Introduction
The signaling pathways related to cell
differentiation and senescence do not function properly in
malignant tumor cells. As a result, tumor cells exhibit
uncontrolled and invasive growth. Differentiation therapy is one
emerging technique for the treatment of malignant cancer (1). Tumor cells originate from normal cells
and many different carcinogenic stimuli can contribute to the
development of cancer. Several key genes responsible for regulating
apoptosis or senescence may undergo loss of function mutations in
various tumors, particularly oral squamous cell carcinoma (OSCC)
(2). The main objectives of
differentiation therapy may be achieved by the bypassing the
mutated pathway, thus tipping the balance from carcinogenesis to
terminal differentiation, or rescuing the senescence machinery.
E2F overexpression in squamous cell carcinoma cells
results in aberrant proliferation and the suppression of the
squamous differentiation program (3,4). E2F
transcription factors promote the expression of genes related to
DNA synthesis and cell cycle progression, resulting in cell
proliferation (5,6). An increase in E2F activity is often
associated with inappropriate cell proliferation and/or apoptosis
(7), whereas a decrease in E2F
activity is generally associated with a reduction in the
proliferation capacity of cells (8). The deregulation of E2F activity
contributes to carcinogenesis (9).
E2Fs are involved in the differentiation of many cell types, such
as myocytes (10), megakaryocytes
(11), and adipocytes (12). The specific factor (Sp)1 to Sp3
ratio has been recognized as an important driving force of
epithelial cell differentiation (1). Dysregulation of the expression of
these factors or the change of the relative Sp1 and Sp3 expression
levels result in dysregulated cell proliferation and
differentiation (1). The direct
downregulation of E2F and upregulation of Sp1 increases the
expression of differentiation specific markers such as
transglutaminase-1 and involucrin (1). Therefore, the expression of E2F
factors and the ratio of Sp1 to Sp3 expression are important
parameters determining the epithelial cell proliferation and
differentiation.
The non-isoprenoid lipid is found in a wide range of
plant and bacterial species. These lipids exert non-specific
antioxidant and antimutagenic effects, and can regulate cell
proliferation (13). Chemical
analogs of these lipids have exhibited anticancer effects in animal
models of colon (14), lung
(15), and pancreatic tumors
(16). 4-hexylresorcinol (4-HR) is
also non-isoprenoid lipid. According to recent reports, 4-HR
inhibits the transglutaminase-2 and NF-κB pathways (17,18),
and 4-HR also shows a synergistic effect with cisplatin in KB cells
(17,18). Interestingly, the cancer cells that
survived 4-HR administration were less active and morphologically
differentiated than the active proliferating control (data not
shown). These findings suggest the possibility that 4-HR may induce
differentiation in tumor cells. We hypothesize that 4-HR induces
the differentiation of tumor cells through the control of cell
cycle-related genes like E2F transcription factors.
The objectives of this study were: i) to evaluate of
the effect of 4-HR on E2F signaling pathway on OSCC cells, and ii)
to evaluate the level of expression of differentiation markers in
OSCC cells after the administration of 4-HR.
Materials and methods
Cell culture
Cell line SCC-9 cells (human tongue carcinoma; ATCC;
Manassas, VA, USA) were maintained in monolayer cultures in DMEM
(Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine
serum containing L-glutamine, vitamins (Life Technologies, Inc.,
Grand Island, NY, USA), and penicillin-streptomycin (Flow
Laboratories, Rockville, MD, USA). The cells were incubated in a
mixture of 5% CO2 and 95% air at 37°C. The cultures were
maintained for no longer than 12 weeks after recovery from frozen
stocks.
Real-time cell electronic sensing
(RT-CES)
RT-CES is a measurement of impedance in a
gold-plated culture dish. Because the cellular membrane is composed
of lipid bilayer, increased cellular growth increases the
impedance. The label-free detection of cell growth via RT-CES was
conducted using an xCELLigence® system (Roche Applied
Science, Penzberg, Germany) and special gold-plated 96-well culture
dishes in which the SCC-9 cells were grown. 4-HR was added at a
concentration of 1, 5 or 10 μg/ml to each well 20 h following the
initial seeding. No 4-HR was added in the control wells. The cell
index values derived from the measured impedance were determined
every hour for 4 days.
Western blot analysis
Whole cells were lysed in ProteoJET™ Mammalian Cell
Lysis Reagent (Thermo Scientific, Waltham, MA, USA) containing
protease inhibitor cocktail (Sigma-Aldrich Inc., St. Louis, MO,
USA). Proteins were separated on 10% SDS-polyacrylamide gels and
transferred to PVDF membranes. Blots were blocked with 5% skim milk
powder in Tris-buffered saline (20 mM Tris-HCl, 137 mM NaCl, pH
7.6) containing 0.1% Tween-20 (TBS-T buffer) for 1 h at room
temperature (RT). Western blot analyses were performed with
anti-E2F1 (Abcam Inc. Cambridge, MA, USA), anti-E2F2 (Abcam Inc.),
anti-E2F3 (Abcam Inc.), anti-E2F4 (Abcam Inc.), anti-E2F5 (Abcam
Inc.), anti-E2F6 (Abcam Inc.), anti-Sp1 (Abcam Inc.), anti-Sp3
(Abcam Inc.), anti-keratin 10 (Santa Cruz Biotechnology, Santa
Cruz, CA, USA), anti-involucrin (SantaCruz Biotechnology), and
anti-β-actin (Sigma-Aldrich Inc.) antibodies. Primary antibodies
were added to the TBS-T buffer at a 1:1000 dilution and incubated
for 90 min at RT prior to incubation with HRP-conjugated secondary
antibodies (1:5000 dilution; Santa Cruz Biotechnology) for 1 h at
room temperature. The proteins were detected using a
chemiluminescence detection system (Bio-Rad, Hercules, CA, USA).
The relative expression ratio of Sp1 to Sp3 was determined using
Image Lab software (Bio-Rad).
DNA microarray analysis, quantitative
reverse transcription polymerase chain reaction (Q-PCR), and
fluorescent immunocytochemistry
DNA microarray analysis was performed by Genomic
Tree Co. (Daejeon, Korea) using Agilent human whole-genome 4×44K
chips (Santa Clara, CA). DNA microarray analysis was conducted
using a previously published method (19). SCC-9 cells were treated with or
without 4-HR (10 μg/ml) for 12 h, and total RNA was then extracted
from the cells using TRI Reagent® as recommended by the
manufacturer (Molecular Research Center Inc., Cincinnati, OH).
Keratin 10 and involucrin antibodies were purchased
from Santa Cruz Biotechnology for fluorescence immunocytochemistry
analysis. Fluorescence immunocytochemistry was conducted using a
previously published method (17).
4′,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei.
For the Q-PCR analysis, SCC-9 cells were treated for
12 h with 0, 1, 5 or 10 μg/ml 4-HR. Total RNA (1 μg) was used as
template for first-strand DNA synthesis using the ImProm-II Reverse
Transcription System (Promega, Madison, WI, USA). The RT-PCR
protocol and primers used to amplify involucrin and keratin 10 were
published previously (20,21).
Results
4-HR decreased E2F2, E2F3, and Sp3
expression and increased Sp1 expression
In previous studies, we found that 4-HR has
anti-tumor effects on several cancer cell lines (17,18).
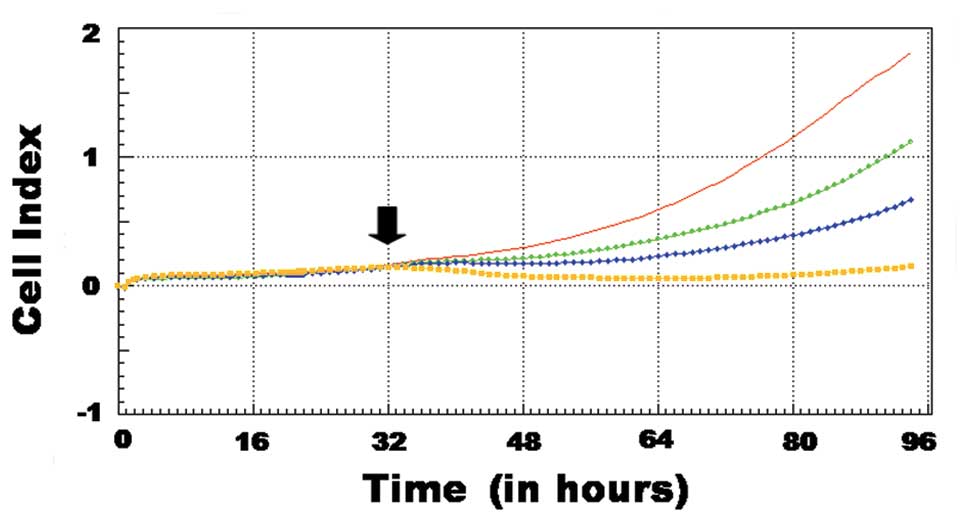
In this study, the anti-proliferative effect of 4-HR on SCC-9 cells
observed in continuous RT-CES-based assays (Fig. 1). The growth of the SCC-9 cells
slowed following the addition of 4-HR. The addition of 10 μg/ml
4-HR induced a prominent inhibitory effect, as determined by the
observed decline in the growth curve (yellow) (Fig. 1). The 4-HR induced growth inhibition
was dose-dependent.
To identify the target genes of 4-HR, a DNA
microarray comparing 4-HR treated and untreated cells was
performed, and differentially expressed genes were selected for
further analysis (Table I). As
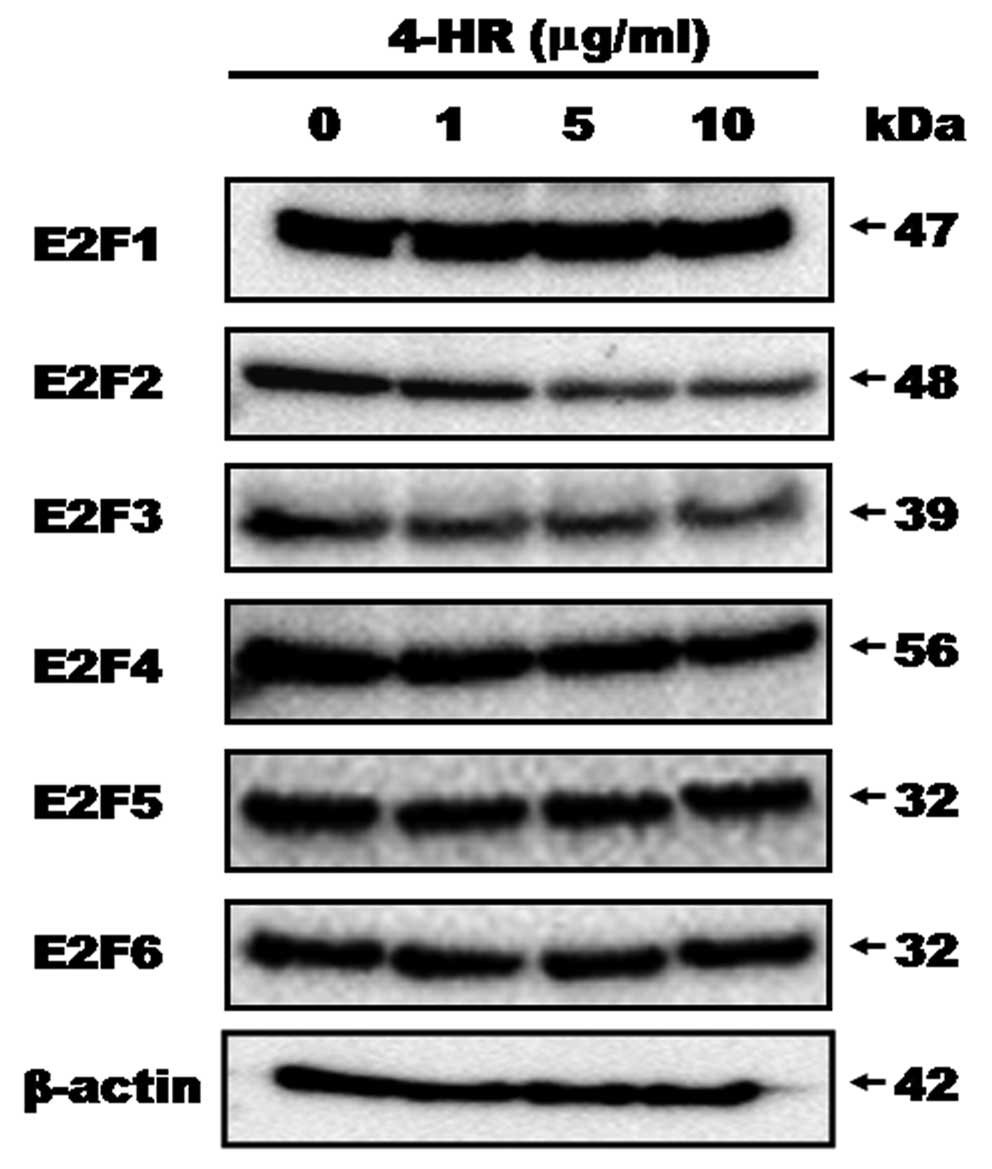
shown in Fig. 1 and Table I, 4-HR decreased the levels of E2F2
and E2F3 expression. As 4-HR concentrations increased from 1 to 10
μg/ml, E2F2 expression was decreased (Fig. 2). The level of E2F3 expression was
also decreased in 4-HR treated cells compared to the untreated
control (Fig. 2). There was no
significant difference in the expression of any other E2F
transcription factors; E2F1, E2F4, E2F5, and E2F6 expression were
all similar in the 4-HR treated cells and the untreated control
cells.
 | Table IThe results of DNA microarray.a |
Table I
The results of DNA microarray.a
| TITLE | GenBank | Fold-ratio |
|---|
| E2F2 | NM_004091 | 0.428 |
| E2F3 | NM_001949 | 0.507 |
| Sp1 | NM_138473 | 1.498 |
| Sp3 | BX648857 | 0.464 |
| Involucrin | NM_005547 | 3.176 |
| Keratin 10 | NM_000421 | 3.213 |
| Keratin 13 | NM_002274 | 2.718 |
| Keratin 14 | NM_000526 | 2.823 |
| Keratin 16 | NM_005557 | 2.635 |
| Keratin 17 | NM_000422 | 2.951 |
| Keratin 18 | NM_000224 | 2.806 |
| Keratin 19 | NM_002276 | 2.841 |
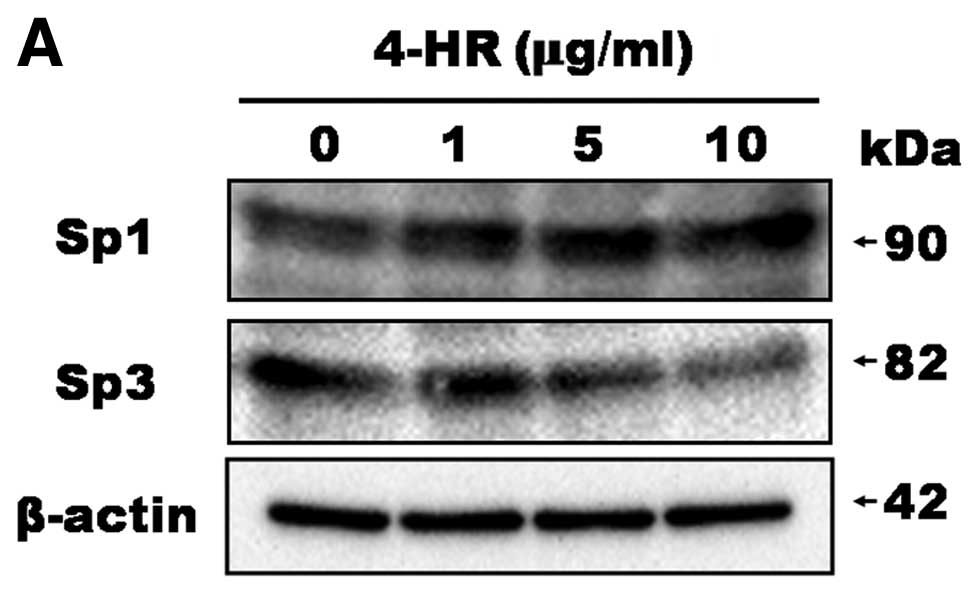
With respect to Sp transcription factors, Sp1
expression was increased by 4-HR treatment (Fig. 3A). However, Sp3 expression was
decreased. DNA microarray analysis also demonstrated a decreased in
the level of Sp3 expression (Table
I). As a result, the Sp1/Sp3 ratio was increased upon 4-HR
treatment in a dose-dependent manner. The Sp1/Sp3 ratios were 0.88,
1.72, 2.70 and 19.95 in the untreated control and in cells treated
with 1, 5 and 10 μg/ml 4-HR, respectively (Fig. 3B). These results indicate that 4-HR
regulates E2F2 and E2F3 expression and the Sp1/Sp3 expression ratio
in SCC-9 cells.
4-HR accelerates SCC-9 cell
differentiation
We also investigated genes that were involved in the
epithelial cell differentiation, as identified by DNA microarray
analyses. DNA microarray analysis revealed that keratins and
involucrin exhibited significantly higher expression at 12 h after
10 μg/ml of 4-HR administration (Table
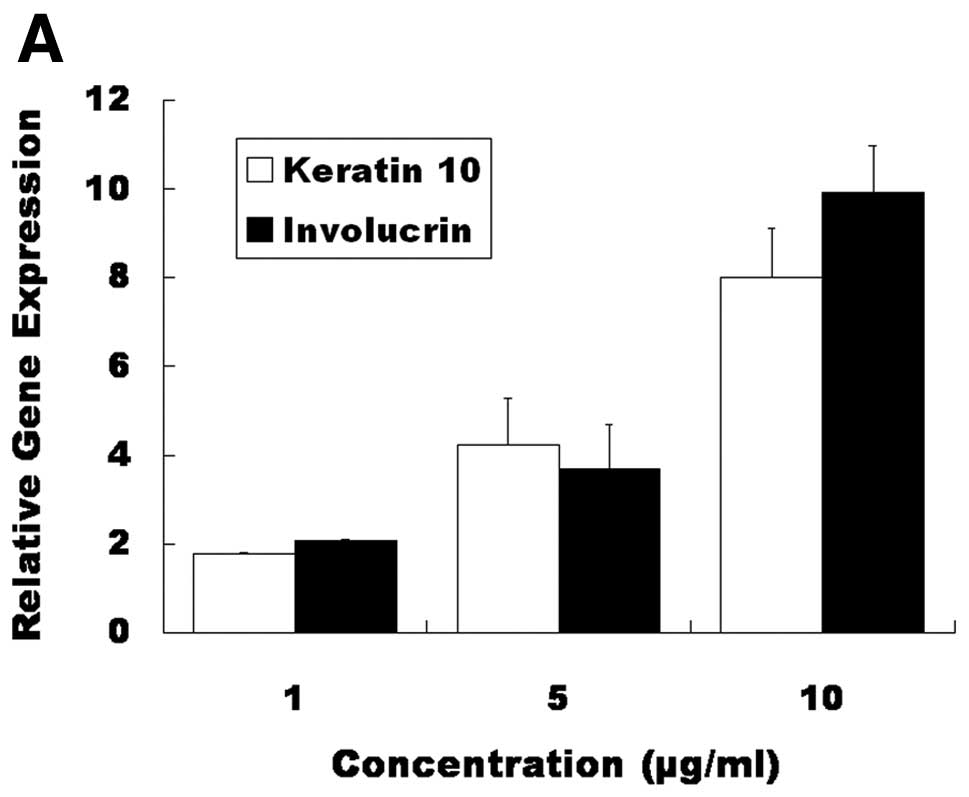
I). The increased expression of keratin 10 and involucrin mRNA
was confirmed by RT-PCR (Fig. 4A).
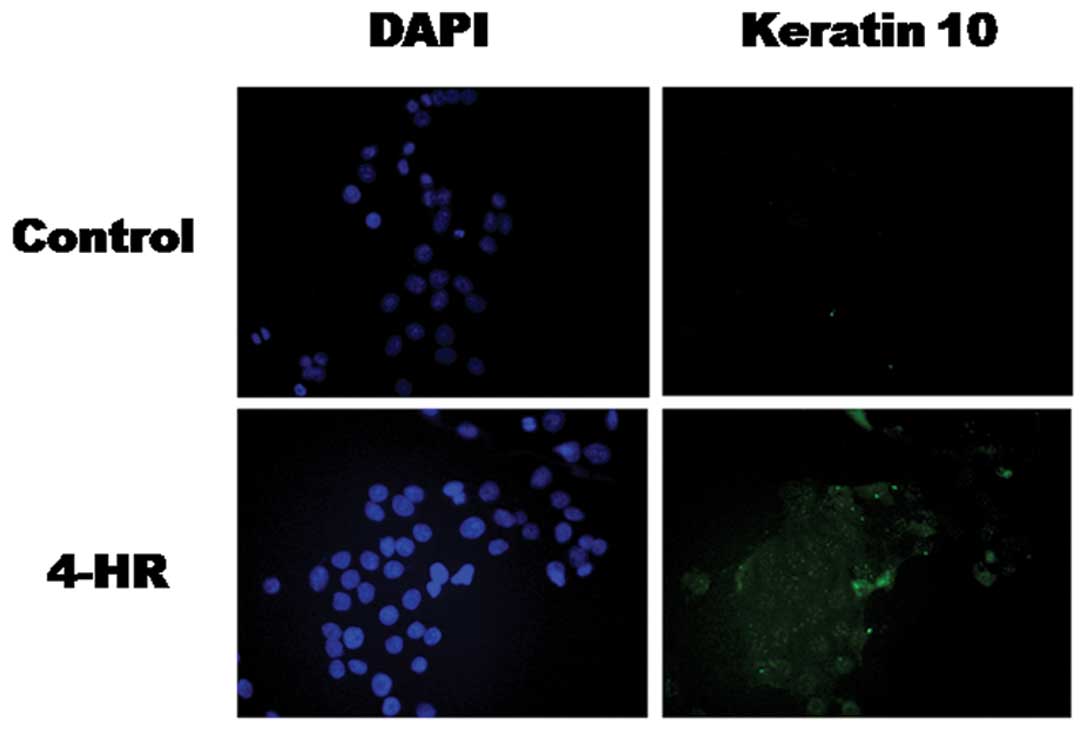
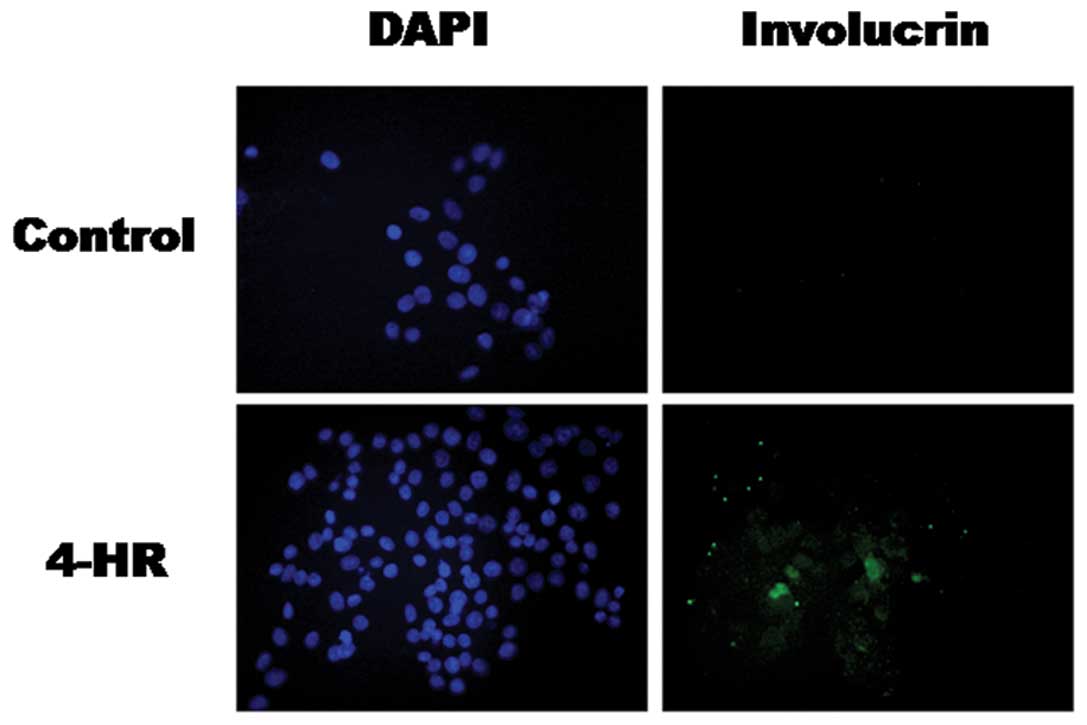
Increased expression of the keratin 10 and involucrin proteins was
confirmed by western blot analysis (Fig. 4B) and immunocytochemistry (Figs. 5 and 6). These results indicate that 4-HR
stimulates the expression of differentiation marker genes in SCC-9
cells.
Discussion
In this study, we demonstrated that 4-HR suppressed
growth (Fig. 1) and stimulated the
expression of differentiation marker genes in SCC-9 cells (Fig. 4). The 4-HR-induced SCC-9 cell
differentiation was mediated by the E2F signaling pathway. E2F2,
E2F3, and Sp3 were suppressed by 4-HR (Figs. 2 and 3). The level of Sp1 expression was
increased by 4-HR treatment (Fig.
3). Indeed, 4-HR increased the expression of the epithelial
cell differentiation markers keratin 10 and involucrin. Taken
together, these data show that 4-HR induced SCC-9 cell
differentiation through the downregulation of E2F2 and upregulation
of Sp1/Sp3 ratio.
Senescence is defined as a permanent cell cycle
arrest induced by oncogene expression and it is an effective
anti-cancer mechanism in vivo(22–24).
Therefore, treatments aiming to inhibit the uncontrolled growth of
cancer cells while simultaneously restoring their normal
differentiation program would be invaluable (1). To develop such ‘differentiation
therapies’, it is important to find candidate drugs that can
control the E2F signaling pathway.
Squamous epithelial cell differentiation is
controlled by Sp1 and E2F (25).
Sp1 can act as a differentiation activator and its activity is
regulated by E2F (1). E2F1, E2F2,
and E2F3 are expressed highly in human osteosarcoma cells, and a
large number of genes with known role in development and
differentiation are consequently deregulated (26). However, the inhibition of E2F alone
or the introduction of Sp1 alone is not sufficient to reinstate
squamous differentiation (1). In
the present study, 4-HR was able to suppress E2F2 and E2F3
expression (Fig. 2). At the same
time, Sp1 expression was increased, and Sp3 expression was
decreased (Fig. 3A). The Sp1 to Sp3
ratio is important in the regulation of cell differentiation, and
the increase in this ratio observed upon 4-HR treatment (Fig. 3) is consistent with the activation
of tumor cell differentiation (1).
In keratinocytes, E2F inhibition is a prerequisite
for differentiation, and E2Fs 1–5 are all able to suppress
differentiation-specific markers (3,4). E2F
suppresses multiple markers of squamous cell differentiation
(3,27). E2F is also able to suppress the
differentiation induced by multiple stimuli (3,27).
Thus, E2F has the capacity to act as an important regulator of
squamous cell differentiation. In this study, the increased
expression of differentiation markers such as keratin 10 and
involucrin was due in part to the combination of the inhibition of
E2F2 and E2F3 and the upregulation of the Sp1/Sp3 ratio by 4-HR
(Fig. 4). However, these
observations were made at the cellular level, and it will be
important to validate these results in vivo. To this end,
systemic administration of 4-HR in xenografted mice has been
performed, and the expression of differentiation markers in the
xenografted tumor mass is currently under investigation (data not
shown).
In conclusion, 4-HR stimulated the differentiation
of SCC-9 cells, potentially through the suppression of the E2F
signaling pathway.
Acknowledgements
This study was supported by a grant from the
Next-Generation BioGreen21 Program ( Center for Nutraceutical &
Pharmaceutical Materials no. PJ009051), Rural Development
Administration, Republic of Korea.
References
|
1
|
Wong CF, Barnes LM, Dahler AL, et al: E2F
suppression and Sp1 overexpression are sufficient to induce the
differentiation-specific marker, transglutaminase type 1, in a
squamous cell carcinoma cell line. Oncogene. 24:3525–3534. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bruckman KC, Schönleben F, Qiu W, Woo VL
and Su GH: Mutational analyses of the BRAF, KRAS, and PIK3CA genes
in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol
Oral Radiol Endod. 110:632–637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wong CF, Barnes LM, Dahler AL, et al: E2F
modulates keratinocyte squamous differentiation: implications for
E2F inhibition in squamous cell carcinoma. J Biol Chem.
278:28516–28522. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong CF, Barnes LM, Smith L, Popa C,
Serewko-Auret MM and Saunders N: E2F6: a member of the E2F family
that does not modulate squamous differentiation. Biochem Biophys
Res Commun. 324:497–503. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Polyak K, Kato JY, Solomon MJ, et al:
p27Kip1, a cyclin-Cdk inhibitor, links transforming
growth factor-beta and contact inhibition to cell cycle arrest. J
Gene Dev. 8:9–22. 1994.
|
|
6
|
DeGregori J, Kowalik T and Nevins JR:
Cellular targets for activation by the E2F1 transcription factor
include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol.
15:4215–4224. 1995.PubMed/NCBI
|
|
7
|
DeGregori J, Leone G, Miron A, Jakoi L and
Nevins JR: Distinct roles for E2F proteins in cell growth control
and apoptosis. Proc Natl Acad Sci USA. 94:7245–7250. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu L, Timmers C, Maiti B, et al: The
E2F1-3 transcription factors are essential for cellular
proliferation. Nature. 414:457–462. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu Z, Zheng S and Yu Q: The E2F family and
the role of E2F1 in apoptosis. Int J Biochem Cell Biol.
41:2389–2397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang J, Helin K, Jin P and Nadal-Ginard B:
Inhibition of in vitro myogenic differentiation by cellular
transcription factor E2F1. Cell Growth Differ. 6:1299–1306.
1995.PubMed/NCBI
|
|
11
|
Guy CT, Zhou W, Kaufman S and Robinson MO:
E2F-1 blocks terminal differentiation and causes proliferation in
transgenic megakaryocytes. Mol Cell Biol. 16:685–693.
1996.PubMed/NCBI
|
|
12
|
Fajas L, Landsberg RL, Huss-Garcia Y,
Sardet C, Lees JA and Auwerx J: E2Fs regulate adipocyte
differentiation. Dev Cell. 3:39–49. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kozubek A and Tyman JHP: Resorcinolic
lipids, the natural non-isoprenoid phenolic amphiphiles and their
biological activity. Chem Rev. 99:1–25. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mutoh M, Takahashi M, Fukuda K, et al:
Suppression of cyclooxygenase-2 promoter-dependent transcription
activity in colon cancer cells by chemopreventive agents with a
resorcin-type structure. Carcinogenesis. 21:959–963. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hasegawa R, Furukawa F, Toyoda K, et al:
Inhibitory effect of antioxidants on
N-bis(2-hydroxypropyl)nitrosamine-induced lung carcinogenesis in
rats. Jpn J Cancer Res. 81:871–877. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maruyama H, Amamura T, Nakae D, et al:
Effect of catechol and its analogs on pancreatic carcinogenesis
initiated by N-nitrosobis(2-oxopropyl)amine in Syrian
hamsters. Carcinogenesis. 12:1331–1334. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim SG, Jeong JH, Park YW, et al:
4-Hexylresorcinol inhibits transglutaminase-2 activity and has
synergistic effects along with cisplatin in KB cells. Oncol Rep.
25:1597–1602. 2011.PubMed/NCBI
|
|
18
|
Kim SG, Lee SW, Park YW, Jeong JH and Choi
JY: 4-hexylresorcinol inhibits NF-κB phosphorylation and has a
synergistic effect with cisplatin in KB cells. Oncol Rep.
26:1527–1532. 2011.PubMed/NCBI
|
|
19
|
Kim JY, Choi JY, Jeong JH, et al: Low
molecular weight silk fibroin increases alkaline phosphatase and
type I collagen expression in MG63 cells. BMB Rep. 43:52–56. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trollinger DR, Cascio WE and Lemasters JJ:
Selective loading of Rhod 2 into mitochondria shows mitochondrial
Ca2+ transients during the contractile cycle in adult
rabbit cardiac myocytes. Biochem Biophy Res Commun. 236:738–742.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wagenblast J, Baghi M, Arnoldner C, et al:
Effect of bortezomib and cetuximab in EGF-stimulated HNSCC.
Anticancer Res. 28:2239–2243. 2008.PubMed/NCBI
|
|
22
|
Braig M, Lee S, Loddenkemper C, et al:
Oncogene-induced senescence as an initial barrier in lymphoma
development. Nature. 436:660–665. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z, Trotman LC, Shaffer D, et al:
Crucial role of p53-dependent cellular senescence in suppression of
Pten-deficient tumorigenesis. Nature. 436:725–730. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Michaloglou C, Vredeveld LC, Soengas MS,
et al: BRAFE600-associated senescence-like cell cycle arrest of
human naevi. Nature. 436:720–724. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hazar-Rethinam M, Cameron SR, Dahler AL,
et al: Loss of E2F7 expression is an early event in squamous
differentiation and causes derepression of the key differentiation
activator Sp1. J Invest Dermatol. 131:1077–1084. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Muller H, Bracken AP, Vernell R, et al:
E2Fs regulate the expression of genes involved in differentiation,
development, proliferation, and apoptosis. Genes Dev. 15:267–285.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dicker AJ, Popa C, Dahler AL, et al: E2F-1
induces proliferation-specific genes and suppresses squamous
differentiation-specific genes in human epidermal keratinocytes.
Oncogene. 19:2887–2894. 2000. View Article : Google Scholar
|