Introduction
Uveal melanoma is the most common primary
intraocular malignant tumor in adults with a high mortality of ~50%
(1). Early metastasis accounts for
the high death rate of uveal melanoma (2). Unfortunately, it is evidenced that
many patients may have subclinical metastasis at the time of
diagnosis (1). Despite the advances
in surgery, radiotherapy and chemotherapy, the 5-year relative
survival rate has not improved from 1973 to 2008 (3). Metastasis of uveal melanoma is a
complex and multistep process, involving increased migratory and
invasive potential of tumor cells. Currently, the molecular
mechanisms of its aggressiveness are still not elucidated (4). Therefore, identifying the crucial
signals that promote invasive and metastatic potential of uveal
melanoma will contribute to provide biomarkers for early prognosis
and targets for treatment.
MicroRNAs (miRNAs) are ~22nt non-coding RNAs that
negatively regulate gene expression at the post-transcriptional
level via direct binding with the 3′ untranslated regions (UTRs) of
target mRNAs. Many miRNAs have been implicated to play important
roles in tumor metastasis, such as miR-9, miR-31, miR-125a, miR-373
(5). Accumulated evidence has shown
the involvement of miR-9 in tumor cell proliferation and tumor
progression (6–8). More attention was paid to miR-9
mediated-tumor cell proliferation; however, the current knowledge
about miR-9 in tumor metastasis is still preliminary. MiR-9 has
been shown to promote tumor metastasis in breast cancer and
colorectal cancer (9,10). Whereas, a recent study showed that
miR-9 suppressed melanoma progression (11). Currently, the role of miR-9 in
metastasis of uveal melanoma and the possible mechanism have not
been reported.
NF-κB (nuclear factor kappaB) is a transcription
factor that plays important roles in cell proliferation, apoptosis,
differentiation, as well as in tumor angiogenesis and metastasis
(12). The mammalian NF-κB family
is composed of five members, NF-κB1 (p50/p105), NF-κB2 (p52/p100),
RelA (p65), RelB and c-Rel. NF-κB regulates gene transcription
activity through forming a complex containing hetero- or
homo-dimers of the five subunits and other adaptor proteins and
binding to the promoter of the responsive gene (13). MMP-2 (matrix metalloproteinase),
MMP-9, and VEGFA (vascular endothelial growth factor A) are some
responsive genes of NF-κB1 involved in tumor invasion and
metastatasis, and their upregulation has been suggested to be
correlated with metastasis of uveal melanoma (14,15).
Bioinformatic analysis reveals a conserved target site for miR-9 in
the NF-κB1-3′-UTR at nucleotides 3405–3412. Moreover, NF-κB1 has
been shown to be a direct target gene of miR-9 in certain types of
cancer (7,11). However, for its role in uveal
melanoma metastasis has not been reported.
In this study, we identified an inverse correlation
between miR-9 expression and uveal melanoma cell invasive
potential. We further addressed miR-9-mediated tumor cell migration
and invasion, and identified NF-κB1 as a direct target of miR-9.
Additionally, we showed MMP-2, MMP-9, and VEGFA, the downstream
signals of NF-κB1, were regulated by miR-9 simultaneously.
Materials and methods
Cell culture and transfection
Paired human uveal melanoma cells with highly
invasive potential (MUM-2B and C918) and the relatively poorly
invasive potential (MUM-2C and OCM-1A) were kindly provided by
Professor Elisabeth A. Seftor (Children’s Memorial Research Center,
Chicago, IL). The highly invasive MUM-2B cells were transfected
with miR-9 mimic (miR-9) or the negative control mimic (miR-con)
(Dharmacon, Lafayette, CO) using Lipofectamine 2000 (Invitrogen)
following the manufacturer’s protocol. The poorly invasive MUM-2C
cells were transfected with miR-9 inhibitor (anti-miR-9) or the
negative control inhibitor (anti-miR-con) as above.
Quantitative real-time polymerase chain
reaction
Total RNA was extracted from cells using Trizol
reagent (Invitrogen). TaqMan microRNA assays (Applied Biosystems
Inc., Foster City, CA) were used to quantify the relative
expression of miR-9. Small nuclear RNA, U6B (Applied Biosystems
Inc.), was treated as normalization control. NF-κB1, MMP-2, MMP-9,
VEGFA, and GAPDH (control) mRNA levels were assessed by SYBR Green
quantitative PCR. Reactions were conducted according to the
manufacturer’s instructions using Power SYBR Green PCR Master Mix
(Applied Biosystems Inc.). Forward (F) and reverse (R) primers were
used as follows: NF-κB1, F: 5′-ACAG CAGATGGCCCATACCT-3′ and R:
5′-CATACATAACGGA AACGAAATCCTCT-3′; MMP-2, F: 5′-TCCCATTTTGATGA
CGATGA-3′ and R: 5′-CCGTACTTGCCATCCTTCTC-3′; MMP-9, F:
5′-CATCGTCATCCAGTTTGGTG-3′ and R: 5′-TCGAAGATGAAGGGGAAGTG-3′;
VEGFA, F: 5′-CCTT GCTGCTCTACCTCCAC-3′ and R: 5′-ATGATTCTGCCC
TCCTCCTT-3′; GAPDH, F: 5′-TGTTCGACAGTCAGC CGC-3′, and R:
5′-GGTGTCTGAGCGATGTGGC-3′. All real-time amplifications were
measured in triplicate and performed with the ABI PRISM 7300
sequence detection system. The fold-change of miR-9, NF-κB1, MMP-2,
MMP-9, and VEGFA mRNA was calculated using the 2-ΔΔCT
method.
Cell proliferation assay
Cell proliferative activity was determined by the
MTT (3-[4,5-dimethyl-thiazol-2-yl]-2,5-diphenyltetrazolium bromide)
assay, according to standard methods (4). Absorbance was measured at 490 nm and
detected using μQuant Universal Microplate Spectrophotometer
(BioTek Instruments, Inc.).
Invasion assay
The invasion assay was performed using
cultrex-24-well membrane invasion chambers (8 μm pore size,
Millipore). Briefly, 40 μl of coating solution (BD matrigel diluted
with medium at the rate of 1:3, BD Biosciences, San Jose, CA) was
placed in each well of the top invasion chamber, and the cells were
starved in serum-free medium. Post-transfection cells (24 h) were
harvested, resuspended in serum-free medium and seeded into the top
chamber at 1×104 cell/well. Complete medium (600 μl )
with 12% FBS was added to the bottom wells of the chambers. The
chambers were incubated at 37°C in an incubator containing 5%
CO2. After 24 h, the non-invading cells were removed
from the top chamber. The invading cells were fixed with methanol
and stained by 0.05% crystal violet solution. The number of
invasive cells on the lower surface of the membrane was counted
under a microscope. The mean of triplicate assays for each
experimental condition is reported.
Migration assay
Cell migration was also measured with
cultrex-24-well membrane invasion chambers, without coating
solution. Briefly, post-transfected cells were harvested and
transferred with 5×103 cells to milicell inserts. After
culture for 16 h, migrated cells were fixed, stained and counted as
above.
MiR-9 target prediction
Computer-based programs were used to predict
potential miR-9 targets. Using ‘hsa-miR-9′ as a search term, we
queried PicTar (http://pictar.mdc-berlin.de/), TargetScan (http://targetscan.org/) and miRBase targets
(http://mirbase.org/). A gene was considered to be
a putative target of miR-9 only if it was predicted by three
programs.
Plasmid construction
Plasmid pcDNA3.1-EGFP was derived from pCDNA3.1(+)
(Invitrogen) by inserting the coding EGFP fragment at
HindIII and BamHI sites. The EGFP fragment was
amplified from pEGFP-N1 (Clontech, CA, USA) with primers of EGFP
forward, 5′-GCAGCCAAGCTTGCCACCA TGTGTAGCAAGGGC-3′, and EGFP
reverse, 5′-CGCGGAT CCTTTACTTGTACAGCTCGTCC-3′. The 3′-untranslated
mRNA sequences of NF-κB1 containing the predicted miR-9 binding
site (nt 3405–3412) were synthesized by Invitrogen with
BamHI and EcoRI enzyme digest sites at 5′ end and 3′
end, respectively. Similar fragments with specifically mutated
miR-9 targeting region were also synthesized. The oligonucleotides
were cloned into pcDNA/EGFP at the BamHI and EcoRI
sites, and named as pcDNA-EGFP-NF-κB1-Wt (Wt-3′-UTR) and
pcDNA-EGFP-NF-κB1-Mut (Mut-3′-UTR) respectively. All constructs
were verified by DNA sequencing.
Fluorescent report assay
MUM-2B cells were seeded in 24-well plates and were
transfected with miR-9 mimic or the negative control mimic for 24
h, and then transfected with wild-type or mutant reporter vectors.
The cells were washed twice with cold PBS, then lyzed with RIPA
lysis buffer (150 mmol/l NaCl, 50 mmol/l Tris-HCl, pH 7.4, 1%
Triton X-100, 0.1% SDS). Afterward protein was harvested,
transferred to 96-well Black Cliniplate (Corning) and then the
fluorescent intensity was measured on a varioskan fluorescence
spectrometer (Thermo). The RFP expression vector, pDsRed-N1
(Clontech), was co-transfected and used for normalizing the
transfection efficiency.
Western blot analysis
Cells were harvested at the end of treatment.
Protein concentration was confirmed by the Bradford assay. Protein
was separated by 10% SDS-PAGE and then transferred to PVDF membrane
using standard procedures. The membrane was incubated with the
primary antibody for NF-κB1 (Sigma, St. Louis, MO) or β-actin
(Boster, Wuhan, China), and then washed, incubated with
HRP-conjugated secondary antibody. Intensity of the bands was
visualized by an enhanced chemiluminescence (ECL, Amersham
Pharmacia Biotech) system and exposed.
Statistical analysis
The data are represented as the mean ± SD. The
difference between the groups was determined by non-parametric
test. Differences with P<0.05 were considered statistically
significant. All the analyses were carried out with the SPSS 9.0
(SPSS Inc., Chicago, IL, USA).
Results
MiR-9 expression negatively correlates
with the invasive potential of uveal melanoma cell lines
To test the possibility that miR-9 was involved in
tumor cell invasion, two paired highly/poorly invasive human uveal
melanoma cell lines were applied (MUM-2B vs MUM-2C, C918 vs
OCM-1A), which had been successfully used in several studies
investigating tumor metastasis (1,16). We
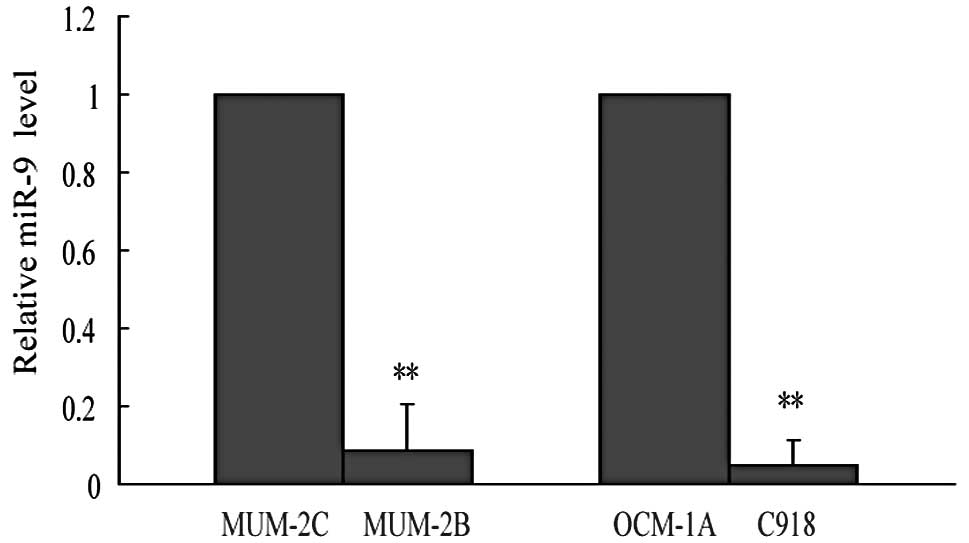
first detected miR-9 expression levels in these cell lines, and our
data showed that miR-9 expression was dramatically reduced by 91%
in highly invasive uveal melanoma MUM-2B cells compared with the
corresponding poorly invasive MUM-2C cells. Another paired
highly/poorly invasive cell lines confirmed the negative
correlation between miR-9 and invasive capability, that miR-9
expression was severely repressed by 95% in highly invasive C918
cells in comparison with poorly invasive OCM-1A cells (Fig. 1). These data indicated that miR-9
might be involved in tumor metastasis and function as a tumor
metastasis suppressor.
Ectopic expression of miR-9 leads to the
reduction of cell migration and invasion
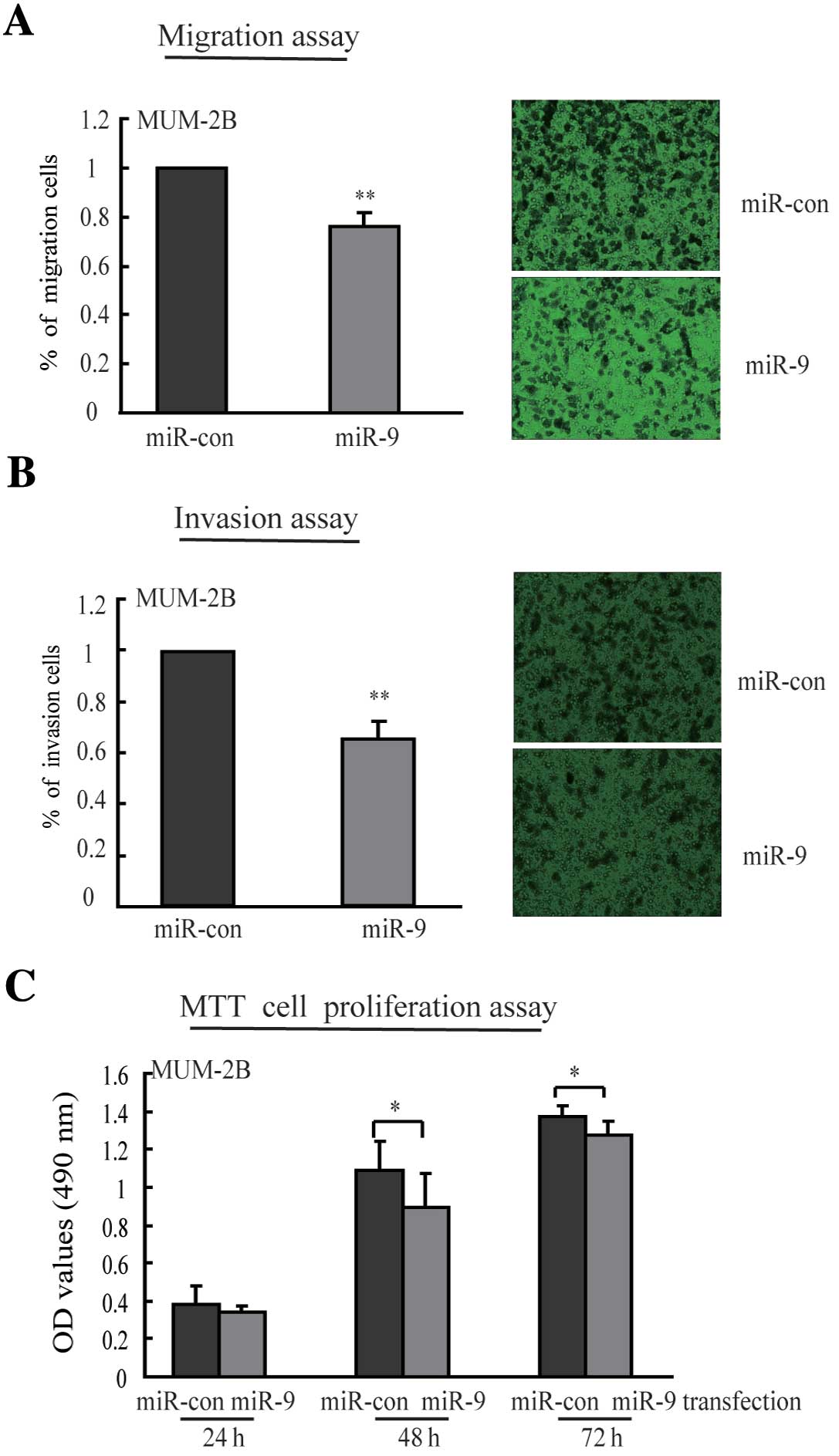
To determine the function of miR-9 on cell migration
and invasion, MUM-2B cells were transfected with miR-9 mimic or
negative mimic, and then evaluated in transwell assays with or
without matrigel coating solution in the upper side of the well
membrane. As shown in Fig. 2A, in
the migration assay, cell numbers migrating to the lower side of
the membrane after miR-9 mimic transfection decreased by 24%
(P<0.01) as compared to negative control mimic transfection.
Re-expression of miR-9, cell numbers invading to the bottom side of
the matrigel coating membrane decreased by 35% (P<0.01)
(Fig. 2B). These results suggest
that ectopic expression of miR-9 repressed migratory and invasive
potential of highly aggressive uveal melanoma cells. In addition,
we observed inhibition of cell proliferation after introduction of
miR-9 for 48 and 72 h, but the effects were relatively minor
(Fig. 2C).
NF-κB1 is a critical downstream target of
miR-9
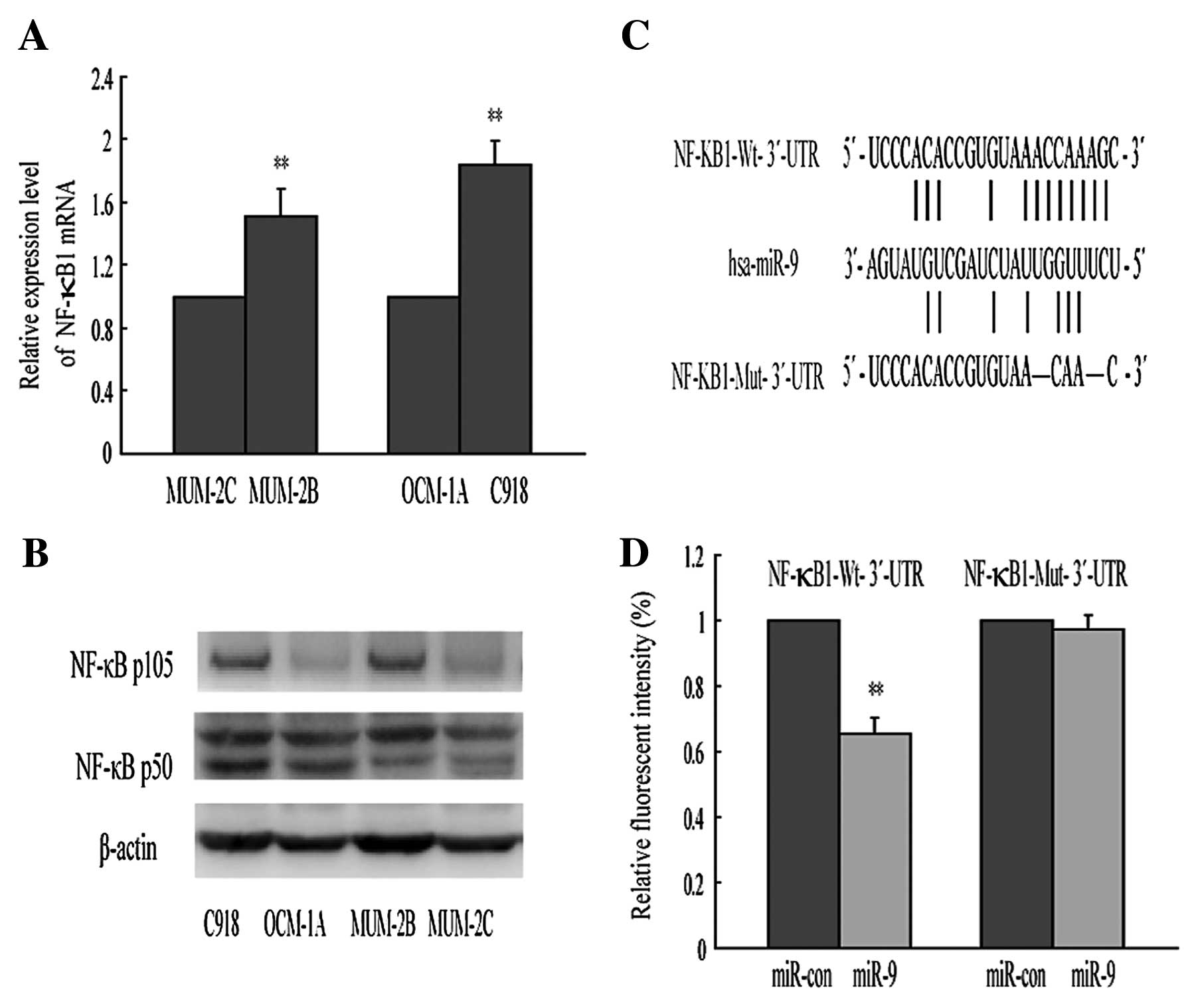
In order to explore the molecular mechanism by which
miR-9 functions as a tumor suppressor, we chose three algorithm
programs to predict the putative target genes. Among these genes,
NF-κB1 was of particular interest, since it is a well-known
oncogene involved in cancer development and upregulated in a
variety of solid tumors (13,17).
In the present study, we found that NF-κB1 mRNA and protein
expression were enhanced in highly invasive cell lines (MUM-2B,
C918) compared with the corresponding poorly invasive cell lines
(MUM-2C, OCM-1A) (Fig. 3A and B),
consistent with previous studies. As shown in Fig. 3C, there is a conserved target site
for miR-9 in the NF-κB1-3′-UTR at nucleotides 3405–3412.
Combination of Figs. 1 and 3 show increased expression of NF-κB1 was
parallel with reduced abundance of miR-9, indicating that NF-κB1
may be a target of miR-9 in the regulation of tumor cell invasion.
To identify NF-κB1 as a direct target of miR-9 in uveal melanoma
cells, we constructed two enhanced green fluorescence protein
(EGFP) reporter vectors containing the 3′-UTRs of NF-κB1. Wt-3′-UTR
vector harbored the target sequence (AACCAAAG) of miR-9 seed region
in NF-κB1. The parallel vector, Mut-3′-UTR, contained the specific
mutation at the miR-9-targeted region to abolish the miR-9 binding
(Fig. 3C). Fig. 3D shows that transient transfection
of MUM-2B cells with miR-9 mimic and Wt- 3′-UTR report vector led
to a significant decrease of fluorescence intensity compared with
the control. However, the fluorescence intensity was unaffected
when Mut-3′-UTR reporter vector and miR-9 mimic were cotransfected.
These results suggest that miR-9 could directly bind to NF-κB1 and
may regulate its expression in post-transcriptional levels.
MiR-9 negatively regulates NF-κB1 mRNA
and protein expression
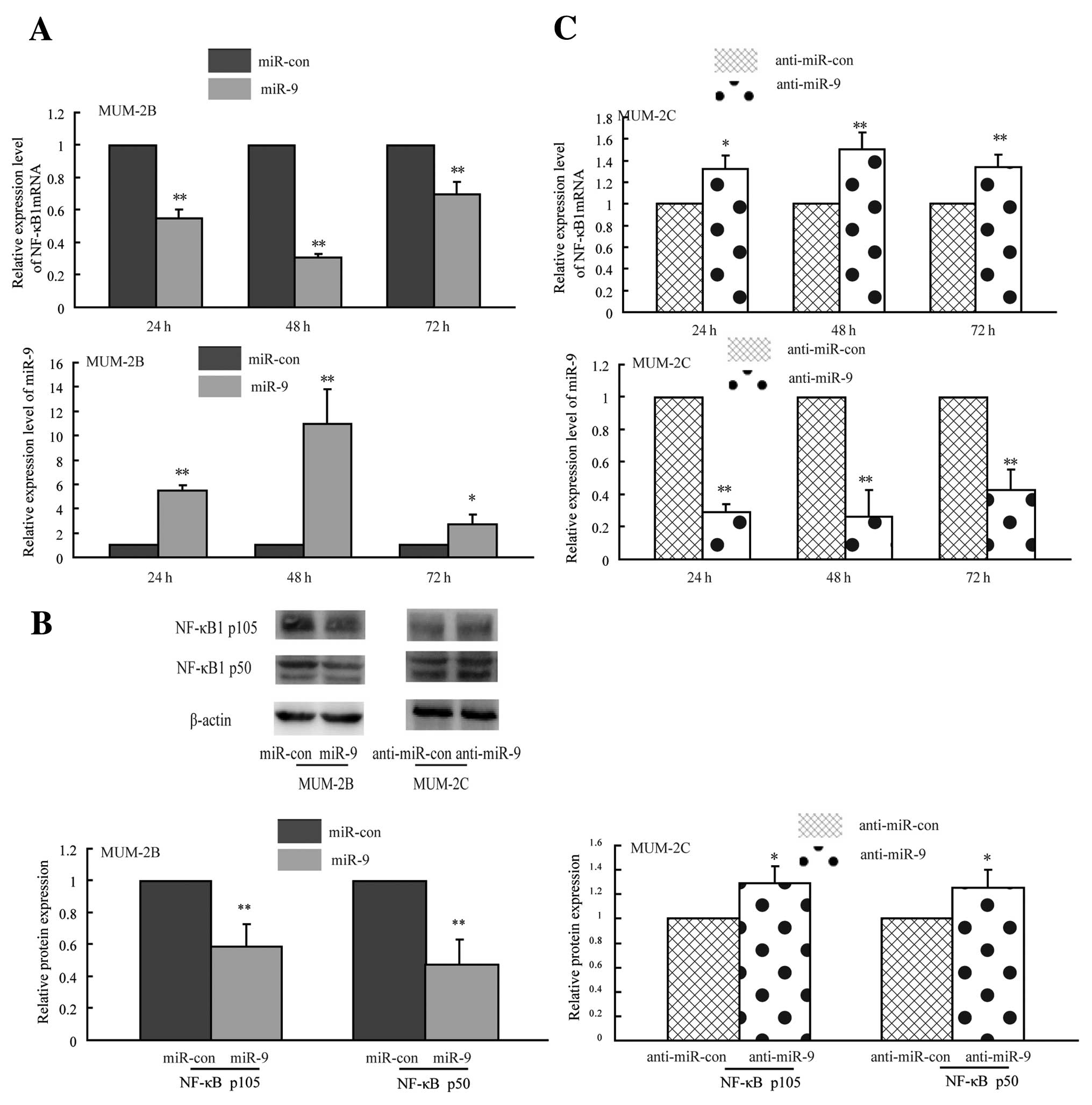
To determine whether miR-9 regulated endogenous
NF-κB1 expression in uveal melanoma cells, we performed gain-loss
of function assays. Briefly, highly invasive MUM-2B cells with low
miR-9 expression were transfected with miR-9 mimic to enhance the
miR-9 function; meanwhile, poorly invasive MUM-2C cells carrying
high miR-9 expression were transfected with miR-9 inhibitor to
block the miR-9 function, then the effects of miR-9 on NF-κB1 mRNA
and protein expression were measured by quantitative PCR and
western blotting. As shown in Fig.
4A, NF-κB1 mRNA level was reduced by 45, 69 and 31% after
MUM-2B cells transfection of miR-9 mimic for 24, 48 and 72 h,
respectively, indicating that miR-9 regulates endogenous NF-κB1
mRNA levels partly through the mechanism of mRNA degradation. The
NF-κB1 gene encodes two functional proteins p105 and p50.
Overexpression of miR-9 in MUM-2B cells led to reduction of NF-κB
p105 and p50 proteins, respectively, by 42 and 52% compared with
the control groups, suggesting that miR-9 regulated NF-κB1 partly
through translational repression and/or mRNA degradation (Fig. 4B). In contrast, blocking the
expression of miR-9 in MUM-2C cells led to 32, 50 and 34% increase
of NF-κB1 mRNA for 24 h, 48 and 72 h (Fig. 4C), meanwhile NF-κB p105 and p50
protein had a 29 and 25% increase after loss of miR-9 expression
(Fig. 4B), supporting the negative
regulatory function of miR-9 on NF-κB1 expression. These data
showed that miR-9 restoration reduced NF-κB1 expression both in
mRNA and protein levels in MUM-2B cells; silencing miR-9 expression
increased NF-κB1 mRNA and protein amounts in MUM-2C cells. These
findings suggest that miR-9 negatively regulates NF-κB1 expression
at post-transcriptional level in highly-poorly invasive uveal
melanoma cells.
MiR-9 indirectly regulates MMP-2, MMP-9,
and VEGFA expression
It has been reported that activated NF-κB
transcription factors resulted in increased expression of MMP-2,
MMP-9 and VEGFA, and contributed to tumor angiogenesis and
metastasis (18,19). Thus, we suspect that miR-9-regulated
NF-κB1 expression may suppress invasive capability of uveal
melanoma cells partly through MMP-2, MMP-9, and VEGFA. We first
detected MMP-2, MMP-9, and VEGFA mRNA expression in two paired
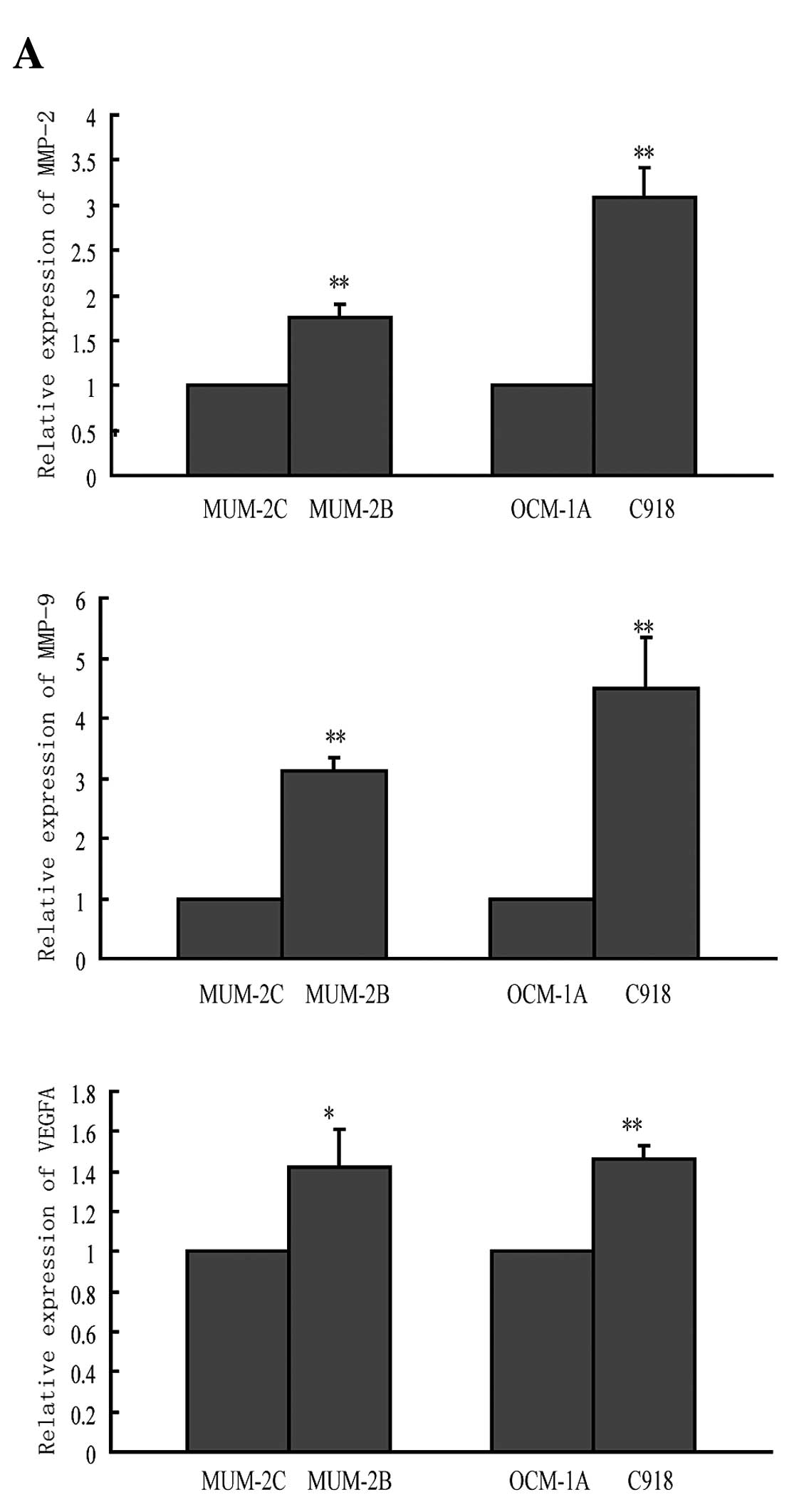
highly-poorly invasive cell lines. Fig.
5A showed increased expression of all three molecules in highly
invasive cell lines (MUM-2B, C918) compared to the corresponding
poorly invasive cell lines (MUM-2C, OCM-1A), similar to NF-κB1
expression. Then, we detected the effect of miR-9 on MMP-2, MMP-9,
and VEGFA mRNA expression. As expected, miR-9 overexpression
decreased mRNA expression levels of all three molecules (Fig. 5B). In contrast, anti-miR-9 increased
their mRNA levels (Fig. 5C). The
alteration was similar to the changes of NF-κB1 expression. Since
in silico analysis did not show any miR-9 binding sites in
the 3′-UTRs of MMP-2, MMP-9, and VEGFA, the suppression is possibly
an indirect effect through NF-κB1. Therefore, reduction of MMP-2,
MMP-9, and VEGFA expression through targeting NF-κB1 is likely to
contribute to the miR-9-mediated suppression of cell invasion.
Discussion
Metastasis is responsible for the high mortality of
uveal melanoma patients. Unfortunately, at the time of diagnosis,
many patients already harbor microscopic metastases, thus it is
urgent to identify early prognostic markers for metastasis
(2). MiRNAs are non-coding small
RNAs that have different expression profiles in different cancer
types and stages. Some miRNAs, such as miR-20a, miR-17a, and
miR-106a have been shown to be up-regulated in uveal melanoma,
while others, like miR-145 and miR-204, to be downregulated
(20). Additionally, miR-34a,
let-7b, and miR-199a expression levels have been suggested to be
correlated with metastasis of uveal melanoma (21,22).
Thus, miRNA expression may represent a highly accurate biomarker
for metastatic risk in uveal melanoma. In this study, we identified
strongly decreased expression of miR-9 in highly invasive uveal
melanoma cells compared with the corresponding poorly invasive
cells. These data indicated that some special miRNAs may be
developed as a new diagnostic marker for progression and metastasis
of uveal melanoma.
Currently, controversy exists regarding the effects
of miR-9 on cancer progression. In most cases, miR-9 functions as a
tumor suppressor, such as in gastric cancer, colorectal cancer,
pancreatic adenocarcinoma, and medulloblastoma (6,7,23–25).
In addition, miR-9 expression levels have been shown to be
negatively correlated with recurrence in ovarian cancer and clear
cell renal cell carcinoma (26,27).
However, miR-9 has been reported as an oncogene in endometrial
cancer and gastric cancer (28,29).
Previous studies also showed miR-9 had opposite roles in cancer
metastasis. MiR-9 has been shown to promote metastasis of breast
cancer and colorectal cancer by targeting E-cadherin and promoting
cell motility (9,10). Recently, Liu et al obtained
different results that miR-9 was downexpressed in metastatic
melanoma and indirectly upregulated E-cadherin through a
NF-κB1-Snail1 pathway, thereby suppressing melanoma proliferation
and metastasis (11). Since the
lack of lymphatics in the eyes, uveal melanoma has a strong
predilection for hematogenous metastasis, thereby rendering uveal
melanoma a unique model for studying the hematogenous dissemination
of cancer (30). Hence, we shed
light on the regulation of miR-9 in hematogenous dissemination. In
our study, we showed miR-9 reduced migration and invasion of highly
invasive uveal melanoma cells. However, we showed that miR-9 had no
significant effect on cell proliferation in uveal melanoma. The
discrepancies in miR-9 effects might be partly explained with
cancer types, stages, cell lines, or even different technical
platforms. It is also possible that miR-9 plays different roles in
tumor development, progression, metastasis and recurrence.
To date, several miR-9 target genes have been
confirmed in different cancer types, such as NF-κB1 in gastric
cancer, E-cadherin in breast cancer, FOXO1 in endometrial cancer,
CDX2 in gastric cancer (7,9,28,29).
As we were pursuing to clarify the mechanism of miR-9 on tumor cell
migration and invasion, NF-κB1 which played vital roles in uveal
melanoma development and progression attracted our interests. Gao
et al reported that the expression of NF-κB p105/p50
dramatically increased in melanoma and was correlated with melanoma
progression (31). Dror et
al detected activation of NF-κB1 and NF-κB2 pathway in both
primary and metastases uveal melanoma (17). Consistent with these reports, we
also observed increased NF-κB1 expression in highly invasive cell
lines in comparison with the relative poorly invasive cell lines.
Furthermore, we also confirmed the post-transcriptional regulation
of NF-κB1 by miR-9 in uveal melanoma. These findings extend our
knowledge of miRNA/NF-κB signaling pathway in uveal melanoma and
provide additional evidence for developing NF-κB to a therapeutic
target.
NF-κB1 facilitates invasion and metastasis partly by
transcriptional regulation of MMP-2, MMP-9 and VEGFA, which are
closely associated with uveal melanoma progression. Vaisanen et
al have indicated MMP-2 as an prognostic marker for high
metastatic risk in uveal melanoma (14). El-Shabrawi et al have shown
that MMP-9 is predominantly expressed in highly aggressive uveal
melanoma cells (32). A recent
report showed that high level of VEGFA was correlated with the
number and location of micrometastases in a murine model of uveal
melanoma (33). In this study, we
observed that MMP-2, MMP-9, and VEGFA altered in the same pattern
as NF-κB1. Thus, NF-κB1-regulated MMP-2, MMP-9, and VEGFA may
contribute to miR-9-suppressed metastasis of uveal melanoma.
In conclusion, our results clearly showed that miR-9
was significantly downregulated in the highly invasive uveal
melanoma cell lines and could suppress cell migration and invasion.
It functioned at least in part through direct targeting NF-κB1
expression and downregulation of its downstream genes such as
MMP-2, MMP-9, and VEGFA. Thus, the anti-metastatic capabilities of
miR-9 seem to behave as a critical safeguard against the
acquisition of metastatic potential. Therefore, rescuing miR-9 may
offer a novel strategy for blocking tumor metastasis.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 30873099, 81102458 and
81172004) and the Priority Academic Program Development of Jiangsu
Higher Education Institutions (PAPD). We thank Professor Elisabeth
A. Seftor for providing the human uveal melanoma cell lines.
References
|
1
|
Seftor EA, Meltzer PS, Kirschmann DA,
Pe’er J, Maniotis AJ, Trent JM, Folberg R and Hendrix MJ: Molecular
determinants of human uveal melanoma invasion and metastasis. Clin
Exp Metastasis. 19:233–246. 2002.
|
|
2
|
Triozzi PL, Eng C and Singh AD: Targeted
therapy for uveal melanoma. Cancer Treat Rev. 34:247–258. 2008.
|
|
3
|
Singh AD, Turell ME and Topham AK: Uveal
melanoma: trends in incidence, treatment, and survival.
Ophthalmology. 118:1881–1885. 2011.
|
|
4
|
Sun Q, Cong R, Yan H, Gu H, Zeng Y, Liu N,
Chen J and Wang B: Genistein inhibits growth of human uveal
melanoma cells and affects microRNA-27a and target gene expression.
Oncol Rep. 22:563–567. 2009.
|
|
5
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009.
|
|
6
|
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC,
Chan WC, et al: Aberrant hypermethylation of miR-9 genes in gastric
cancer. Epigenetics. 6:1189–1197. 2011.
|
|
7
|
Wan HY, Guo LM, Liu T, Liu M, Li X and
Tang H: Regulation of the transcription factor NF-kappaB1 by
microRNA-9 in human gastric adenocarcinoma. Mol Cancer.
9:162010.
|
|
8
|
Hu X, Schwarz JK, Lewis JS, Huettner PC,
Rader JS, Deasy JO, et al: A microRNA expression signature for
cervical cancer prognosis. Cancer Res. 70:1441–1448. 2010.
|
|
9
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, et al: miR-9, a MYC/MYCN-activated microRNA, regulates
E-cadherin and cancer metastasis. Nat Cell Biol. 12:247–256.
2010.
|
|
10
|
Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng
S and Ge W: MicroRNA-9 up-regulation is involved in colorectal
cancer metastasis via promoting cell motility. Med Oncol.
29:1037–1043. 2012.
|
|
11
|
Liu S, Kumar SM, Lu H, Liu A, Yang R,
Pushparajan A, Guo W and Xu X: MicroRNA-9 up-regulates E-cadherin
through inhibition of NF-κB1-Snail1 pathway in melanoma. J Pathol.
226:61–72. 2012.
|
|
12
|
Amiri KI and Richmond A: Role of nuclear
factor-kappa B in melanoma. Cancer Metastasis Rev. 24:301–313.
2005.
|
|
13
|
Pereira SG and Oakley F: Nuclear
factor-kappaB1: regulation and function. Int J Biochem Cell Biol.
40:1425–1430. 2008.
|
|
14
|
Vaisanen A, Kallioinen M, von Dickhoff K,
Laatikainen L, Hoyhtya M and Turpeenniemi-Hujanen T: Matrix
metalloproteinase-2 (MMP-2) immunoreactive protein - a new
prognostic marker in uveal melanoma? J Pathol. 188:56–62. 1999.
|
|
15
|
Sahin A, Kiratli H, Soylemezoglu F, Tezel
GG, Bilgic S and Saracbasi O: Expression of vascular endothelial
growth factor-A, matrix metalloproteinase-9, and extravascular
matrix patterns and their correlations with clinicopathologic
parameters in posterior uveal melanomas. Jpn J Ophthalmol.
51:325–331. 2007.
|
|
16
|
Hendrix MJ, Seftor EA, Meltzer PS, Gardner
LM, Hess AR, et al: Expression and functional significance of
VE-cadherin in aggressive human melanoma cells: role in
vasculogenic mimicry. Proc Natl Acad Sci USA. 98:8018–8023.
2001.
|
|
17
|
Dror R, Lederman M, Umezawa K, Barak V,
Pe’er J and Chowers I: Characterizing the involvement of the
nuclear factor-kappa B (NF kappa B) transcription factor in uveal
melanoma. Invest Ophthalmol Vis Sci. 51:1811–1816. 2010.
|
|
18
|
Yan C and Boyd DD: Regulation of matrix
metalloproteinase gene expression. J Cell Physiol. 211:19–26.
2007.
|
|
19
|
Karst AM, Gao K, Nelson CC and Li G:
Nuclear factor kappa B subunit p50 promotes melanoma angiogenesis
by upregulating interleukin-6 expression. Int J Cancer.
124:494–501. 2009.
|
|
20
|
Yang C and Wei W: The miRNA expression
profile of the uveal melanoma. Sci China Life Sci. 54:351–358.
2011.
|
|
21
|
Yan D, Zhou X, Chen X, Hu DN, Dong XD,
Wang J, et al: MicroRNA-34a inhibits uveal melanoma cell
proliferation and migration through downregulation of c-Met. Invest
Ophthalmol Vis Sci. 50:1559–1565. 2009.
|
|
22
|
Worley LA, Long MD, Onken MD and Harbour
JW: Micro-RNAs associated with metastasis in uveal melanoma
identified by multiplexed microarray profiling. Melanoma Res.
18:184–190. 2008.
|
|
23
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, et al: Epigenetic regulation of microRNA
expression in colorectal cancer. Int J Cancer. 125:2737–2743.
2009.
|
|
24
|
Omura N, Li CP, Li A, Hong SM, Walter K,
Jimeno A, et al: Genome-wide profiling of methylated promoters in
pancreatic adenocarcinoma. Cancer Biol Ther. 7:1146–1156. 2008.
|
|
25
|
Ferretti E, De Smaele E, Po A, Di
Marcotullio L, Tosi E, Espinola MS, et al: MicroRNA profiling in
human medulloblastoma. Int J Cancer. 124:568–577. 2009.
|
|
26
|
Laios A, O’Toole S, Flavin R, Martin C,
Kelly L, Ring M, et al: Potential role of miR-9 and miR-223 in
recurrent ovarian cancer. Mol Cancer. 7:352008.
|
|
27
|
Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W,
Tamboli P, et al: Hsa-miR-9 methylation status is associated with
cancer development and metastatic recurrence in patients with clear
cell renal cell carcinoma. Oncogene. 29:5724–5728. 2010.
|
|
28
|
Myatt SS, Wang J, Monteiro LJ, Christian
M, Ho KK, Fusi L, et al: Definition of microRNAs that repress
expression of the tumor suppressor gene FOXO1 in endometrial
cancer. Cancer Res. 70:367–377. 2010.
|
|
29
|
Rotkrua P, Akiyama Y, Hashimoto Y, Otsubo
T and Yuasa Y: MiR-9 downregulates CDX2 expression in gastric
cancer cells. Int J Cancer. 129:2611–2620. 2011.
|
|
30
|
Wöll E, Bedikian A and Legha SS: Uveal
melanoma: natural history and treatment options for metastatic
disease. Melanoma Res. 9:575–581. 1999.
|
|
31
|
Gao K, Dai DL, Martinka M and Li G:
Prognostic significance of nuclear factor-kappaB p105/p50 in human
melanoma and its role in cell migration. Cancer Res. 66:8382–8388.
2006.
|
|
32
|
El-Shabrawi Y, Ardjomand N and Radner H:
MMP-9 is predominantly expressed in epithelioid and not spindle
cell uveal melanoma. J Pathol. 194:201–206. 2001.
|
|
33
|
Crosby MB, Yang H, Gao W, Zhang L and
Grossniklaus HE: Serum vascular endothelial growth factor (VEGF)
levels correlate with number and location of micrometastases in a
murine model of uveal melanoma. Br J Ophthalmol. 95:112–117.
2011.
|