Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common malignancy worldwide and causes nearly one million deaths a
year (1). Despite the development
of several modalities for the treatment of HCC (2–7),
including transcatheter arterial embolization, percutaneous
ablation, surgical resection, liver transplantation and molecular
targeted medicine, the prognosis of patients with HCC still remains
relatively poor. One of the major factors responsible for these
unsatisfactory outcomes is the high frequency of intrahepatic
recurrence after curative treatment (5,8).
Intrahepatic recurrence is the result of two mechanisms:
intrahepatic metastasis originating from the primary cancer, and a
second primary cancer arising through multicentric carcinogenesis.
Intrahepatic metastasis may correlate with early recurrence and
poor prognosis, whereas multicentric carcinogenesis is associated
with a relatively good prognosis (9–11).
With the aim of controlling intrahepatic recurrence of HCC, various
studies have investigated the molecular mechanisms underlying
intrahepatic metastasis (12–17),
which frequently occurs at an advanced disease stage, presumably
through tumor cell dispersal via the portal vein; there is a strong
statistical correlation between the presence of intrahepatic
metastasis and the frequency of vascular invasion (18).
Tumor invasiveness may be considered a phenomenon of
cell motility. In fact, tumor cell motility plays a central role in
carcinoma cell dissemination, and the cytoskeleton, a key structure
of the cell machinery, is continuously remodeled during cell
movement. In this context, several molecules related to the
microtubule (MT)- and actin-dependent dynamics of tumor cells have
been investigated as possible predictors of intrahepatic metastasis
of HCCs, or targets of preventive therapy. Highly dynamic MTs are
distributed randomly throughout the cell periphery, and the less
dynamic ones are located between the nucleus and the leading edge
of the cell (19). A previous study
has revealed that overexpression of HDAC6 increases cell
motility, suggesting that deacetylation of at least one cytoplasmic
HDAC protein enhances motility (20). It has also been shown that in
HDAC6-inhibited cells, MT dynamics are decreased, leading to
an increase of focal adhesion accumulation, and thus a decrease in
cell motility (21). Moreover,
HDAC6 protein can also interact with a different substrate,
cortactin, in vivo and in vitro, and both HDAC6
catalytic domains are necessary for the interaction. Cortactin is
an acetylated protein found in areas of dynamic actin assembly,
such as the leading edge of migrating cells (22). This protein was originally
identified as a substrate of Src tyrosine kinase, and plays
a role in regulating cell motility. Disruption of HDAC6 leads to
hyperacetylation of cortactin and prevents its translocation to the
cell periphery, blocks association with F-actin, and impairs cell
motility (23). We recently
demonstrated that disruption of the HDAC6/NACC1 (nucleus
accumbens associated 1) deacetylation system markedly downregulated
cell motility through MT and cortactin deacetylation (24). Thus, HDAC6 may act as a mediator
between actin- and tubulin-associated proteins to regulate cell
motility.
Although overexpression of HDAC6 and its
relationship with invasion and metastasis have been documented in
several malignancies (24–26), it has not been documented in HCCs.
The present study examined the expression of HDAC6 in HCC
cultured cells and primary tumors, and investigated its association
with migration and invasion activities in vitro and in
vivo.
Materials and methods
Cell culture
HCC cell lines were obtained from HSRRB (Health
Science Research Resources Bank, Osaka, Japan; HLF, PLC/PRF/5) and
IDAC (Institute of Department, Aging and Cancer, Tohoku University,
Sendai, Japan; Hep3B). Two human normal hepatocyte cell lines,
Hc-cells (Applied Cell Biology Research Institute, Human Hepatocyte
Cell culture #3716, Kirkland, WA, USA) and WRL-68 (American Type
Culture Collection, Rockville, MD, USA), were obtained commercially
and maintained under the recommended conditions.
Surgical specimens and
immunohistochemistry
Immunohistochemistry for HDAC6 protein expression
was performed on tumor samples from 70 patients with HCC treated
between 2006 and 2011 at the Department of Surgery, School of
Medicine, Iwate Medical University, Morioka, Japan. The patient
characteristics are summarized in Table
I. Permission for the study was obtained from the Institutional
Review Board (School of Medicine, Iwate Medical University,
Morioka, Japan) and written consent was obtained from all patients
before surgery.
 | Table ICharacteristics of 70 patients with
hepatocellular carcinoma. |
Table I
Characteristics of 70 patients with
hepatocellular carcinoma.
| Factor | No. of patients
(%) |
|---|
| Gender |
| Male | 52 (74) |
| Female | 18 (26) |
| Age (years) |
| Mean (range) | 64.2 (39–81) |
| <65 | 33 (47) |
| ≥65 | 37 (53) |
| Clinical stage |
| I | 9 (13) |
| II | 31 (45) |
| III | 26 (38) |
| IVA | 4 (4) |
| No. of tumors |
| Single | 50 (71) |
| Multiple | 20 (29) |
| Tumor diameter
(cm) |
| <3 | 24 (35) |
| ≥3 | 46 (65) |
| Lymph node
status |
| Yes | 2 (3) |
| None | 68 (97) |
| Virus
infection |
| HBV | 28 (40) |
| HCV | 21 (30) |
| HBV + HCV | 1 (1) |
| Non infection | 20 (29) |
| Vascular
invasion |
| Presence | 20 (28) |
| Absence | 50 (72) |
| Intrahepatic
metastasis |
| Presence | 19 (27) |
| Absence | 51 (73) |
Surgical specimens were fixed in 10% buffered
formalin solution and embedded in paraffin wax, and two or more
blocks were made for immunohistochemistry. Sections 4 μm thick were
cut, and stained with hematoxylin and eosin. Serial sections were
stained with the avidin-biotin system on a Ventana automated
immunostainer with the Ventana immunohistochemistry detection
system (Ventana Medical Systems, Tucson, AZ, USA), in accordance
with the manufacturer’s manual. An anti-HDAC6 antibody (H-300,
diluted 1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA) was
used as the primary antibody.
Western blotting
All cell lines were cultured to 70–80% confluence on
10-cm Petri dishes. Cold PBS was added, and the cells were removed
from the dishes by scraping. After removal of the supernatants, the
cell pellet was dissolved in cell lysis buffer [50 mM Tris-HCl, pH
8.0/150 mM NaCl/1 mM EDTA, pH 8.0/1% Triton X-100/0.1% sodium
deoxycholate/0.1% SDS/1 mM PMSF/10 mM NaF/2 mM
Na3VO4/1× protease inhibitor complete (Roche
Diagnostics GmbH, Mannheim, Germany)]. Cell samples containing
equal amounts of protein were mixed with 5× sample buffer, and
heated for 5 min at 95°C. Protein was electrophoresed on 4–12%
Nu-PAGE for 45 min at 200 V constant voltage, and then transferred
onto polyvinylidene difluoride membranes (Invitrogen, Carlsbad, CA,
USA) for 1 h at 30 V constant voltage. Membranes were blocked with
5% blocking reagent (Cell Signaling Technology, Danvers, MA, USA)
in 1× Tris-buffered saline/Tween-20 buffer for 1 h at room
temperature, and immunostained overnight with a primary antibody
(HDAC6 H-300, diluted 1:250, Santa Cruz Biotechnology) at 4°C. The
membranes were then rinsed with Tris-buffered saline/Tween-20 and
incubated with horseradish HRP-conjugated secondary antibodies
[anti-rabbit or -mouse IgG (diluted 1:5,000, GE Healthcare, Little
Chalfont, UK)] for 1 h at room temperature. Signals were detected
with ECL Prime (GE Healthcare) and ChemiDoc XRS (Bio-Rad
Laboratories, Hercules, CA, USA). The intensity of the detected
signals was measured using 1-D analysis software (Quantity One,
Bio-Rad Laboratories). For normalization of the target,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH, diluted 1:100;
Covance, Princeton, NJ, USA) was used as an internal control.
RNA isolation and reverse transcriptase
quantitative PCR
Total RNA was extracted with TRIzol reagent
(Invitrogen), and transcribed to cDNA using a SuperScript III
first-strand synthesis system (Invitrogen). For quantitative
evaluation of relevant mRNAs, we used Custom TaqMan Gene Expression
assays (HDAC6, Hs00195869_ m1; Invitrogen), and an ABI PRISM 7500
instrument (Invitrogen). For normalization of the target, GAPDH
(Invitrogen) was used as an internal control. All reactions (each
containing 3 templates) were run in triplicate, and average fold
differences were calculated by normalizing the relative expression
(ΔΔCt values) according to ABI User Bulletin #2.
siRNA knockdown of the HDAC6 gene
For silencing of HDAC6 gene mRNA, three
predesigned HDAC6-specific siRNA sequences (#1, s19459; #2,
s19760; #3, s19461; Silencer Select siRNA, Invitrogen), and control
non-specific human siRNAs (Silencer Select Pre-designed siRNA
Negative Control #1, 4390843; #2, 4390844, Invitrogen) were used.
siRNA transfection was performed using Lipofectamine RNAiMAX
Reagent (Invitrogen) in accordance with the manufacturer’s
instructions.
Scratch assay
Confluent monolayer cells were scratched to create a
wound, and then 0, 24 and 48 h later, three different fields of
each wound were photographed with a phase-contrast microscope.
Three independent experiments were performed. Measurements of the
width of each wound were performed under each experimental
condition. At the start of the experiment, the wound size was
measured and scored as 100%. After 24 and 48 h, the width of the
residual wound was measured and the average percentage of wound
closure was calculated by using the free web software ImageJ
(http://rsb.info.nih.gov/ij).
Matrigel invasion assay
The cell invasion assay was performed using a
BioCoat Matrigel invasion chamber (Becton-Dickinson, Bedford, MA,
USA) in accordance with the protocol provided by the manufacturer.
After cells in log-phase growth had been incubated with serum-free
medium for 12 h, they were detached using trypsin-EDTA. Resuspended
cells were added to each chamber at a density of 5×104
cells in 500 μl, and allowed to invade the Matrigel for 24 h at
37°C under a 5% CO2 atmosphere. Cells that had not
penetrated the filter were wiped out with cotton swabs, and cells
that had migrated to the lower surface of the filter were stained
with Quick-Diff stain kit (Symex International Reagents, Co., Ltd.,
Hyogo, Japan). After two washes with water, the chambers were
allowed to air-dry, and the number of invading cells was counted
using a light microscope. The degree of invasion was expressed as
the average number of migrated cells bound per microscopic field
over four fields per assay, and as averages for triplicate
experiments.
Statistical analysis
All data are presented as means ± standard error.
Correlations between HDAC6 protein expression and
clinicopathological data were analyzed by Fisher’s exact test or
Kruskal-Wallis test. Mann-Whitney U test for non-parametric samples
was used for analyses of biological experimental data. The level of
significance was considered to be P<0.05.
Results
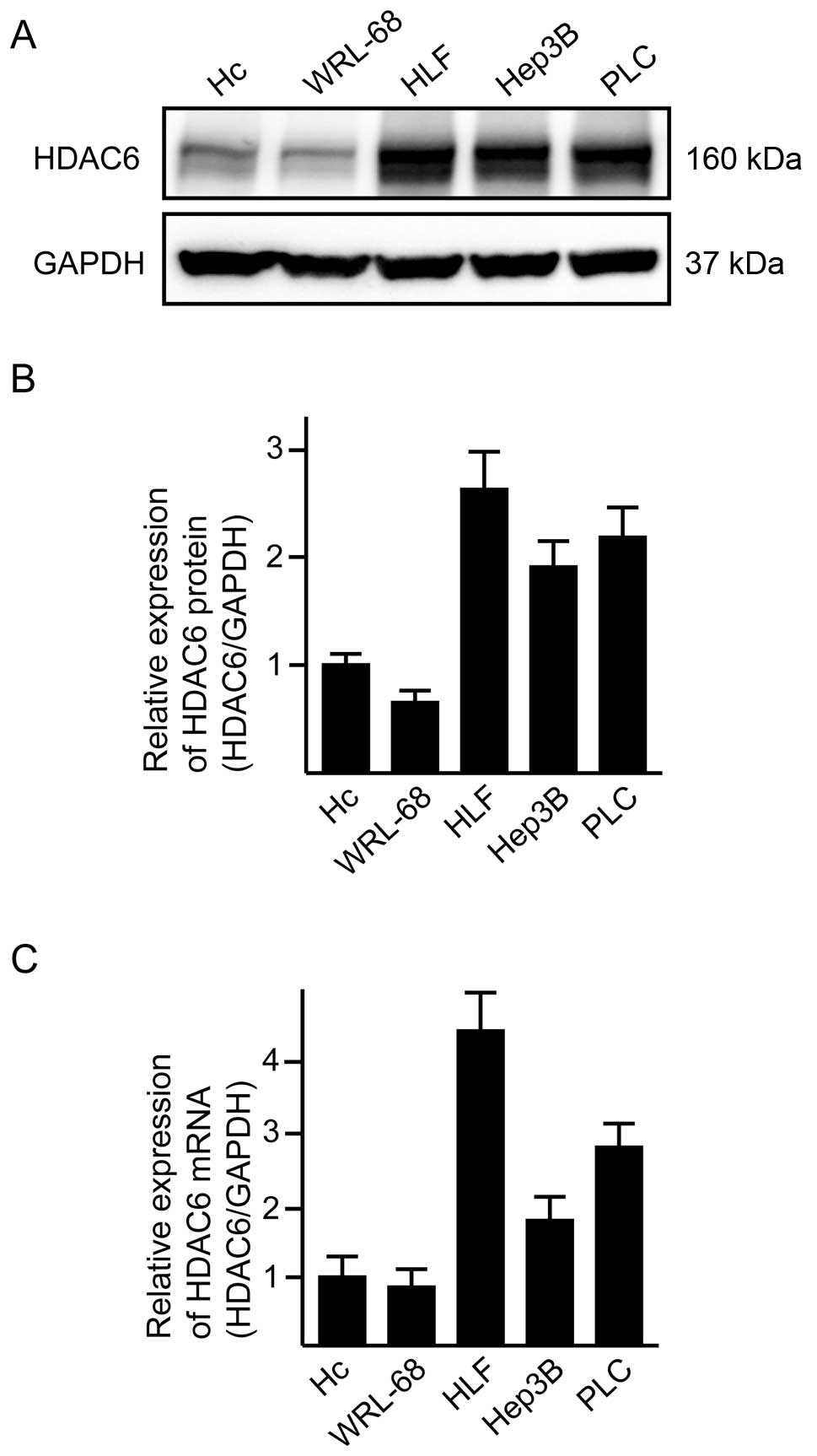
Expression of the HDAC6 gene in cell
lines and efficiency of knockdown by treatment with siRNA
We first examined HDAC6 mRNA/protein expression in
three HCC cell lines and two primary-cultured normal hepatocyte
lines (Fig. 1). Under the
recommended culture conditions at 60–70% confluency, all of the HCC
cell lines exhibited overexpression of HDAC6 mRNA/protein in
comparison with normal hepatocytes (Fig. 1). We evaluated the knockdown
efficiency of HDAC6-siRNAs (#1, #2 and #3; 10 nM) in one HCC
cell line (PCL, Fig. 2). In
comparison with negative control siRNA, all the siRNAs caused
80–90% downregulation of HDAC6 expression (Fig. 2).
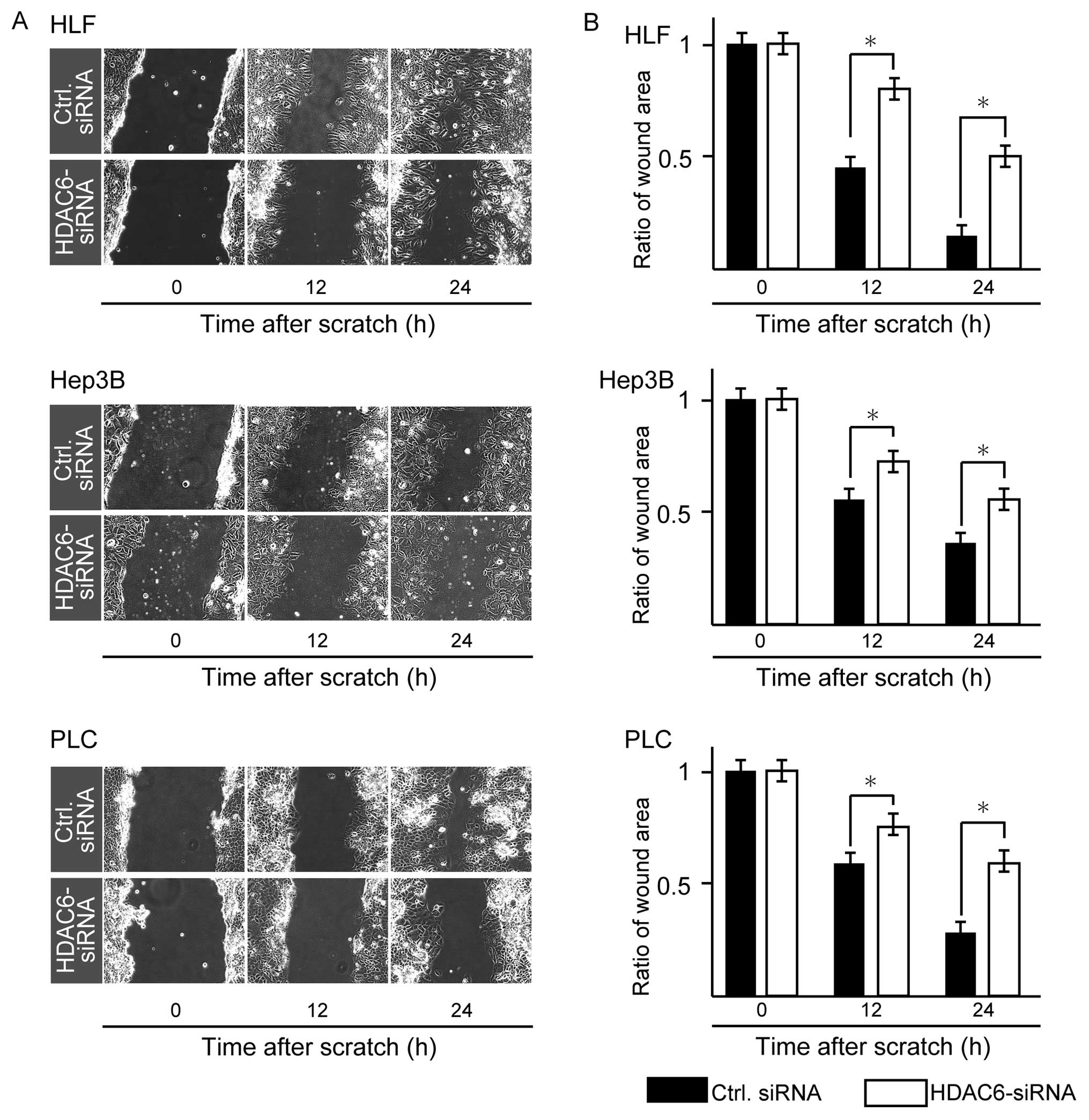
Migration and invasion activities induced
by treatment with HDAC6-siRNA in HCC cell lines
Using #1 HDAC6-siRNA, we then examined the
migration activities of the three HCC cell lines by the scratch
assay. HDAC6 knockdown significantly decreased tumor cell
migration activities of all three lines in comparison with the
negative control at 24 and 48 h (P<0.05, Mann-Whitney U-test;
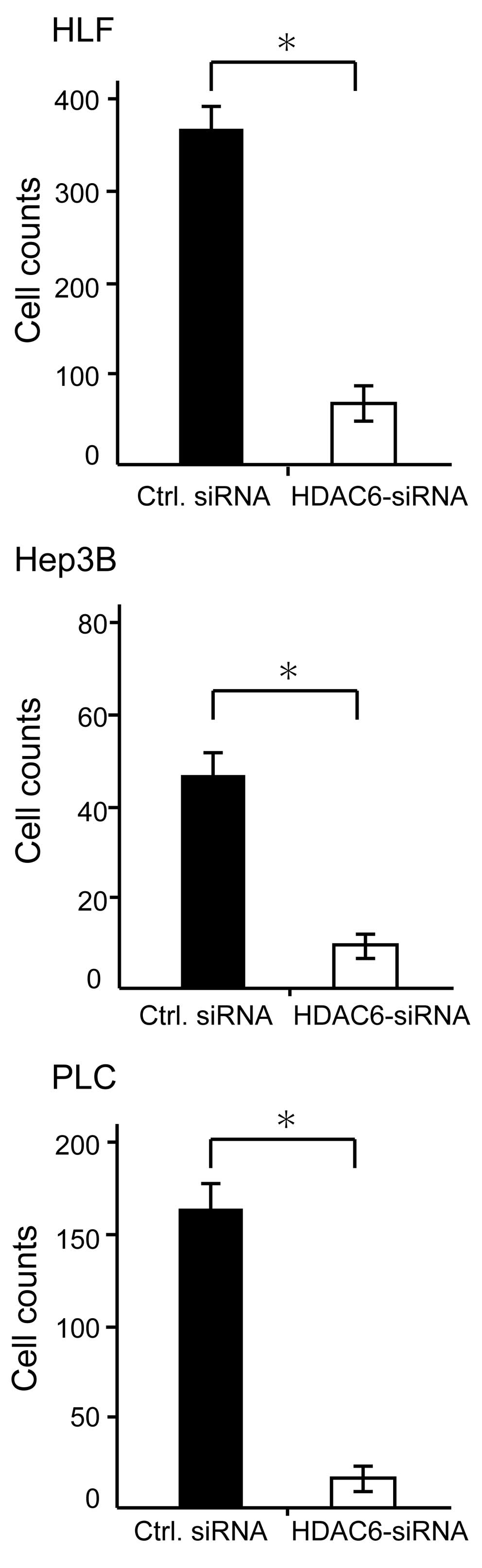
Fig. 3). To determine whether HDAC6
knockdown decreased the invasiveness of HCC cells, we performed the
Matrigel invasion assay. Treatments with HDAC6-siRNA
significantly suppressed the invasiveness of all HCC cell lines in
comparison with control siRNA treatment (Fig. 4).
Immunohistochemistry of HDAC6 protein,
and relationship between HDAC6 expression and clinicopathological
variables in primary HCCs
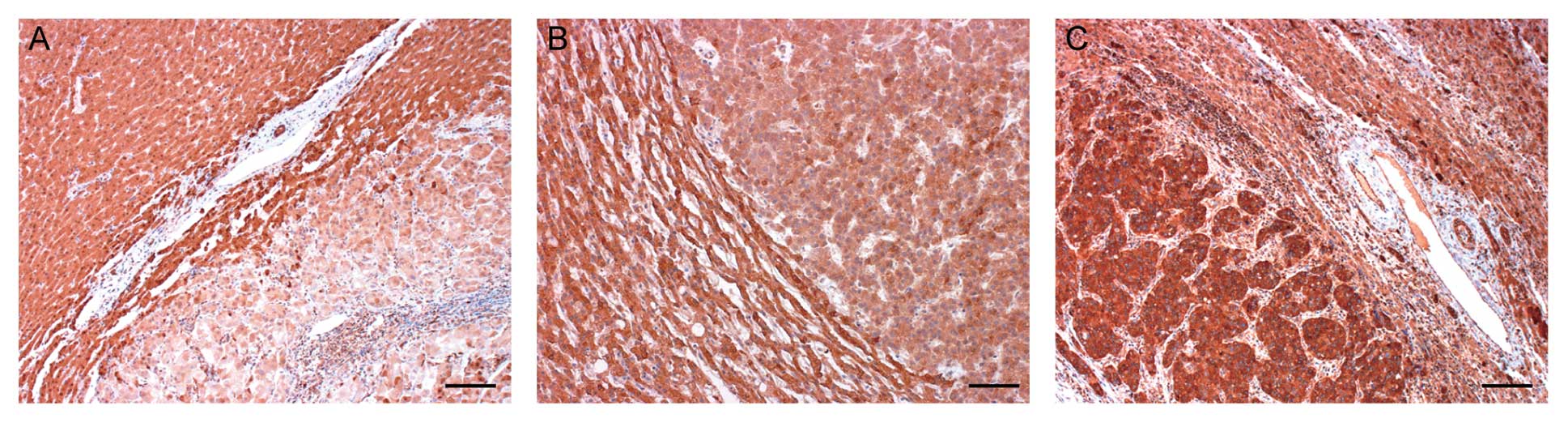
We immunohistochemically examined HDAC6 protein
expression in 70 patients with primary HCCs. Two independent
pathologists performed the assessment of immunohistochemical
staining. Immunoreactivity for HDAC6 was diffusely positive in both
tumor cells and surrounding normal hepatocytes. Overexpression of
HDAC6 protein to a level higher than that in the corresponding
normal hepatocytes was observed in 14 (20%, Fig. 5C) of the 70 primary HCCs. Lower
immunoreactivity for HDAC6 protein was found in 21 (30%, Fig. 5A) of the 70, and the remaining 35
(50%, Fig. 5B) exhibited
immunoreactivity equal to that of the corresponding normal
hepatocytes. Table II summarizes
the relationship between HDAC6 immunoreactivity and
clinicopathological variables in all cases. Overexpression of HDAC6
protein was significantly correlated with high clinical stage,
number of tumors, vascular invasion, and intrahepatic metastasis
(P<0.05) (Table II).
 | Table IIClinicopathological variables of
patients according to HDAC6 expression. |
Table II
Clinicopathological variables of
patients according to HDAC6 expression.
| | Immunoreactivity of
HDAC6 | |
|---|
| |
| |
|---|
| Factor | No. of
patients | 0, +1, +2 (%) | +3 (%) | P-value |
|---|
| Gender |
| Male | 52 | 42 (60) | 10 (14) | 0.513a |
| Female | 18 | 14 (20) | 4 (6) | |
| Age (years) |
| <65 | 33 | 27 (39) | 6 (9) | 0.719a |
| ≥65 | 37 | 29 (41) | 8 (11) | |
| Clinical stage |
| I, II | 40 | 36 (51) | 4 (6) | 0.017a |
| III, IV | 30 | 20 (29) | 10 (14) | |
| No. of tumors |
| Single | 50 | 43 (62) | 7 (10) | 0.047a |
| Multiple | 20 | 13 (18) | 7 (10) | |
| Tumor diameter
(cm) |
| <3 | 24 | 20 (29) | 4 (6) | 0.433a |
| ≥3 | 46 | 36 (51) | 10 (14) | |
| Virus
infection |
| HBV | 28 | 22 (31) | 6 (9) | 0.960b |
| HCV | 21 | 17 (24) | 4 (6) | |
| HBV+HCV | 1 | 1 (1) | 0 (0) | |
| No infection | 20 | 6 (9) | 14 (20) | |
| Vascular
invasion |
| Presence | 20 | 12 (17) | 8 (11) | 0.008a |
| Absence | 50 | 44 (63) | 6 (9) | |
| Intrahepatic
metastasis |
| Presence | 19 | 12 (17) | 7 (10) | 0.031a |
| Absence | 51 | 44 (63) | 7 (10) | |
Discussion
Several growth factors and cytokines such as
transforming growth factor (27),
platelet-derived growth factor (28), epidermal growth factor (29), hepatocyte growth factor (30), and extracellular matrix (31) secreted into the microenvironment
surrounding tumor cells are involved in their migration and
invasion. These secreted proteins and their related intracellular
signaling cascades induce epithelial mesenchymal transition and
accelerate the migration/invasion activity of HCC, resulting in
intrahepatic metastasis. Moreover, several adhesion molecules and
their related proteins such as E-cadherin, ROCK2 and CD24 also are
involved (32–34). Apart from proteins secreted into the
microenvironment of tumor cells and adhesion molecules,
transcriptional factors such as p300 and
Snail(35,36) also contribute to the acquisition of
metastatic potential by HCC cells. Our present study showed that
overexpression of HDAC6, which affects both MT- and actin-dependent
cell migration mechanisms, contributed to acceleration of
migration/invasion activity in HCC cell lines in vitro and
in vivo. In particular, the results of immunostaining of
primary HCCs were well correlated with intrahepatic metastasis.
HDAC6 was thus suggested to be a newly characterized key
player in the control of intrahepatic metastasis.
HDAC6 is a unique protein of the histone
deacetylase family, and can affect the function of cytoplasmic
non-histone proteins. It is a key regulator of many aspects of
cancer biology such as the cell cycle, cell migration, drug
resistance and autophagy, thereby making HDAC6 an attractive
target for cancer therapy (37).
Although HDAC6 protein is expressed in the liver as well as the
heart, kidney, testis, brain, and pancreas (38), there has been little information
about the significance of HDAC6 in HCC tumorigenesis. It is
well known that HDAC inhibitors, such as TSA and SAHA, block
invasive cell motility, and therefore it is anticipated that they
might be applicable for control of intrahepatic metastasis.
However, most of these molecules act by altering gene expression
via hyperacetylated HDAC nuclear substrates, such as histones or
transcription factors, and the spectrum of targeted molecules is
broad. Therefore, development of inhibitors that are more selective
in targeting intrahepatic metastasis is warranted. An HDAC6
inhibitor known as tubacin (tubulin acetylation inducer) was
isolated through a multidimensional chemical genetic screen of
7,392 small molecules and a cell-based assay targeting the
acetylation activity of proteins other than histones (39,40).
Unlike other histone deacetylase inhibitors, tubacin was found to
inhibit the deacetylation of MT in mammalian cells without
affecting the level of histone acetylation, gene expression, or
cell cycle progression (13,14).
Furthermore, using scratch assay and trans-Matrigel migration
assays, another group has demonstrated that NK84-mediated
inhibition of HDAC6 in ovarian cancer cell lines retarded
cell spreading and inhibited cell migration, respectively (41). Recently, the effectiveness of
combination therapy using an HDAC6-selective inhibitor
(ACY-1215) and bortezomib has been demonstrated in a preclinical
trial (42). Thus, a new class of
agents targeting HDAC6 is currently being developed, and
their efficacy is being tested.
The functions of HDAC6 in cell
migration/invasion activity may depend on decacetylation activity
targeting α-tubulin and cortactin. Both proteins are also
deacetylated by SIRT2, which belongs to another class of the
histone deacetylase family. Using SIRT2-specific siRNA
combined with tubacin treatment, Zuo et al(25) have demonstrated that cell migratory
and invasive abilities can be dramatically suppressed. Moreover,
SIRT2-deficient mice show gender-specific tumorigenesis, females
primarily developing mammary tumors, and males developing more HCCs
(43). The significance of SIRT2
should therefore be examined in human HCC tumorigenesis, including
its relationship with intrahepatic metastasis.
Acknowledgements
This study was supported in part by Grants-in-Aid
for Scientific Research (22390071), the MIAST project, and a
Grant-in-Aid for Strategic Medical Science Research from the
Ministry of Education, Culture, Sports, Science and Technology of
Japan.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2
|
Takahara T, Nitta H, Hasegawa Y, Itou N,
Takahashi M and Wakabayashi G: Using sorafenib for recurrent
hepatocellular carcinoma after liver transplantation - interactions
between calcineurin inhibitor: two case reports. Transplant Proc.
43:2800–2805. 2011. View Article : Google Scholar
|
|
3
|
Nitta H, Sasaki A, Fujita T, et al:
Laparoscopy-assisted major liver resections employing a hanging
technique: the original procedure. Ann Surg. 251:450–453. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sasaki A, Nitta H, Otsuka K, Takahara T,
Nishizuka S and Wakabayashi G: Ten-year experience of totally
laparoscopic liver resection in a single institution. Br J Surg.
96:274–279. 2009.PubMed/NCBI
|
|
5
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar
|
|
6
|
Salhab M and Canelo R: An overview of
evidence-based management of hepatocellular carcinoma: a
meta-analysis. J Cancer Res Ther. 7:463–475. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomas MB, Jaffe D, Choti MM, et al:
Hepatocellular carcinoma: consensus recommendations of the National
Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol.
28:3994–4005. 2010. View Article : Google Scholar
|
|
8
|
Izumi N, Asahina Y, Noguchi O, et al: Risk
factors for distant recurrence of hepatocellular carcinoma in the
liver after complete coagulation by microwave or radiofrequency
ablation. Cancer. 91:949–956. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Poon RT, Fan ST, Ng IO, Lo CM, Liu CL and
Wong J: Different risk factors and prognosis for early and late
intrahepatic recurrence after resection of hepatocellular
carcinoma. Cancer. 89:500–507. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arii S, Monden K, Niwano M, et al: Results
of surgical treatment for recurrent hepatocellular carcinoma;
comparison of outcome among patients with multicentric
carcinogenesis, intrahepatic metastasis, and extrahepatic
recurrence. J Hepatobiliary Pancreat Surg. 5:86–92. 1998.
View Article : Google Scholar
|
|
11
|
Miyata R, Tanimoto A, Wakabayashi G, et
al: Accuracy of preoperative prediction of microinvasion of portal
vein in hepatocellular carcinoma using superparamagnetic iron
oxide-enhanced magnetic resonance imaging and computed tomography
during hepatic angiography. J Gastroenterol. 41:987–995. 2006.
View Article : Google Scholar
|
|
12
|
Ma WL, Hsu CL, Yeh CC, et al: Hepatic
androgen receptor suppresses hepatocellular carcinoma metastasis
through modulation of cell migration and anoikis. Hepatology. Feb
9–2012.(Epub ahead of print).
|
|
13
|
Yamazaki K, Masugi Y and Sakamoto M:
Molecular pathogenesis of hepatocellular carcinoma: altering
transforming growth factor-beta signaling in hepatocarcinogenesis.
Dig Dis. 29:284–288. 2011. View Article : Google Scholar
|
|
14
|
Zheng F, Liao YJ, Cai MY, et al: The
putative tumour suppressor microRNA-124 modulates hepatocellular
carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut.
61:278–289. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu J, Chen Y, Cao J, et al: p28(GANK)
overexpression accelerates hepatocellular carcinoma invasiveness
and metastasis via phosphoinositol 3-kinase/AKT/hypoxia-inducible
factor-1alpha pathways. Hepatology. 53:181–192. 2012. View Article : Google Scholar
|
|
16
|
Yao J, Liang L, Huang S, et al:
MicroRNA-30d promotes tumor invasion and metastasis by targeting
Galphai2 in hepatocellular carcinoma. Hepatology. 51:846–856.
2010.PubMed/NCBI
|
|
17
|
Mazzocca A, Liotta F and Carloni V:
Tetraspanin CD81-regulated cell motility plays a critical role in
intrahepatic metastasis of hepatocellular carcinoma.
Gastroenterology. 135:244–256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakon M, Nagano H, Nakamori S, et al:
Intrahepatic recurrences of hepatocellular carcinoma after
hepatectomy: analysis based on tumor hemodynamics. Arch Surg.
137:94–99. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gundersen GG and Bulinski JC: Selective
stabilization of microtubules oriented toward the direction of cell
migration. Proc Natl Acad Sci USA. 85:5946–5950. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hubbert C, Guardiola A, Shao R, et al:
HDAC6 is a microtubule-associated deacetylase. Nature. 417:455–458.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tran AD, Marmo TP, Salam AA, et al: HDAC6
deacetylation of tubulin modulates dynamics of cellular adhesions.
J Cell Sci. 120:1469–1479. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu H and Parsons JT: Cortactin, an
80/85-kilodalton pp60src substrate, is a filamentous actin-binding
protein enriched in the cell cortex. J Cell Biol. 120:1417–1426.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luxton GW and Gundersen GG: HDAC6-pack:
cortactin acetylation joins the brew. Dev Cell. 13:161–162. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsunoda K, Oikawa H, Tada H, et al:
Nucleus accumbens-associated 1 contributes to cortactin
deacetylation and augments the migration of melanoma cells. J
Invest Dermatol. 131:1710–1719. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zuo Q, Wu W, Li X, Zhao L and Chen W:
HDAC6 and SIRT2 promote bladder cancer cell migration and invasion
by targeting cortactin. Oncol Rep. 27:819–824. 2012.PubMed/NCBI
|
|
26
|
Park SY, Jun JA, Jeong KJ, et al: Histone
deacetylases 1, 6 and 8 are critical for invasion in breast cancer.
Oncol Rep. 25:1677–1681. 2011.PubMed/NCBI
|
|
27
|
Joshi A and Cao D: TGF-beta signaling,
tumor microenvironment and tumor progression: the butterfly effect.
Front Biosci. 15:180–194. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu J, Ustach C and Kim HR:
Platelet-derived growth factor signaling and human cancer. J
Biochem Mol Biol. 36:49–59. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu X and Kang Y: Epidermal growth factor
signalling and bone metastasis. Br J Cancer. 102:457–461. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou HY, Pon YL and Wong AS: HGF/MET
signaling in ovarian cancer. Curr Mol Med. 8:469–480. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang JD, Nakamura I and Roberts LR: The
tumor microenvironment in hepatocellular carcinoma: current status
and therapeutic targets. Semin Cancer Biol. 21:35–43. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang XR, Xu Y, Yu B, et al: CD24 is a
novel predictor for poor prognosis of hepatocellular carcinoma
after surgery. Clin Cancer Res. 15:5518–5527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Inayoshi J, Ichida T, Sugitani S, et al:
Gross appearance of hepatocellular carcinoma reflects E-cadherin
expression and risk of early recurrence after surgical treatment. J
Gastroenterol Hepatol. 18:673–677. 2003. View Article : Google Scholar
|
|
34
|
Wong CC, Wong CM, Tung EK, Man K and Ng
IO: Rho-kinase 2 is frequently overexpressed in hepatocellular
carcinoma and involved in tumor invasion. Hepatology. 49:1583–1594.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yokomizo C, Yamaguchi K, Itoh Y, et al:
High expression of p300 in HCC predicts shortened overall survival
in association with enhanced epithelial mesenchymal transition of
HCC cells. Cancer Lett. 310:140–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Miyoshi A, Kitajima Y, Kido S, et al:
Snail accelerates cancer invasion by upregulating MMP expression
and is associated with poor prognosis of hepatocellular carcinoma.
Br J Cancer. 92:252–258. 2005.PubMed/NCBI
|
|
37
|
Aldana-Masangkay GI and Sakamoto KM: The
role of HDAC6 in cancer. J Biomed Biotechnol. 2011:8758242011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Grozinger CM, Hassig CA and Schreiber SL:
Three proteins define a class of human histone deacetylases related
to yeast Hda1p. Proc Natl Acad Sci USA. 96:4868–4873. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Haggarty SJ, Koeller KM, Wong JC,
Grozinger CM and Schreiber SL: Domain-selective small-molecule
inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin
deacetylation. Proc Natl Acad Sci USA. 100:4389–4394. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Haggarty SJ, Koeller KM, Wong JC, Butcher
RA and Schreiber SL: Multidimensional chemical genetic analysis of
diversity-oriented synthesis-derived deacetylase inhibitors using
cell-based assays. Chem Biol. 10:383–396. 2003. View Article : Google Scholar
|
|
41
|
Bazzaro M, Lin Z, Santillan A, et al:
Ubiquitin proteasome system stress underlies synergistic killing of
ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor.
Clin Cancer Res. 14:7340–7347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Santo L, Hideshima T, Kung AL, et al:
Preclinical activity, pharmacodynamic and pharmacokinetic
properties of a selective HDAC6 inhibitor, ACY-1215, in combination
with bortezomib in multiple myeloma. Blood. 119:2579–2589. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim HS, Vassilopoulos A, Wang RH, et al:
SIRT2 maintains genome integrity and suppresses tumorigenesis
through regulating APC/C activity. Cancer Cell. 20:487–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|