Introduction
Metastasis proceeds through a multi-step process
that can be divided into dissemination and colonization stages
(1–3). Dissemination frequently occurs at
early stages where disseminated tumor cells (DTCs) are detected in
the peripheral blood or in the bone marrow (4–9). This
is further supported by a high recurrence rate in certain types of
cancers (10–12), providing a rationale for systemic
adjuvant chemotherapy (13,14). In contrast, colonization mechanisms
are later activated at the metastatic site (15,16),
frequently after several years of dormancy (9). Colonization mechanisms can be acquired
de novo in the primary tumors, and these clones might be
selected, evolved and expanded at the metastatic sites (17). Another possibility is that these
characteristics do not exist in the primary tumors but are induced
by interaction with the metastatic microenvironments (18,19),
as supported by altered drug responsiveness when co-cultured with
stromal cells (20–22) or when grafted to different organs
(23). Regardless of when the
colonization mechanisms evolve, they are activated in a metastatic
site-specific manner. Recently, we conducted a longitudinal
comparison during colon-to-lung metastasis to identify the
molecular mechanisms of colonization at the gene expression level
(24). In the present study, we
performed similar expression analyses with additional genes and
samples including colon-to-liver metastases and compared them with
those from colon-to-lung metastases.
The two most frequent target organs of metastatic
colorectal cancers (mCRCs) are the liver and the lung (25,26),
but the underlying metastatic tropism and the mechanisms
determining which organ to metastasize remain controversial. In
Paget’s seed and soil hypothesis, metastatic clones are selected
for their ability to grow in distant microenvironments (27–30),
as supported by punctuated parallel evolution (15,16,31).
On the other hand, clinicians frequently observe microscopic portal
vein thrombi trapped in the microvasculature of the liver,
suggesting the importance of the anatomy of the blood flow for
hepatic tropism (25,32,33).
The former hypothesis suggests that the newly activated
colonization mechanism in the distant microenvironment is the key
element during metastasis, whereas the latter suggests that the
dissemination route plays a key role. Gene expression analyses of
over 200 genes with known functions in metastasis, chemokine
signals and angiogenesis might be sufficient to reveal the
activated pathways. Nonetheless, relevant clinical samples are
difficult to obtain because of the rarity of metastasectomy and
decreasing integrity of the samples during the latent years.
Fortunately, current trends toward more aggressive metastasectomy
(34) may generate more clinical
samples in the future.
The liver and the lung have different tissue
structures, resident cell types (35,36)
and growth stimulants that are released. For example, IGF1,
IGF2, HGF, MST (HGF-like) activator and
RARRES2 are expressed at significantly higher levels in the
liver, whereas GDF10, CSH1, BTG2,
LTBP2, VWF, CSF3, IL6, FIGF,
HBEGF, endothelin1, LTF, WIFP2 and
VIM are higher in the lung (37). Hepatectomy has been reported to
stimulate not only regeneration of the liver, but also the growth
of colorectal cancers in animal experiments (38), indicating that wound healing signals
in the liver might be conducive to mCRC cell growth. Indeed, HGF
and its receptor MET were identified as prognostic markers for
hepatic metastasis in CRCs (39,40).
In lung cancers, the prediction of metastasis with primary tumor
gene expression signatures has resulted in limited success
(41), raising the possibility that
the metastatic potential of lung cancer may not be predetermined in
the primary tumor. Taken together, it is likely that different
colonization mechanisms are activated in different
microenvironments, and here we provide molecular evidence
suggesting that multiple distinct colonization mechanisms are
activated in the lung in contrast to the liver that requires little
change to be colonized by colorectal cancers.
Materials and methods
Patient and sample selection
Twelve pairs of primary colon tumor samples and
matched hepatic or pulmonary metastases were collected from 12
patients (Table I) by surgical
resection conducted at Samsung Medical Center between 2001 and
2008. The institutional review board approved the study protocol
and prior consent was obtained from all patients. A clinical
coordinator reviewed all medical records. Tumor specimens were
stained with hematoxylin and eosin (H&E) and examined by a
pathologist to remove necrotic tissue and/or intervening stromal
tissues and were classified according to the World Health
Organization histopathological criteria. After necrotic regions
were removed and the tumor masses were snap-frozen and stored in
liquid nitrogen. Specimens were examined by pathologists to select
the specimens with >90% cancer cell content for RNA extraction
and gene expression analyses.
 | Table ISummary of clinical information. |
Table I
Summary of clinical information.
| | | Primary tumor | Metastatic
tumor |
|---|
| | |
|
|
|---|
| Case | Gender | Age (years) | Size (cm) | pStagea | Primary site | Sizeb (cm) | Metastatic
site |
|---|
| 1 | M | 72 | 7.7 | III | S colon | 3.5 | Liver |
| 2 | M | 47 | 10.0 | III | Lower rectum | 4.3 | Liver |
| 3 | F | 66 | 2.7 | III | Lower rectum | 2.7 | Liver |
| 4 | F | 69 | 6.0 | III | Colon | 1.7 | Liver |
| 5 | F | 70 | 4.8 | III | Colon | 2.7 | Liver |
| 6 | M | 59 | 9.0 | III | Colon | 4.0 | Liver |
| 7 | F | 36 | 4.8 | III | Lower rectum | 2.0 | Lung |
| 8 | F | 49 | 4.4 | III | Lower rectum | 3.4 | Lung |
| 9 | F | 57 | 5.2 | III | Upper rectum | 3.0 | Lung |
| 10 | M | 65 | 3.5 | I | Lower rectum | 3.2 | Lung |
| 11 | M | 70 | 3.5 | II | Upper rectum | 1.8 | Lung |
| 12 | F | 53 | 5.2 | III | S-colon | 1.0 | Lung |
Gene expression profiling using the
RT2 ProfilerTM PCR array
To extract total-RNA from the tumor specimens, the
NucleoSpin RNA kit was used following the manufacturer’s protocol.
Only RNA samples with a RIN value >7.0 were used as templates
for cDNA synthesis using the RT2 First Strand kit (Cat.
no. C-03; SABiosciences). Human tumor metastasis (PAHS-028),
chemokines (PAHS-022) and angiogenesis (PAHS-024) finder
RT2 profiler PCR arrays and RT2
SYBR-Green/Rox PCR Master mix (APMM012C and PA-012-24;
SABiosciences) were used to quantitatively analyze the gene
expression levels of 84 metastasis genes (APC, BRMS1, CCL7,
CD44, CDH1, CDH11, CDH6, CDKN2A, CHD4, COL4A2, CST7, CTBP1, CTNNA1,
CTSK, CTSL1, CXCL12, CXCR4, DENR, EPHB2, ETV4, EWSR1, FAT, FGFR4,
FLT4, FN1, FXYD5, GNRH1, KISS1R, HGF, HPSE, HRAS, HTATIP2, IGF1,
IL18, IL1B, IL8RB, ITGA7, ITGB3, CD82, KISS1, KRAS, RPSA, MCAM,
MDM2, MET, METAP2, MGAT5, MMP10, MMP11, MMP13, MMP2, MMP3, MMP7,
MMP9, MTA1, MTSS1, MYC, MYCL1, NF2, NME1, NME2, NME4, NR4A3, PLAUR,
PNN, PTEN, RB1, RORB, SET, SMAD2, SMAD4, SRC, SSTR2, SYK, TCF20,
TGFB1, TIMP2, TIMP3, TIMP4, TNFSF10, TP53, TRPM1, TSHR, VEGFA),
84 chemokines and receptor genes (CCL1, CCL11, CCL13, CCL15,
CCL16, CCL17, CCL18, CCL19, CCL2, CCL3, CCL4, CCL5, CCL7, CCL8,
CXCL1, CXCL10, CXCL11, CXCL12, CXCL13, CXCL2, CXCL3, CXCL5, CXCL6,
CXCL9, CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR10,
CCRL1, CCRL2, CXCR3, CXCR4, CXCR6, CYFIP2, APLNR, BDNF, C5, C5AR1
GPR77, CCBP2, CKLF, CMTM1, CMTM2, CMTM3, CMTM4, CMKLR1, CSF3,
CX3CL1, CX3CR1, TYMP, GDF5, GPR31, HCAR1, HIF1A, IL13, IL16, IL18,
IL1A, IL4, IL8, CXCR1, LTB4R, MMP2, MMP7, MYD88, NFKB1, AIMP1,
SDF2, SLIT2, TCP10, TLR2, TLR4, TNF, TNFRSF1A, TNFSF14, TREM1, VHL,
XCL1, XCR1), 84 angiogenesis genes (ANGPT1, ANGPT2, ANPEP,
TYMP, EREG, FGF1, FGF2, FIGF, FLT1, JAG1, KDR, LAMA5, NRP1, NRP2,
PGF, PLXDC1, STAB1, VEGFA, VEGFC, ANGPTL3, BAI1, COL4A3, IL8,
LAMA5, NRP1, NRP2, STAB1, ANGPTL4, PECAM1, PF4, PROK2, SERPINF1,
TNFAIP2, HAND2, SPHK1, CCL11, CCL2, CXCL1, CXCL10, CXCL3, CXCL5,
CXCL6, CXCL9, IFNA1, IFNB1, IFNG, IL1B, IL6, MDK, TNF, S1PR1,
EFNA1, EFNA3, EFNB2, EGF, EPHB4, FGFR3, HGF, IGF1, ITGB3, PDGFA,
TEK, TGFA, TGFB1, TGFB2, TGFBR1, CCL11, CDH5, COL18A1, S1PR1, ENG,
ITGAV, ITGB3, THBS1, THBS2, LECT1, LEP, MMP2, MMP9, PLAU, PLG,
TIMP1, TIMP2, TIMP3, AKT1, HIF1A, HPSE, ID1, ID3, NOTCH4,
PTGS1). Five housekeeping genes (B2M, HPRT1, RPL13A, GAPDH,
ACTB) and MMP2 were triplicated, and twenty-seven genes were d
(CCL7, CXCL12, CXCR4, HGF, IGF1, IL18, IL1B, ITGB3, MMP2, MMP3,
MMP9, TGFB1, TIMP2, TIMP3, VEGFA, TYMP, CCL11, CCL2, CXCL1, CXCL10,
CXCL3, CXCL5, CXCL6, CXCL9, TIMP2, TIMP3, HIF1A, HPSE).
Negative controls for contamination were included in the 96-well
PCR arrays. RT-PCR was conducted using an ABI 7300 Real Time PCR
system (Applied Biosystems).
Data analysis
Ct values >35 were considered to indicate either
reaction failure or absence of expression and were excluded from
the data analyses. The amount of sample loaded per well was
estimated and normalized according to the expression levels of six
housekeeping genes. Differential gene expression was estimated as
follows: ΔCt = Ct(lung metastases) − Ct(colon
primary) and fold change =2(−ΔCt). Quantitative
analyses including hierarchical clustering, heat map analyses and
volcano plots of the array data for the primary colon and
metastatic tumors were conducted using Web-based PCR array data
analysis software (http://www.sabiosciences.com). The metastatic
signature tropism was selected by progressive removal of redundant
genes from the metastasis arrays for the maintenance of the
original clustering pattern. Removal of any single gene from the
17-gene signature disrupted the original clustering pattern.
Results
Quantitative gene expression
The gene expression level of 224 genes was
determined quantitatively for a total of 24 CRC tumors, 12 pairs of
primary-to-metastatic tumors including six colon-to-liver (no. 1–6)
pairs and six colon-to-lung (no. 7–12) pairs. Ct values were
normalized against the deviation from the median of the five
housekeeping genes. The Ct values for 27 duplicate or 6 triplicate
genes were highly reproducible for assayable genes (41) with activated expression. For those
genes with expression at basal levels or lower, Ct values >35
were collectively considered as no expression and were excluded
from further analysis. Among the assayable genes, the average
correlation between duplicates was 0.92.
Metastatic tropism: hepatic vs. pulmonary
CRC metastases of CRCs
To overview the relative similarities or differences
among the samples, an unsupervised two-dimensional hierarchical
clustering was conducted with the Ct values. In the metastasis
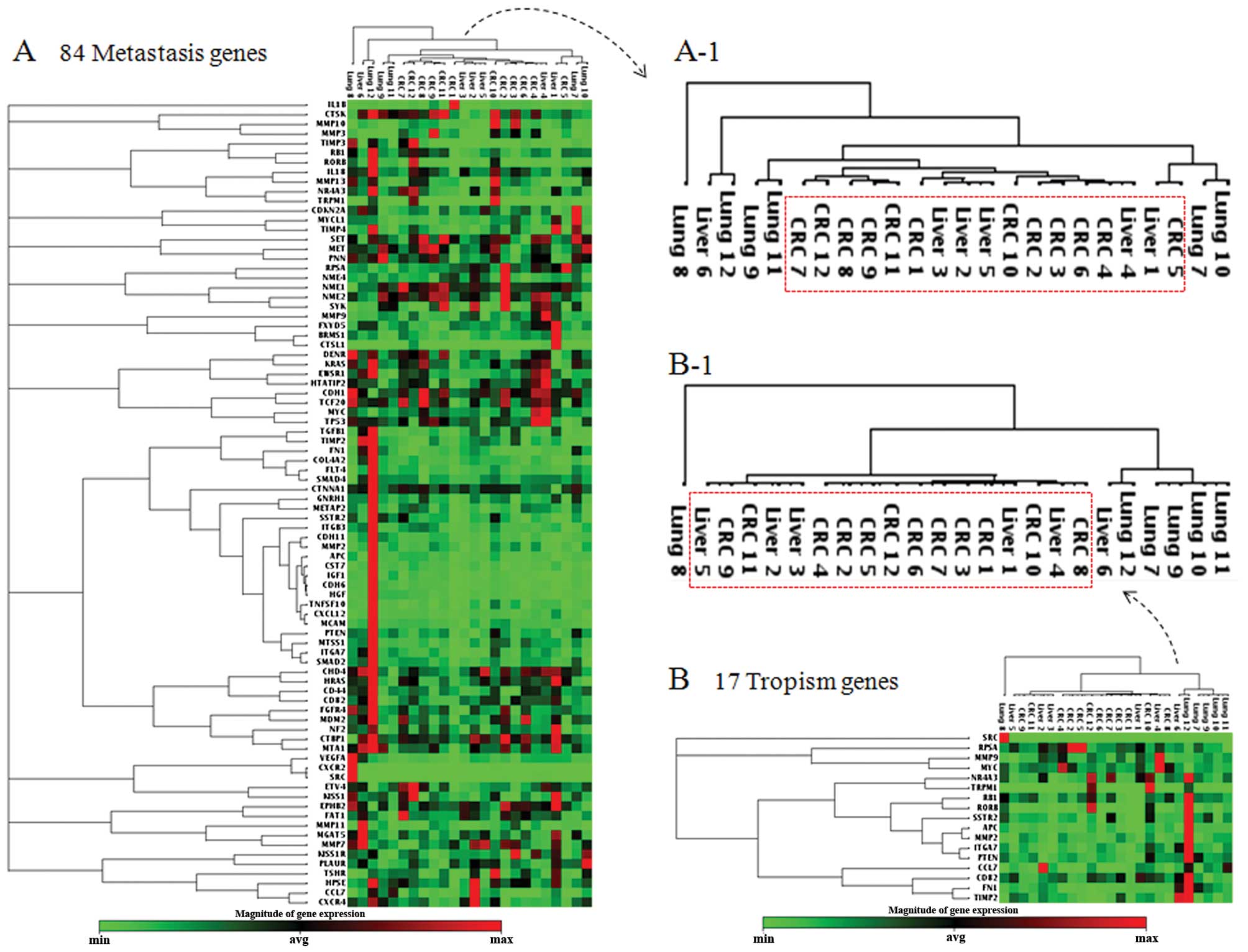
array data, all hepatic metastases formed closely related clusters
with the primary CRCs with one exception of Liver 6 (Fig. 1A). The clinical information was
examined to determine the possibility of Liver 6 being a secondary
metastasis from pulmonary metastasis, but no pulmonary metastasis
was detected in that patient. Among the six cases of hepatic
metastases, only one original tumor pair was clustered together
(CRC4 and Liver 4) and the rest clustered with a tumor from a
different patient, indicating that the similarities between the
hepatic metastases and the primary CRCs were greater than the
person-to-person diversities in the tested samples. In contrast to
the hepatic metastases, the pulmonary metastases were highly
diversified from one another and from the primary colorectal tumors
(Fig. 1A). To identify potential
markers for tropism, heat map analyses were performed with the ΔCt
values between the hepatic and pulmonary metastases (data not
shown). Forty-six genes were differentially regulated
(>1.5-fold) when the hepatic and pulmonary metastases were
compared. From the listed genes, those with similar expression
patterns were removed to finalize the 17-gene tropism signature
(Table II). To confirm that the
tropism signature was sufficient to distinguish the hepatic from
the pulmonary metastases, all of the 24 tumor samples were analyzed
again by unsupervised hierarchical clustering (Fig. 1B). The 17-gene signature not only
distinguished the liver metastases from the lung metastases, but
also clustered primary CRCs with the hepatic metastases,
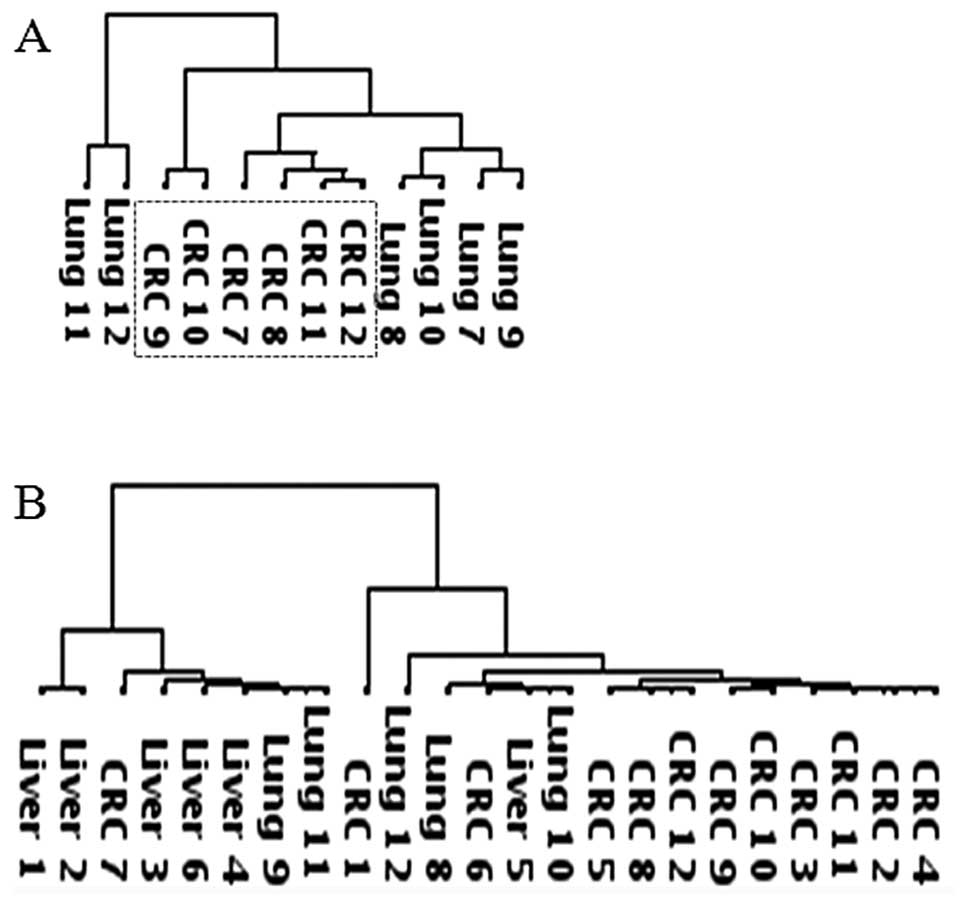
reconstituting the clustergram of 84 metastasis genes. Chemokine
array data were obtained only for the pulmonary metastases and
their primary CRCs due to a shortage of RNA samples. Hierarchical
clustering of the chemokine array data also showed that the
pulmonary metastases were diversified from the primary CRCs
(Fig. 2A). Tropism genes could not
be identified from the hepatic metastases due to the lack of
chemokine array data. Nonetheless, expression profiling of the
17-tropism genes was sufficient to distinguish the hepatic and
pulmonary CRC metastases and to show similarity between the hepatic
and primary CRCs in the tested samples.
 | Table IImCRC tropism signature. |
Table II
mCRC tropism signature.
| No. | Gene symbol | Fold change |
|---|
| 1 | APC | 6.23 |
| 2 | MMP2 | 4.23 |
| 3 | RORB | 4.01 |
| 4 | SSTR2 | 3.56 |
| 5 | ITGA7 | 2.84 |
| 6 | RB1 | 2.58 |
| 7 | PTEN | 1.92 |
| 8 | TRPM1 | 1.92 |
| 9 | NR4A3 | 1.83 |
| 10 | SRC | 1.53 |
| 11 | FN1 | 1.52 |
| 12 | TIMP2 | −1.56 |
| 13 | MMP9 | −1.59 |
| 14 | CD82 | −1.71 |
| 15 | RPSA | −1.72 |
| 16 | CCL7 | −1.93 |
| 17 | MYC | −2.02 |
Overall activation of angiogenic pathways
in pulmonary metastases
To examine the possible role of angiogenic pathways
in late steps of colonization, supporting the growth of
micrometastases into a detectible size, angiogenesis array data
were obtained for all 24 tumor samples. Unlike the metastasis array
and the chemokine array data, hierarchical clustering of the
angiogenesis array data did not group the pulmonary metastases and
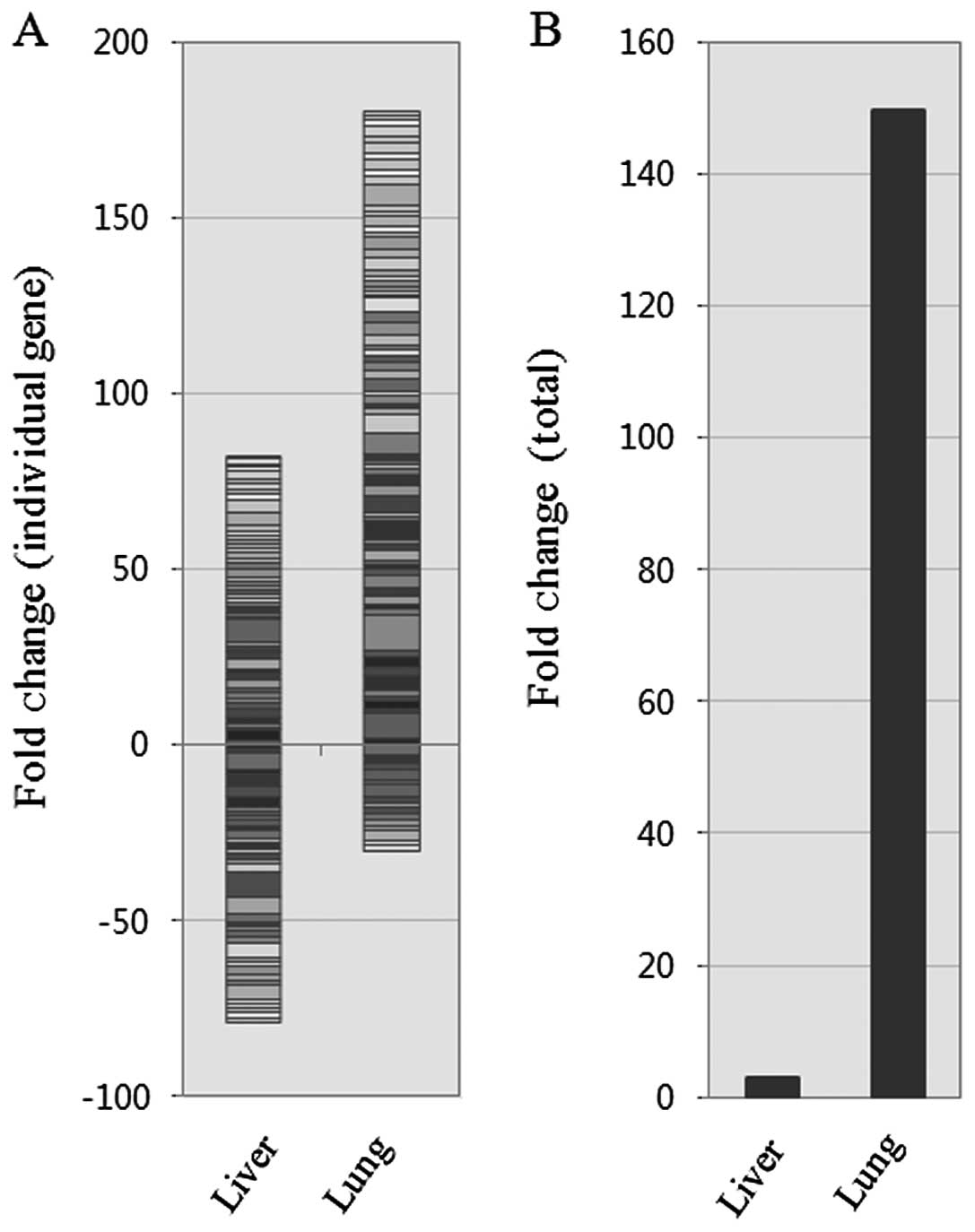
the hepatic metastases into separate clusters (Fig. 2B). However, when overall gene
expression was examined between these groups, the total fold change
was significantly upregulated (~150-fold) during colon-to-lung
metastasis compared to colon-to-liver metastasis (Fig. 3). These data indicate that the
diversity of angiogenic pathways is activated in the tested samples
of pulmonary metastases.
Discussion
In metastatic cases, primary tumor surgical
specimens are already equipped with dissemination mechanisms;
therefore, longitudinal comparison with actively colonizing
metastatic tumors would unveil the colonization mechanisms of the
micrometastases. In this study we compared six pairs of
colon-to-lung metastases with colon-to-liver metastases to
understand how colonization mechanisms differ in the liver and the
lung, the two most frequent target organs for metastatic CRCs.
Unsupervised hierarchical clustering of the
metastasis array data revealed that the pulmonary and hepatic
metastases were intrinsically different, with the exception of
Liver 6 that clustered with Lung 12. All 84 metastasis array genes
were not required for distinction between the two groups. Among the
differentially expressed genes, a minimal 17-gene tropism signature
was determined and was shown to separately cluster these two sample
groups (Fig. 1). Interestingly,
clustergrams with either 84 metastasis genes or 17 tropism genes
demonstrated that all primary CRCs were similar to the hepatic
metastases. These data suggest that the divergence of pulmonary
metastases was not an over-interpretation of technical artifacts,
such as major stromal contamination or random/uncontrolled events
that can occur in any collection of samples. In addition,
significant divergence did not occur in the primary tumors. If
divergence of metastatic potential is manifested in the primary
tumors, gene expression profiles of the primary tumors should be
able to predict recurrence, which is not the case in lung cancers
(41). Lastly, the liver and colon
might have such similar microenvironments that little change is
needed for CRCs to cross-colonize between them, which is consistent
with previous reports (42) and a
classical view of passive transport to amiable microenvironments in
the liver via the portal vein (32,33).
Although the number of samples tested was too small to exert a
definitive conclusion, the divergence of pulmonary metastases from
one another raises the possibility that there might be multiple
distinct metastatic microenvironments in the lung.
Angiogenesis needs to be activated for both primary
and metastatic tumors to grow into a detectible size, and
consequently not specific to either metastatic colonization or
tropism. Our data showed that the expression profiles of
angiogenesis-related genes were not specific to the tumors formed
in the liver, the lung or the colorectal microenvironments.
Nonetheless, overall expression in the pulmonary metastases was
~150-fold higher than those in the hepatic metastases and the
primary CRCs (Fig. 3). This might
be an adaptive change to the relatively sterile microenvironments
of the lung compared with the strong regenerative potentials found
in the liver and the digestive track.
In this study we examined differentially activated
genes during colon-to-liver and colon-to-lung metastases to learn
that pulmonary metastases were highly diversified in contrary to
hepatic metastases that were indistinguishable from primary CRCs
and identified a 17-gene tropism signature. Our data suggest that
pulmonary metastases need different therapeutics from hepatic
metastases and primary CRCs; however, further clinical studies with
larger sample sizes are required to validate this conclusion.
Acknowledgements
This study was supported by grants from the Samsung
Biomedical Research Institute (no. SBRI C-A6-411, C-A7-802), the
National Research Foundation of Korea (NRF) funded by the Korean
government (MEST) (no. R01-2006-000-11114-0, and NTX2091113) and
the Seoul R&BD Program (SS100010).
References
|
1
|
Nguyen DX, Paula DB and Massagué J:
Metastasis: from dissemination to organ-specific colonization. Nat
Rev Cancer. 9:274–284. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fidler IJ: Critical factors in the biology
of human cancer metastasis: twenty-eighth G.H.A. Clowes memorial
award lecture. Cancer Res. 50:6130–6138. 1990.PubMed/NCBI
|
|
3
|
Talmadge JE and Fidler IJ: AACR centennial
series: the biology of cancer metastasis: historical perspective.
Cancer Res. 70:5649–5669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pantel K and Brakenhoff RH: Dissecting the
metastatic cascade. Nat Rev Cancer. 4:448–456. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Braun S, Vogl FD, Naume B, et al: A pooled
analysis of bone marrow micrometastasis in breast cancer. N Engl J
Med. 353:793–802. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hüsemann Y, Geigl JB, Schubert F, et al:
Systemic spread is an early step in breast cancer. Cancer Cell.
13:58–68. 2008.
|
|
8
|
Lin H, Balic M, Zheng S, Datar R and Cote
RJ: Disseminated and circulating tumor cells: Role in effective
cancer management. Crit Rev Oncol Hematol. 77:1–11. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Flatmark K, Borgen E, Nesland JM, et al:
Disseminated tumor cells as a prognostic biomarker in colorectal
cancer. Br J Cancer. 104:1434–1439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sugimura H, Nichols FC, Yang P, et al:
Survival after recurrent non small-cell lung cancer after complete
pulmonary resection. Ann Thorac Surg. 83:409–418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feld R, Rubinstein LV and Weisenberger TH:
Sites of recurrence in resected stage I non-small-cell lung cancer:
a guide for future studies. J Clin Oncol. 2:1352–1358.
1984.PubMed/NCBI
|
|
12
|
Gutt R, Liauw SL and Weichselbaum RR:
Adjuvant radiotherapy for resected pancreatic cancer: a lack of
benefit or a lack of adequate trials? Nat Clin Pract Gastroenterol
Hepatol. 6:38–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schabel FM Jr: Rationale for adjuvant
chemotherapy. Cancer. 39:2875–2882. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Visbal AL, Leighl NB, Feld R and Shepherd
FA: Adjuvant chemotherapy for early-stage non-small cell lung
cancer. Chest. 128:2933–2943. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Navin N, Kendall J, Troge J, et al: Tumour
evolution inferred by single-cell sequencing. Nature. 472:90–94.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Campbell PJ, Yachida S, Mudie LJ, et al:
The patterns and dynamics of genomic instability in metastatic
pancreatic cancer. Nature. 467:1109–1113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Greaves M and Maley CC: Clonal evolution
in cancer. Nature. 481:306–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Malanchi I, Santamaria-Martinez A, Susanto
E, et al: Interactions between cancer stem cells and their niche
govern metastatic colonization. Nature. 481:85–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Navab R, Strumpf D, Bandarchi B, et al:
Prognostic gene-expression signature of carcinoma-associated
fibroblasts in non-small cell lung cancer. Proc Natl Acad Sci USA.
108:7160–7165. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mink SR, Vashistha S, Zhang W, et al:
Cancer-associated fibroblasts derived from EGFR-TKI-resistant
tumors reverse EGFR pathway inhibition by EGFR-TKIs. Mol Cancer
Res. 8:809–820. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang W, Li Q, Yamada T, et al: Crosstalk
to stromal fibroblasts induces resistance of lung cancer to
epidermal growth factor receptor tyrosine kinase inhibitors. Clin
Cancer Res. 15:6630–6638. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Serebriiskii I, Castelló-Cros R, Lamb A,
Golemis EA and Cukierman E: Fibroblast-derived 3D matrix
differentially regulates the growth and drug-responsiveness of
human cancer cells. Matrix Biol. 27:573–585. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wilmanns C, Fan D, O’Brian CA, Bucana CD
and Fidler IJ: Orthotopic and ectopic organ environments
differentially influence the sensitivity of murine colon carcinoma
cells to doxorubicin and 5-fluorouracil. Int J Cancer. 52:98–104.
1992. View Article : Google Scholar
|
|
24
|
Kim S-H, Choi S, Cho YB, et al:
Differential gene expression during colon-to-lung metastasis. Oncol
Rep. 25:629–36. 2011.PubMed/NCBI
|
|
25
|
Gilson N, Honoré O, Detry O, et al:
Surgical management of hepatic metastases of colorectal origin.
Acta Gastroenterol Belg. 72:321–326. 2009.PubMed/NCBI
|
|
26
|
Kjeldsen BJ, Kronborg O, Fenger C and
Jorgensen OD: The pattern of recurrent colorectal cancer in a
prospective randomized study and the characteristics of diagnostic
tests. Int J Colorectal Dis. 12:329–334. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fidler IJ and Kripke ML: Genomic analysis
of primary tumors does not address the prevalence of metastatic
cells in the population. Nat Genet. 34:232003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jones S, Chen WD, Parmigiani G, et al:
Comparative lesion sequencing provides insights into tumor
evolution. Proc Natl Acad Sci USA. 105:4283–4288. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fidler IJ: The pathogenesis of cancer
metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003.
|
|
30
|
Psaila B and Lyden D: The metastatic
niche: adapting the foreign soil. Nat Rev Cancer. 9:285–293. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Klein CA: Parallel progression of primary
tumours and metastases. Nat Rev Cancer. 9:302–312. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gupta GP and Massague J: Cancer
metastasis: building a frame-work. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamamoto J, Sugihara K, Kosuge T, et al:
Pathologic support for limited hepatectomy in the treatment of
liver metastasis from colorectal cancer. Ann Surg. 221:74–78. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park JS, Kim HK, Choi YS, et al: Outcomes
after repeated resection for recurrent pulmonary metastases from
colorectal cancer. Ann Oncol. 21:1285–1289. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Suriawinata AA and Thung NS: Liver.
Histology for Pathologists. Mills SE: 3rd edition. Lippincott
Williams & Wilkins; pp. 685–703. 2007
|
|
36
|
Colby TV, Leslie KO and Yousem SA: Lung.
Histology for Pathologists. Mills SE: 3rd edition. Lippincott
Williams & Wilkins; pp. 473–504. 2007
|
|
37
|
Stelzer G, Inger A, Olender T, et al:
GeneDecks: paralog hunting and gene-set distillation with GeneCards
annotation. OMICS. 13:477–487. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fidler IJ: Critical factors in the biology
of human cancer metastasis: twenty-eighth G.H.A. Clowes memorial
award lecture. Cancer Res. 50:6130–8. 1990.PubMed/NCBI
|
|
39
|
Kopitz C, Gerg M, Bandapalli OR, et al:
Tissue inhibitor of metalloproteinases-1 promotes liver metastasis
by induction of hepatocyte growth factor signaling. Cancer Res.
67:8615–8623. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stein U, Walther W, Arlt F, et al: MACC1,
a newly identified key regulator of HGF-MET signaling, predicts
colon cancer metastasis. Nat Med. 15:59–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee ES, Son DS, Kim S-H, et al: Prediction
of recurrence-free survival in postoperative non-small cell lung
cancer patients by using an integrated model of clinical
information and gene expression. Clin Cancer Res. 14:7397–7404.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koh KH, Rhee H, Kang HJ, et al:
Differential gene expression profiles of metastases in paired
primary and metastatic colorectal carcinomas. Oncology. 75:92–101.
2008. View Article : Google Scholar : PubMed/NCBI
|