Introduction
Gonadotropin-releasing hormone (GnRH) as a part of
the hypothalamic-pituitary-gonadal axis is the key hormone in the
control of reproductive functions. It induces the release of
gonadotropines, follicle-stimulating hormone (FSH) and luteinizing
hormone (LH), from the anterior pituitary gland. These hormones
have stimulating effects on the gonades resulting in an increased
production of estrogens, gestagens and androgens. GnRH acts by
binding to its specific high-affinity receptor localized on the
gonadotrope cells. The GnRH receptor, which belongs to the family
of 7TM domain receptors, regulates gene expression through
G-protein coupled signal cascades, involving phospholipases and
adenylate cyclase (1–3).
Given that estrogen exposure is a known risk factor
for breast cancer, the examination of genetic variants in genes
encoding for proteins of steroid hormone metabolism regulation
attracted high attention (4–8).
Genetic variations of GnRHR gene, which might affect GnRH receptor
levels or the structure of this receptor, could influence the
disease rate or cancer progression by differential regulation of
steroid hormone synthesis. Besides this hypothesis of indirect
effects on tumor progress via steroide hormones, a direct effect by
extrapituitary GnRHR receptor signaling in gynecological cancer
cells has been shown (9,10). The majority of human breast cancers
express GnRHR and evidence is growing for an estrogen-independent
intrinsic regulation of breast cancer cell proliferation mediated
by GnRHR (11). Thus, GnRHR has
been suggested to be an interesting therapeutic target for breast
cancer therapy (12–16).
In this phenotype-genotype association study, we
tested whether three SNPs in the GnRHR gene might be associated
with breast cancer risk or with histopathological characteristics
of the tumor. For this purpose, we genotyped 565 DNA samples from
women with or without breast cancer and analyzed the genotype
frequency, allele frequency and allele positivity in both groups
and in histopathological sub-groups.
Patients and methods
Patients
Blood samples from 254 Caucasian women with sporadic
breast cancer and 311 age-matched Caucasian women without any
malignancy were included in this retrospective study. The
histopathological characteristics of the patients are shown in
Table I. Whole blood or DNA samples
of breast cancer case participants were provided by the Institute
of Pathology, University of Regensburg, in an anonymous and
randomized manner or have been collected at the department of
Obstetrics and Gynecology, University of Regensburg, between 2005
and 2007. Control subjects were selected from the same geographic
origin as the cases, the Oberpfalz area of Bavaria, Germany.
Inclusion criterion for the control subjects was the absence of any
clinical relevant malignancy at the beginning of the study. The
study was approved by the Ethics Committee of the University of
Regensburg and informed consent of the patients was collected.
 | Table IHistopathological characteristics and
receptor status of breast cancer cases included in this study
(n=254). |
Table I
Histopathological characteristics and
receptor status of breast cancer cases included in this study
(n=254).
| Characteristic | Patient (n) |
|---|
| Tumor size |
| pT1 | 146 |
| pT2 | 86 |
| pT3 | 8 |
| pT4 | 10 |
| pTa | 4 |
| Histological
grade |
| G1 | 31 |
| G2 | 137 |
| G3 | 83 |
| Ga | 3 |
| Nodal status |
| N0 | 156 |
| N1–3 | 82 |
| Na | 16 |
| ERα status |
| Negative | 40 |
| Positive | 204 |
| Intermediateb | 7 |
| ERa | 3 |
| PR status |
| Negative | 83 |
| Positive | 149 |
| Intermediateb | 20 |
| PRa | 2 |
| Her2 status |
| Negative | 195 |
| Positive | 41 |
| Her2a | 18 |
SNP analysis
Three SNPs in the GnRHR gene were identified using
the web sites www.genecards.org and http://www.ncbi.nlm.nih.gov/SNP. The basis of SNP
selection was their possible biological relevance. All chosen SNPs
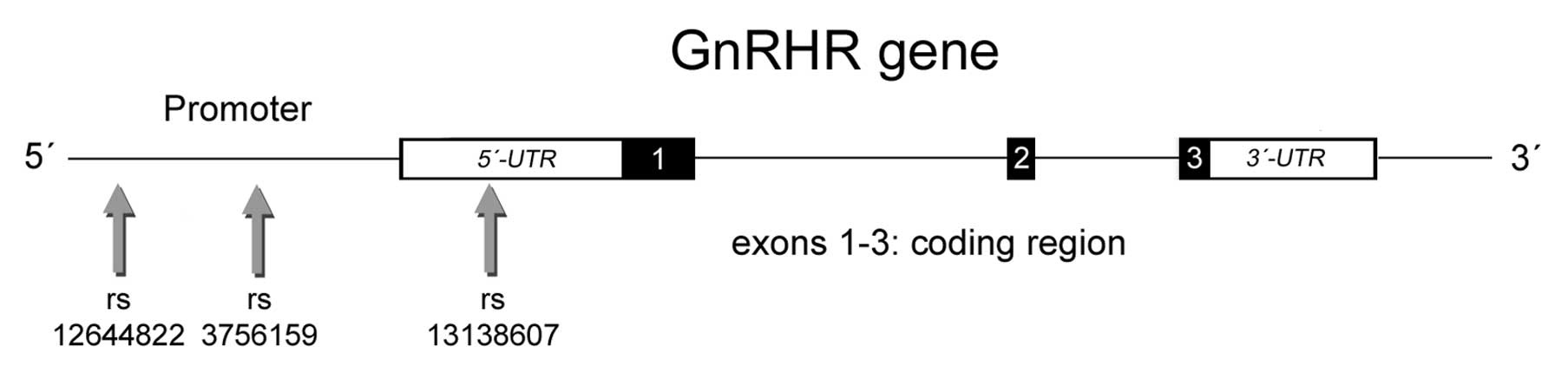
are located in regulatory regions of GnRHR gene. The polymorphism
rs13138607 is located at position 68304145 on chromosome 4 in the
5′ untranslated region of GnRHR gene, in an exonic splicing
enhancer binding site (Fig. 1). SNP
rs12644822 at position 68305130 is located in the 5′ region near
the GnRHR gene. Polymorphism rs3756159 at position 68305073 also
located in the 5′ region near the gene is part of a transcription
factor binding site. Reported HapMap genotype frequencies for SNP
rs13138607, from the web site http://www.ncbi.nlm.nih.gov/SNP, were 0.298 (T/T),
0.351 (C/C) and 0.351 (T/C), in a population of 114 individuals
from UT, USA with ancestors in Western and Northern Europe (CEU).
Published frequencies for rs12644822 were 0.217 (T/T), 0.450 (C/C)
and 0.333 (T/C) in the HapMap CEU population, and 0.288 (T/T),
0.339 (C/C) and 0.373 (T/C) for SNP rs3756159 in the same
population.
Genomic DNA was extracted from 100 μl EDTA-blood
after addition of 300 μl lysis buffer [1% v/v Triton-X, 0.32M
Sucrose, 0.01M Tris (pH 7.5) and 5 mM MgCl2) and 2-fold
centrifugation (13,000 × g] for 30 sec. Pellet was resuspended in
50 μl PCR buffer (GoTaq buffer, Promega, Madison, WI, USA)
containing 0.5% Tween-20 and 10 mAnson units proteinase K (Merck,
Darmstadt, Germany) followed by incubation at 50°C overnight and
finally heat inactivation of the enzyme for 10 min at 95°C. The
genomic DNA-containing lysate was subjected to a tetra-primer ARMS
PCR approach (17) allowing
allele-specific amplification using the PCR primers listed in
Table II (synthesized at Metabion,
Martinsried, Germany). For this purpose, to 100 ng of genomic DNA,
2 μl of 5× GoTaq buffer, 0.2 μl of dNTP Mix (10 mM) (Fermentas, St.
Leon-Rot, Germany), 0.2 μl of each PCR primer (10 μM) and 0.5 units
GoTaq polymerase (Promega) were added and PCR reaction was carried
out using a T1 thermocycler (Biometra, Göttingen, Germany). The PCR
program for SNP analysis was 10 min 94°C followed by 38 PCR cycles
(rs13138607 and rs3756159) respectively 37 cycles (rs12644822) of
94°C (30 sec), 60°C (30 sec) and 72°C (60 sec), followed by a final
extension for 5 min step at 72°C. PCR products were analyzed by
1.5% agarose gel electrophoresis. Allele-specific PCR product sizes
for SNP rs13138607 were 187/248 bp (C/T), 197/263 bp (C/T) for
rs12644822 and 112/149 bp (C/T) for SNP rs3756159. As a control for
genotyping in each PCR run three previously characterized samples
representing the heterozygous and the two homozygous genotypes in
addition to the unknown samples were analyzed.
 | Table IIOligonucleotides used for SNP
genotyping by means of tetra primer PCR. |
Table II
Oligonucleotides used for SNP
genotyping by means of tetra primer PCR.
| SNP | Primer | Sequence (5→3′) |
|---|
| rs12644822 | 822-T |
AAGTAGAGGGCGAATGATGTTTGCCA |
| 822-C |
ATTCAACAACTTAGAGCTCCTCAAAGGC |
| 822–1 |
TGTCTCTGCTTCACCTTCCTCAACCATA |
| 822-2 |
AGAATGCCTTGAAGGGATTTGGGAAATA |
| rs3756159 | 159-T |
CCGACTTTCATAGCCACACCCTGAAT |
| 159-C |
CACAACATGAAAGGTATAAAGCCCTCCAG |
| 159-1 |
TTCAACAACTTAGAGCTCCTCAAATGCG |
| 159-2 |
TGTAGCATACAGAGAATGCCTTGAAGGG |
| rs13138607 | 607-T |
TTGTGACCATAAAATTTTTACCTCCA |
| 607-C |
TTCTCCTAGATGAGTCAGAACTTAGTTTTTAC |
| 607-1 |
TATTTGTATGTCTTTCCAATGGTTATCC |
| 607-2 |
CTGAGCTCCTTTTTGACTGTCACTATAT |
Statistical analysis
Hardy-Weinberg equilibrium was estimated by the
Fisher’s exact test and the χ2 test, and all values were
subjected to one-way ANOVA to achieve homogeneity of variance.
Statistical tests for association (95% CI, confidence interval) and
for significance were carried out using SPSS for Windows 8.0 (SPSS,
Inc., Chicago, IL, USA). Afterwards tests for deviation from
Hardy-Weinberg equilibrium were conducted, allele frequency, allele
positivity and genotype frequencies were determined. Odds ratio
(OR) was calculated using the more frequent homozygous genotypes as
reference group.
Results
Although neither analysis of genotype and allele
frequency nor of allele positivity of GnRHR SNPS rs13138607,
rs3756159 and rs12644822 revealed significant differences between
the cancer and the control group (Table III), we observed differences in
SNP frequencies after sub-grouping the breast cancer cases with
regard to their tumor grading.
 | Table IIIAnalysis of GnRHR SNP frequencies in
breast cancer cases and controls (healthy women). |
Table III
Analysis of GnRHR SNP frequencies in
breast cancer cases and controls (healthy women).
| Genotype
frequency | Allele frequency | Allele
positivity |
|---|
|
|
|
|
|---|
| CC | TT | CT | C | T | C | T |
|---|
| SNP rs12644822 |
| Cases (n=240) | 0.35 | 0.12 | 0.54 | 0.61 | 0.39 | 0.88 | 0.65 |
| Controls
(n=243) | 0.36 | 0.14 | 0.50 | 0.61 | 0.39 | 0.86 | 0.64 |
| P-value | | 0.621 | 0.605 | | 0.860 | | 0.443 |
| SNP rs3756159 |
| Cases (n=241) | 0.21 | 0.20 | 0.59 | 0.50 | 0.50 | 0.80 | 0.79 |
| Controls
(n=244) | 0.26 | 0.24 | 0.50 | 0.51 | 0.49 | 0.76 | 0.74 |
| P-value | | 0.878 | 0.140 | | 0.773 | | 0.305 |
| SNP rs13138607 |
| Cases (n=238 | 0.24 | 0.24 | 0.52 | 0.50 | 0.50 | 0.76 | 0.76 |
| Controls
(n=243) | 0.26 | 0.23 | 0.51 | 0.52 | 0.48 | 0.77 | 0.74 |
| P-value | | 0.652 | 0.820 | | 0.663 | | 0.694 |
Homozygous analysis of SNP rs13138607, located in an
exonic splicing enhancer binding site of GnRHR gene, demonstrated
that the CC genotype was less frequent in patients with poorly
differentiated (G3) tumors (18%) than in patients with better
differentiated (G1 and G2) tumors (27%, OR, 2.65, P=0.017) or in
healthy women (26%, OR, 2.03, P=0.04) (Table IV). Analysis of allele frequency
and allele positivity of this SNP also showed the C allele to be
less frequent in G3 tumors than in G1 and G2 tumors (frequency 42
vs. 54%, OR, 1.61, P=0.017, positivity 66 vs. 80%, OR, 2.05,
P=0.021) and than in healthy women (OR, 1.48, P=0.038).
 | Table IVSubgroup analysis of GnRHR SNP
frequencies. |
Table IV
Subgroup analysis of GnRHR SNP
frequencies.
| Genotype
frequency | Allele
frequency | Allele
positivity |
|---|
|
|
|
|
|---|
| CC | TT | CT | C | T | C | T |
|---|
| SNP rs12644822 |
| G1+2 (n=163) | 0.29 | 0.13 | 0.58 | 0.58 | 0.42 | 0.87 | 0.71 |
| G3 (n=74) | 0.46 | 0.09 | 0.45 | 0.68 | 0.32 | 0.91 | 0.54 |
| P-value | | 0.120 | 0.019 | | 0.039 | | 0.013 |
| OR | | | 2.018 | | 1.538 | | 2.037 |
| 95% CI | | | 1.12–3.65 | | 1.02–2.32 | | 0.28–0.87 |
| SNP rs3756159 |
| G1+2 (n=161) | 0.23 | 0.16 | 0.61 | 0.53 | 0.47 | 0.84 | 0.77 |
| Controls
(n=311) | 0.23 | 0.26 | 0.51 | 0.49 | 0.51 | 0.74 | 0.74 |
| P-value | | 0.1293 | 0.447 | | 0.1707 | 0.0324 | 0.9669 |
| OR | | | | | | 1.682 | |
| 95% CI | | | | | | 0.34–0.91 | |
| SNP rs13138607 |
| G3 (n=74) | 0.18 | 0.34 | 0.49 | 0.42 | 0.58 | 0.66 | 0.82 |
| Controls
(n=311) | 0.26 | 0.23 | 0.51 | 0.52 | 0.48 | 0.77 | 0.74 |
| P-value | | 0.04 | 0.3511 | | 0.038 | 0.053 | 0.1403 |
| OR | | 2.03 | | | 1.482 | | |
| 95% CI | | 1.03–4.72 | | | 1.02–2.15 | | |
| G1+2 (n=161) | 0.27 | 0.20 | 0.53 | 0.54 | 0.46 | 0.80 | 0.73 |
| G3 (n=74) | 0.18 | 0.34 | 0.49 | 0.42 | 0.58 | 0.66 | 0.82 |
| P-value | | 0.017 | 0.333 | | 0.017 | 0.021 | 0.1048 |
| OR | | 2.65 | | | 1.61 | 2.05 | |
| 95% CI | | 0.17–0.85 | | | 0.42–0.92 | 0.26–0.90 | |
Comparing the grading subgroups for SNP rs3756159,
we found the C-allele positivity to be increased in the G1+G2 group
when compared to the group of women without any malignancy (OR,
1.682, P=0.324).
Examining SNP rs12644822, a higher incidence of
homozygote CC vs. the heterozygote genotype was found in poorly
differentiated tumors (G3) in comparison to well or moderately
differentiated tumors (G1 and G2) (OR, 2.018, P=0.019).
Additionally, both T-allele frequency and T-allele positivity of
SNP rs12644822 were decreased in G3 tumors (Table III).
Discussion
Since discovery of common genetic variants such as
single-nucleotide polymorphisms, genotype-phenotype association
studies have explored their impact as susceptibility factors
predisposing individuals to a variety of diseases. SNPs located in
exon regions of a gene may alter protein structure and function or
may influence gene expression levels when localized in gene
regulatory regions. In this study, we selected SNPs with a
potential relevance for gene expression, located in an exonic
splicing enhancer binding site or in the 5′-promoter region of
GnRHR gene.
Significant association of common alleles with an
etiologically complex disease like breast cancer was shown in
numerous studies (18–21). SNPs in genes whose products are
known to be involved in malign pathophysiological processes
attracted the main attention. In hormone-dependent cancer such as
breast cancer, particularly SNPs in genes involved in estrogen
biosynthesis and signaling or hormones of the
hypothalamic-pituitary-gonadal axis are of great interest (22). In previous SNP studies on GNRH
receptor gene, certain polymorphisms have been found to be
associated with polycystic ovary syndrome, but did not associate
with idiopathic hypogonadotropic hypogonadism (23–26).
With regard to breast cancer, recently an SNP in the GnRH gene was
reported to be significantly associated with disease-free survival
and negative nodal status of premenopausal breast cancer patients
(27). In contrast, another large
breast cancer study on SNPs in the GnRH and GnRHR gene could not
find a statistically significant association with breast cancer
risk (26). Only one previous study
examined SNPs in the 5′-region of GnRHR gene, but did not associate
it with breast cancer risk, but with onset of menarche (28).
To the best of our knowledge, this is the first
study examining the relevance of three SNPs in the 5′-regulatory
region of GnRHR for breast cancer susceptibility. In our
population, the frequencies of the tested SNPs did not differ
between women with breast cancer and the control collective, but we
observed some differences between grading subgroups. However, due
to the performed multiple comparison testing, the obtained P-values
between 0.1 and 0.5 do not express statistical significance, but
only reflect a trend towards significance. Thus, the results
suggesting the T-allele of rs12644822 to be less frequent in G3
tumors, and C-allele positivity of rs3756159 to be increased in G1
and G2 tumors, needs to be confirmed on a larger patient
collective. The same is true for the observations on frequency of
SNP rs13138607, suggesting an association both of the TT genotype
and of T allele with G3 tumors.
In conclusion, we did not observe an association
between the tested SNPs in the 5′-regulatory region of GnRHR gene
and breast cancer susceptibility. Our data demonstrating a
statistical trend for association of GnRHR alleles with tumor
grading might encourage further studies on larger study
populations.
References
|
1
|
Chi L, Zhou W, Prikhozhan A, et al:
Cloning and characterization of the human GnRH receptor. Mol Cell
Endocrinol. 91:R1–R6. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lariviere S, Garrel G, Simon V, et al:
Gonadotropin-releasing hormone couples to 3′,5′-cyclic
adenosine-5′-monophosphate pathway through novel protein kinase
Cdelta and -epsilon in LbetaT2 gonadotrope cells. Endocrinology.
148:1099–1107. 2007.
|
|
3
|
Limonta P, Moretti RM, Marelli MM and
Motta M: The biology of gonadotropin hormone-releasing hormone:
role in the control of tumor growth and progression in humans.
Front Neuroendocrinol. 24:279–295. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee E, Schumacher F, Lewinger JP, et al:
The association of polymorphisms in hormone metabolism pathway
genes, menopausal hormone therapy, and breast cancer risk: a nested
case-control study in the California Teachers Study cohort. Breast
Cancer Res. 13:R372011. View
Article : Google Scholar
|
|
5
|
Udler MS, Azzato EM, Healey CS, et al:
Common germline polymorphisms in COMT, CYP19A1, ESR1, PGR, SULT1E1
and STS and survival after a diagnosis of breast cancer. Int J
Cancer. 125:2687–2696. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang L, Gu L, Qian B, et al: Association
of genetic polymorphisms of ER-alpha and the estradiol-synthesizing
enzyme genes CYP17 and CYP19 with breast cancer risk in Chinese
women. Breast Cancer Res Treat. 114:327–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Diergaarde B, Potter JD, Jupe ER, et al:
Polymorphisms in genes involved in sex hormone metabolism, estrogen
plus progestin hormone therapy use, and risk of postmenopausal
breast cancer. Cancer Epidemiol Biomarkers Prev. 17:1751–1759.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ralph DA, Zhao LP, Aston CE, et al:
Age-specific association of steroid hormone pathway gene
polymorphisms with breast cancer risk. Cancer. 109:1940–1948. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Emons G, Ortmann O, Becker M, et al: High
affinity binding and direct antiproliferative effects of LHRH
analogues in human ovarian cancer cell lines. Cancer Res.
53:5439–5446. 1993.PubMed/NCBI
|
|
10
|
Emons G, Schroder B, Ortmann O, Westphalen
S, Schulz KD and Schally AV: High affinity binding and direct
antiproliferative effects of luteinizing hormone-releasing hormone
analogs in human endometrial cancer cell lines. J Clin Endocrinol
Metab. 77:1458–1464. 1993.
|
|
11
|
Cheung LW, Leung PC and Wong AS:
Gonadotropin-releasing hormone promotes ovarian cancer cell
invasiveness through c-Jun NH2-terminal kinase-mediated activation
of matrix metalloproteinase (MMP)-2 and MMP-9. Cancer Res.
66:10902–10910. 2006. View Article : Google Scholar
|
|
12
|
Fost C, Duwe F, Hellriegel M, Schweyer S,
Emons G and Grundker C: Targeted chemotherapy for triple-negative
breast cancers via LHRH receptor. Oncol Rep. 25:1481–1487.
2011.PubMed/NCBI
|
|
13
|
Schubert A, Hawighorst T, Emons G and
Gründker C: Agonists and antagonists of GnRH-I and -II reduce
metastasis formation by triple-negative human breast cancer cells
in vivo. Breast Cancer Res Treat. 130:783–790. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Torrisi R, Bagnardi V, Rotmensz N, et al:
Letrozole plus GnRH analogue as preoperative and adjuvant therapy
in premenopausal women with ER positive locally advanced breast
cancer. Breast Cancer Res Treat. 126:431–441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grundker C, Fost C, Fister S, Nolte N,
Gunthert AR and Emons G: Gonadotropin-releasing hormone type II
antagonist induces apoptosis in MCF-7 and triple-negative
MDA-MB-231 human breast cancer cells in vitro and in vivo. Breast
Cancer Res. 12:R492010. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Emons G, Kaufmann M, Gorchev G, et al:
Dose escalation and pharmacokinetic study of AEZS-108 (AN-152), an
LHRH agonist linked to doxorubicin, in women with LHRH
receptor-positive tumors. Gynecol Oncol. 119:457–461. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye S, Dhillon S, Ke X, Collins AR and Day
IN: An efficient procedure for genotyping single nucleotide
polymorphisms. Nucleic Acids Res. 29:E88. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harlid S, Ivarsson MI, Butt S, et al:
Combined effect of low-penetrant SNPs on breast cancer risk. Br J
Cancer. 106:389–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pelletier C, Speed WC, Paranjape T, et al:
Rare BRCA1 haplotypes including 3′UTR SNPs associated with breast
cancer risk. Cell Cycle. 10:90–99. 2011.
|
|
20
|
Han W, Kang SY, Kang D, et al: Multiplex
genotyping of 1107 SNPs from 232 candidate genes identified an
association between IL1A polymorphism and breast cancer risk. Oncol
Rep. 23:763–769. 2010.PubMed/NCBI
|
|
21
|
Hamacher R, Diersch S, Scheibel M, et al:
Interleukin 1 beta gene promoter SNPs are associated with risk of
pancreatic cancer. Cytokine. 46:182–186. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park IH, Lee YS, Lee KS, et al: Single
nucleotide polymorphisms of CYP19A1 predict clinical outcomes and
adverse events associated with letrozole in patients with
metastatic breast cancer. Cancer Chemother Pharmacol. 68:1263–1271.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Q, Yang G, Wang Y, et al: Common
genetic variation in the 3′-untranslated region of
gonadotropin-releasing hormone receptor regulates gene expression
in cells and is associated with thyroid function, insulin secretion
as well as insulin sensitivity in polycystic ovary syndrome
patients. Hum Genet. 129:553–561. 2011.
|
|
24
|
Vagenakis GA, Sgourou A, Papachatzopoulou
A, Kourounis G, Papavassiliou AG and Georgopoulos NA: The
gonadotropin-releasing hormone (GnRH)-1 gene, the GnRH receptor
gene, and their promoters in patients with idiopathic
hypogonadotropic hypogonadism with or without resistance to GnRH
action. Fertil Steril. 84:1762–1765. 2005. View Article : Google Scholar
|
|
25
|
Lanfranco F, Gromoll J, von Eckardstein S,
Herding EM, Nieschlag E and Simoni M: Role of sequence variations
of the GnRH receptor and G protein-coupled receptor 54 gene in male
idiopathic hypogonadotropic hypogonadism. Eur J Endocrinol.
153:845–852. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valkenburg O, Uitterlinden AG, Piersma D,
et al: Genetic polymorphisms of GnRH and gonadotrophic hormone
receptors affect the phenotype of polycystic ovary syndrome. Hum
Reprod. 24:2014–2022. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Piersma D, Themmen AP, Look MP, et al:
GnRH and LHR gene variants predict adverse outcome in premenopausal
breast cancer patients. Breast Cancer Res. 9:R512007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nanao K and Hasegawa Y: Polymorphisms at
the 5′ end of the human gonadotropin-releasing hormone receptor
gene are not associated with the timing of menarche in Japanese
girls. Eur J Endocrinol. 143:555–556. 2000.
|