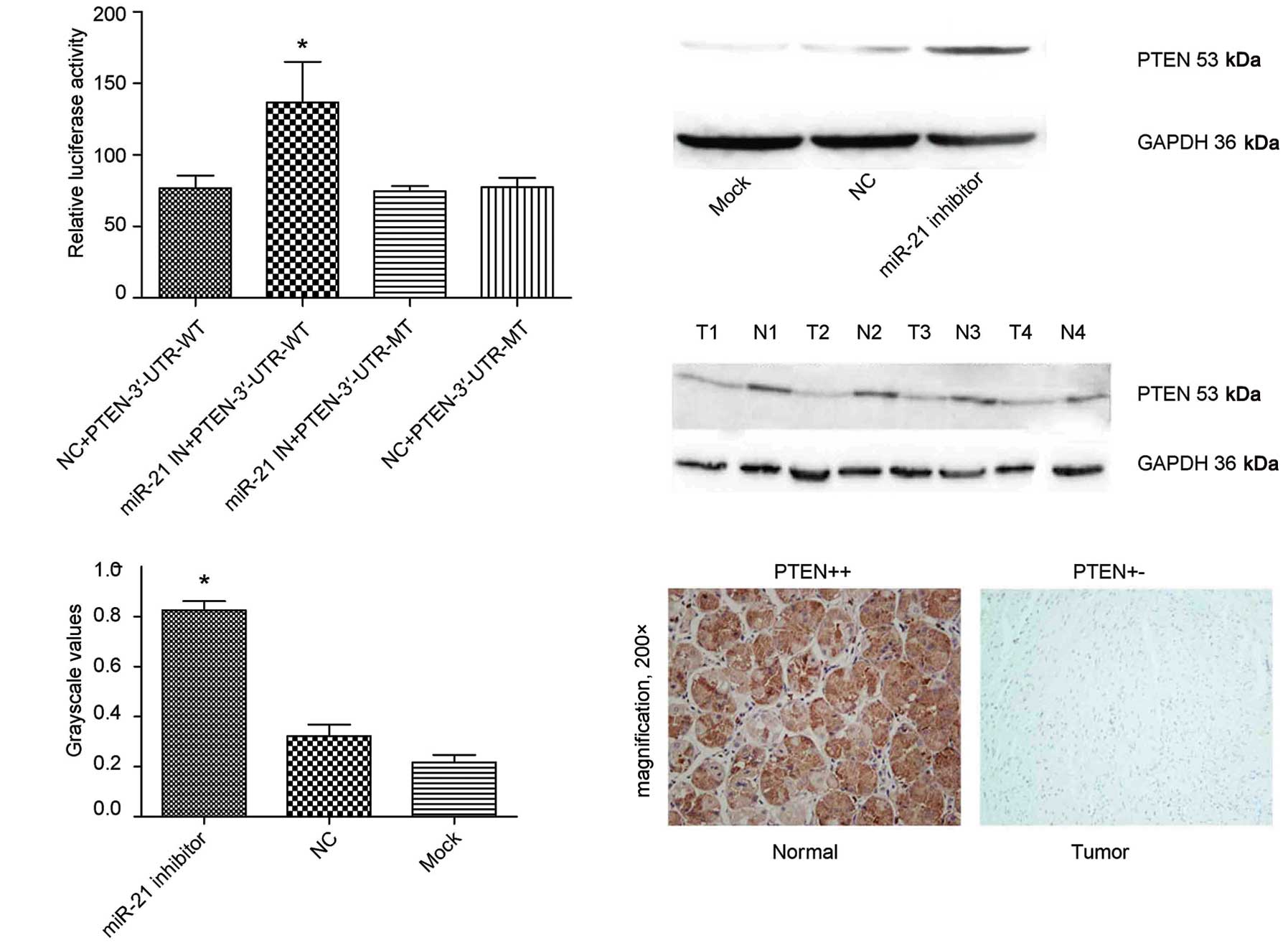
PTEN is a target of miR-21
Because miR-21 has a pivotal function in gastric
cancer, we investigated how this miRNA exerts its role in gastric
cancer. We searched for further information regarding its potential
target genes that exhibit anti-oncogene properties (13–15);
among these genes, PTEN plays a crucial role in the signaling
pathways regulating cell adhesion, proliferation, and migration.
Therefore, we were able to confirm whether PTEN was also the
authentic target gene of miR-21 in gastric cancer. To
experimentally validate whether PTEN was a target gene of miR-21 in
gastric cancer, a region of the PTEN-3′-UTR mRNA was cloned down
stream of the Firefly luciferase gene in the pMIR-REPORT luciferase
vector. This reporter construct (pMIR/PTEN) was co-transfected into
BGC-823 cells with pRL-TK (containing the Renilla luciferase gene
to normalize for transfection differences) and miR-21 inhibitor or
control. A statistically significant increase of Firefly luciferase
activity was observed in BGC-823 cells co-transfected with miR-21
inhibitor and pMIR/PTEN compared with BGC-823 cells co-transfected
with control and pMIR/PTEN (Fig.
5A). The resulting construct, pMIR/PTEN/mut, was co-transfected
into BGC-823 cells with miR-21 inhibitor or control, and the
luciferase activity was measured. Importantly, miR-21 could no
longer increase Firefly luciferase activity of pMIR/PTEN/mut
(Fig. 5A). These results were
confirmed by the following western blot analysis and
immunohistochemistry (Fig. 5B-E).
Taken together, these results indicate that PTEN-3′-UTR carries the
direct binding sites of miR-21.
Discussion
One of the best ways to understand miRNA function is
the elucidation of functional targets, and this usually involves
analysis of changes in target proteins following either a gain or
loss of function of the specific miRNA. Our results indicate that
miR-21 may regulate cell proliferation, apoptosis, and invasiveness
by targeting PTEN. These results are somewhat similar to the
findings in other types of cancer. However, there is no direct
evidence to support a complex correlation among miRNAs, altered
cell phenotype resulting from ectopic miRNA treatments, and the
many targets of the miRNAs. We found an increase expression of a
target gene (PTEN) that resulted from miR-21 inhibition;
additionally, we found the inhibition of gastric cancer cell
proliferation and the acceleration of apoptosis via anti-miR-21.
Thus, we demonstrated that miR-21 generally handles different
gastric cancer cell phenotypes by affecting the target, and that
one target may contribute to several gastric cancer phenotypes
under the control of miR-21. PTEN is a well-known tumor suppressor
in multiple cancers, including HCC, and it affects the Akt and ERK
signaling pathways (25–27). These pathways are linked to cell
survival, proliferation, differentiation, cell migration, and
invasion. Hence, this tumor suppressor could either be directly
regulated by miR-21 or indirectly through the effect of miR-21 on
PTEN. Although PTEN as a target gene of miR-21 has been validated
in hepatocellular cancer, breast cancer, and non-small cell lung
cancer (5,28,29),
Hatley et al (5) confirmed
that PTEN was not regulated by niR-21 in non-small cell lung
cancer. Additionally, Medina et al (30) demonstrated that through
over-expression, miR-21 could lead to pre-B-cell lymphoma, however,
Hatley et al (5) suggested
that miR-21 high-expression was not sufficient in the non-small
cell lung cancer tumorigenesis model. Thus, how do we determine
whether miR-21 is tissue specific? We observed an increase of PTEN
expression in gastric cancer cells treated with anti-miR-21 as
compared to untreated cells or cells treated with the
scrambled-sequence oligonucleotide. These findings further
demonstrate that miR-21, along with its known targets and a few
associated genes, forms a complex regulatory network that plays an
important role in gastric cancer formation and progression.
Therefore, we believe that miR-21 is at the core of a tumorigenic
regulatory network at a non-coding RNA level, and may be a new
potential target for gastric cancer therapy. Taken together, our
findings suggest that the phenotypes of gastric cancer cells
(uncontrolled proliferation, increased survival, and invasiveness)
are at least partly the result of miR-21 regulation of PTEN.
Consequently, suppression of miR-21 may be a novel approach for the
treatment of gastric cancer.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 30872476), the Science and
Technology Commission of Shanghai Municipality (no. 10jc1411100,
09DZ1950100, 09DZ2260200), the Shanghai Key Discipline (S30204) and
Key Projects in the National Science and Technology Pillar Program
of China (no. 2008BA152B03).
References
|
1
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hornstein E, Mansfield JH, Yekta S, et al:
The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sempere LF, Freemantle S, Pitha-Rowe I, et
al: Expression profiling of mammalian microRNAs uncovers a subset
of brain-expressed microRNAs with possible roles in murine and
human neuronal differentiation. Genome Biol. 5:R132004. View Article : Google Scholar
|
|
4
|
Zhao Y, Samal E and Srivastava D: Serum
response factor regulates a muscle-specific microRNA that targets
Hand2 during cardiogenesis. Nature. 436:214–220. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hatley ME, Patrick DM, Garcia MR, et al:
Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21.
Cancer Cell. 18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang P, Zou F, Zhang X, et al: MicroRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu H, Li Y, Gu J, et al: Antisense
oligonucleotide against miR-21 inhibits migration and induces
apoptosis in leukemic K562 cells. Leuk Lymphoma. 51:694–701. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2010.
|
|
9
|
Dicken BJ, Bigam DL, Cass C, et al:
Gastric adenocarcinoma: review and considerations for future
directions. Ann Surg. 24:27–39. 2010.
|
|
10
|
Hundahl SA, Phillips JL and Menck HR: The
National Cancer Data Base report on poor survival of US gastric
carcinoma patients treated with gastrectomy. Cancer. 88:921–932.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheung HH, Davis AJ, Lee TL, et al:
Methylation of an intronic region regulates miR-199a in testicular
tumor malignancy. Oncogene. 30:3404–3415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Yan Z, Zhang J, et al:
Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for
recurrence risk in gastric cancer patients following surgical
resection. Ann Oncol. 22:2257–2266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Si ML, Zhu S, Wu H, et al: miR-21-mediated
tumor growth. Oncogene. 26:2799–2803. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metallopro-teinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiao B, Guo J, Miao Y, et al: Detection of
miR-106a in gastric carcinoma and its clinical significance. Clin
Chim Acta. 400:97–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Du Y, Xu Y, Ding L, et al: Down-regulation
of miR-141 in gastric cancer and its involvement in cell growth. J
Gastroenterol. 44:556–561. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takagi T, Iio A, Nakagawa Y, et al:
Decreased expression of microRNA-143 and -145 in human gastric
cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao C, Zhang Z, Liu W, et al: Reduced
microRNA-218 expression is associated with high nuclear factor
kappa B activation in gastric cancer. Cancer. 116:41–49.
2010.PubMed/NCBI
|
|
22
|
Zhang Y, Guo J, Li D, et al:
Down-regulation of miR-31 expression in gastric cancer tissues and
its clinical significance. Med Oncol. 27:685–689. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang HH, Wang XJ, Li GX, et al: Detection
of let-7a microRNA by real-time PCR in gastric carcinoma. World J
Gastroenterol. 13:2883–2888. 2007.PubMed/NCBI
|
|
24
|
Petrocca F, Visone R, Onelli MR, et al:
E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest
and apoptosis in gastric cancer. Cancer Cell. 13:272–286. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu LT, Chang HC, Chiang LC, et al:
Histone deacetylase inhibitor up-regulates RECK to inhibit MMP-2
activation and cancer cell invasion. Cancer Res. 63:3069–3072.
2003.PubMed/NCBI
|
|
26
|
Yu J, Zhang SS, Saito K, et al: PTEN
regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 28:21–33. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
To MD, Perez-Losada J, Mao JH, et al:
Crosstalk between Pten and Ras signaling pathways in tumor
development. Cell Cycle. 4:1185–1188. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng F, Henson R, Wehbe-Janek H, et al:
MicroRNA-21 regulates expression of the PTEN tumor suppressor gene
in human hepatocellular cancer. Gastroenterology. 133:647–658.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qi L, Bart J, Tan LP, et al: Expression of
miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia
of the breast in relation to ductal carcinoma in situ and invasive
carcinoma. BMC Cancer. 9:1632009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Medina PP, Nolde M, Slack FJ, et al:
OncomiR addiction in an in vivo model of microRNA-21-induced
pre-B-cell lymphoma. Nature. 467:86–90. 2010. View Article : Google Scholar : PubMed/NCBI
|