Introduction
Hepatocellular carcinoma (HCC) is one of the major
global health problems. It accounts for 80–90% of primary liver
cancers, is the fifth most common malignancy (1,2), and
the third most common cause of cancer-related death in the world
(3,4). The prognosis of primary liver cancer
is very poor, as evident by the fact that the number of annual
deaths (~600,000) is almost equal to the number of new cases
diagnosed annually. The median survival rates were 80% (range
63–97%) at 1 year, 70% (34–78%) at 3 years and 50% (17–69%) at 5
years (5). To date, surgical
resection is still the best option for treatment and cure of HCC.
After resection, the 5-year survival rate of HCC reaches to ~50%
with the early disease, although these patients will have a high
recurrence rate (6). However, only
<15% of the patients are eligible for surgery (7) because most are diagnosed at the
advanced stages of the disease when the tumor has invaded and
metastasized to other organs in the body. Most recently, other
therapy choices have become available for treating or curing the
primary liver cancer, such as liver transplantation (8), radiofrequency (9), chemoembolization (10), and gene and immune therapy (11); however, each treatment option has
its own advantages and limitations. Thus, the early detection of
primary liver cancer remains an important approach for prolonging
the survival of patients (12),
while cancer prevention may offer another possibility for reducing
primary liver cancer incidence.
To this end, a number of biomarkers have been
discovered during the past three decades, such as α-fetoprotein
[AFP; (13)], Des-γ-carboxy
prothrombin (14), α-l-fucosidase
(15,16), and glypican-3 [GPC-3; (17,18)].
Using these biomarkers, primary liver cancer can be diagnosed
early. However, their sensitivity and specificity are still limited
(e.g., the sensitivity of AFP for detecting early stage primary
liver cancer is only between 30 and 60%) (19–22).
Although a combination of AFP with other biomarkers is able to
improve early detection of primary liver cancer, the diagnostic
accuracy for primary liver cancer is still low, especially with
tumors <3 cm in size (22–26).
Therefore, their utility has been questioned, and many experts in
the field consider them to be ‘obsolete’ (27). Thus, more reliable and useful
biomarkers are urgently needed.
Our study focused on structural maintenance of
chromosome 4 (SMC4) protein, which is a core subunit of condensin I
and II, large protein complexes, involved in chromosome
condensation. Previous studies have shown that SMC4 is a
chromosomal ATPase that is highly conserved from bacteria to human,
and plays a fundamental role in many aspects of higher-order
chromosome organization and dynamics (28). Indeed, a previous study demonstrated
that SMC4 protein might be useful in early detection of ovarian
cancer (29), while another study
showed that SMC4 regulated expression of the P53 pathway gene in
breast cancer cells, which may be related to the changes in
chromosome stability (30).
However, the role and function of SMC4 in primary liver cancer
remain unknown, this study aimed to determine expression of SMC4
mRNA and protein for association with clinicopathological feature
of the primary liver cancer and then to investigate the role of
SMC4 in primary liver cancer.
Materials and methods
Cell lines and culture
Human hepatocellular carcinoma Bel-7405, smmc-7721,
and H7402 cell lines and human normal liver HL-7702, OSG-7701, and
BH-HC1142 cell lines were purchased from Chinese Academy of
Sciences (Shanghai, China). The normal cells HL-7702,OSG-7701,
BH-HC1142 were used as controls. Cells were all grown in Dulbecco’s
modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (Cyclone, Logan City, UT,
USA) in a humidified incubator with 5% CO2 at 37°C.
Patients and tumor tissues
A total of 72 tissue samples from patients with
primary liver cancer were obtained from Daping Hospital and
Research Institute of Surgery, The Third Military Medical
University between September 2009 and December 2010. The
corresponding normal liver tissues were also collected from these
patients and used as controls. These tissue specimens were
immediately snap-frozen and kept at −80°C until use. The patients
were pathologically confirmed with primary liver cancer and no
patients received chemotherapy or radiotherapy before surgery. This
study was approved by the Ethics Committee of Daping Hospital and
Research Institute of Surgery, the Third Military Medical
University, Chongqing, China (no. ChiCTR-DDT-11001845), and all
patients consented to participate in the study.
RNA isolation and real-time reverse
transcription-PCR
Total RNA was isolated from the tissues and cells
using a TRIzol reagent (Invitrogen) according to the manufacturer’s
instructions. The isolated RNA was then pretreated with RNase-free
DNase at 37°C for 30 min and then subjected to reverse
transcription into cDNA by incubating 2 mg RNA, 1 ml oligo(dT), and
11 ml DEPC-treated H2O at 70°C for 5 min, followed by
addition of 4 ml 5X reaction buffer, 2 ml dNTP mixture, 0.5 ml
ribonuclease inhibitor, and 1 ml RevertAid™ M-MulV reverse
transcriptase. The reaction was incubated at 42°C for 60 min. For
PCR amplification of SMC4 cDNA, SMC4 primers were first designed
using the Primer Express software, version 2.0 (Applied Biosystems)
and synthesized by GenePharma Co. (Shanghai, China) (Table I). PCR mixtures were then denatured
at 95°C for 5 min, followed by 40 cycles of 94°C for 20 sec, 61°C
for 20 sec, and 72°C for 20 sec, and a final extension at 72°C for
5 min. Data were normalized to the geometric mean of the
house-keeping gene, β-actin (Sangon Biotech, Shanghai, China) and
expressed as the control.
 | Table IPrimer sequences used qRT-PCR. |
Table I
Primer sequences used qRT-PCR.
| Forward primer | Reverse primer |
|---|
| SMC4 |
5′-GAGAAAATTCTGGGACCTTT-3′ |
5′-TCTGAATGTCCTTGTGTTCA-3′ |
| β-actin |
5′-CATTAAGGAGAAGCTGTGCT-3′ |
5′-GTTGAAGGTAGTTTCGTGGA-3′ |
Protein extraction and western blot
analysis
For extraction of total cellular proteins from the
tissues and cells, the samples were lysed and clarified by
centrifugation in a RIPA buffer (Beyotime, Shanghai, China). Next,
50 μg of protein extracts were separated on 8% SDS-PAGE gels and
electrophoretically transferred to nitrocellulose membranes at 100
V for 90 min. The membranes were then blotted for 1–2 h with the
following antibodies and dilutions: the goat anti-SMC4 polyclonal
antibody at a dilution of 1:200 (Santa Cruz Biotechnologies, Santa
Cruz, CA, USA), an anti-goat secondary antibody at a dilution of
1:400 (Santa Cruz Biotechnologies). The nitrocellulose membranes
were incubated with ECL western blotting detection reagents from
Beyotime and exposed to X-ray film to detect the positive protein
band. β-actin (Santa Cruz Biotechnologies) was used as the
control.
Immunohistochemistry
Immunohistochemical staining was performed to detect
expression of SMC4 protein in primary liver cancer tissue
specimens. Briefly, deparaffinized 5-μm paraffin sections were
baked at 65°C for 30 min, submerged in the
ethylenediaminetetraacetic acid (EDTA) antigenic retrieval buffer,
and microwaved for antigenic retrieval. After blocking non-specific
binding with 20% normal serum in PBS for 1 h, the sections were
incubated with the goat anti-SMC4 polyclonal antibody at a 1:200
dilution (Santa Cruz Biotechnologies) overnight at 4°C. The next
day, the sections were washed three times with PBS and then
incubated with anti-goat secondary antibody at a 1:400 dilution
(Santa Cruz Biotechnologies), followed by further incubation with a
streptavidin-horseradish peroxidase complex (Beyotime) for 30 min
in the dark. Next, the sections were developed for color using DAB
Plus as a chromogen. To quantify SMC4 protein expression, we used
IHC score systems described previously (31). The intensity of SMC4 immunoreaction
was scored as: 0, negative; 1, weak; 2, moderate; and 3, strong.
The percentage of tumor cell stained were scored as 0, negative; 1,
<10% tumor cells stained; 2, 10–50% tumor cells stained; and 3,
>50% tumor cells stained. The staining intensity and percentage
of staining were then multiplied to produce a SMC4 staining index,
and the data ranged from 0 to 9. The expression levels of SMC4 were
graded accorded to the following scoring criteria: grade 0 (score
0); grade 1 (scores 1–3); grade 2 (scores 4–6); and grade 3 (scores
7–9). Specimens with grade 1 were considered low expression,
whereas grades 2 and 3 were defined as high expression.
Construction of SMC4 siRNA and
transfection
To knockdown SMC4 expression, we first designed and
synthesized 3 pairs of siRNA fragments and one pair of a negative
control (Table II). We then chose
a cell line with high SMC4 expression, H-7402, for SMC4 siRNA
transfection. H-7402 cells were grown and SMC4 siRNA plus
Lipofectamine 2000 were added to each well of cell cultures, and
then kept in a 5% CO2, 37°C incubator for 72 h. Next,
real-time reverse transcription-PCR and western blotting were
performed to analyze the reduced SMC4 expression before any other
experiments were done.
 | Table IIThe sequences of SMC4 siRNA and
negative control siRNA. |
Table II
The sequences of SMC4 siRNA and
negative control siRNA.
| DNA sequences |
|---|
| Homo-2217 |
5′-GCCCAAGAAUGUGUAAACUTT-3′ |
|
5′-AGUUUACACAUUCUUGGGCTT-3′ |
| Homo-830 |
5′-GGCCUGCAGAGAUAAUACUTT-3′ |
|
5′-AGUAUUAUCUCUGCAGGCCTT-3′ |
| Homo-1051 |
5′-GGCUAAAUGAACCUAUUAATT-3′ |
|
5′-UUAAUAGGUUCAUUUAGCCTT-3′ |
| Negative | 5′-UUC UCC GAA CGU
GUC AGG UTT-3′ |
| 5′-ACG UGA CAC GUU
CGG AGA ATT-3′ |
Cell viability MTT assay
Cells were plated into a 96-well culture plate and
kept in a 5% CO2, 37°C incubator for 24 h. The next day,
50 μl of 1X MTT was added to each cell culture well, incubated for
4 h at 37°C, followed by addition of 150 μl of dimethyl sulfoxide
(DMSO). The optical density (OD) values of cell cultures were
measured at a wavelength of 570 nm using an enzyme-labeled
instrument. Cell survival rate was then calculated as % =
(ODsample - ODblank)/ (ODcontrol -
ODblank) × 100%.
Statistical analysis
All statistical analyses were carried out using the
SPSS 13.0 statistical software package (SPSS, Chicago, IL). The
t-test was used to analyze the difference in SMC4 expression
between normal and cancer tissues. The Mann-Whitney U test was
performed to analyze the association between SMC4 expression and
the clinicopathological data from the patients. P<0.05 was
considered statistically significant.
Results
Expression of SMC4 protein in
hepatocellular carcinoma cell lines
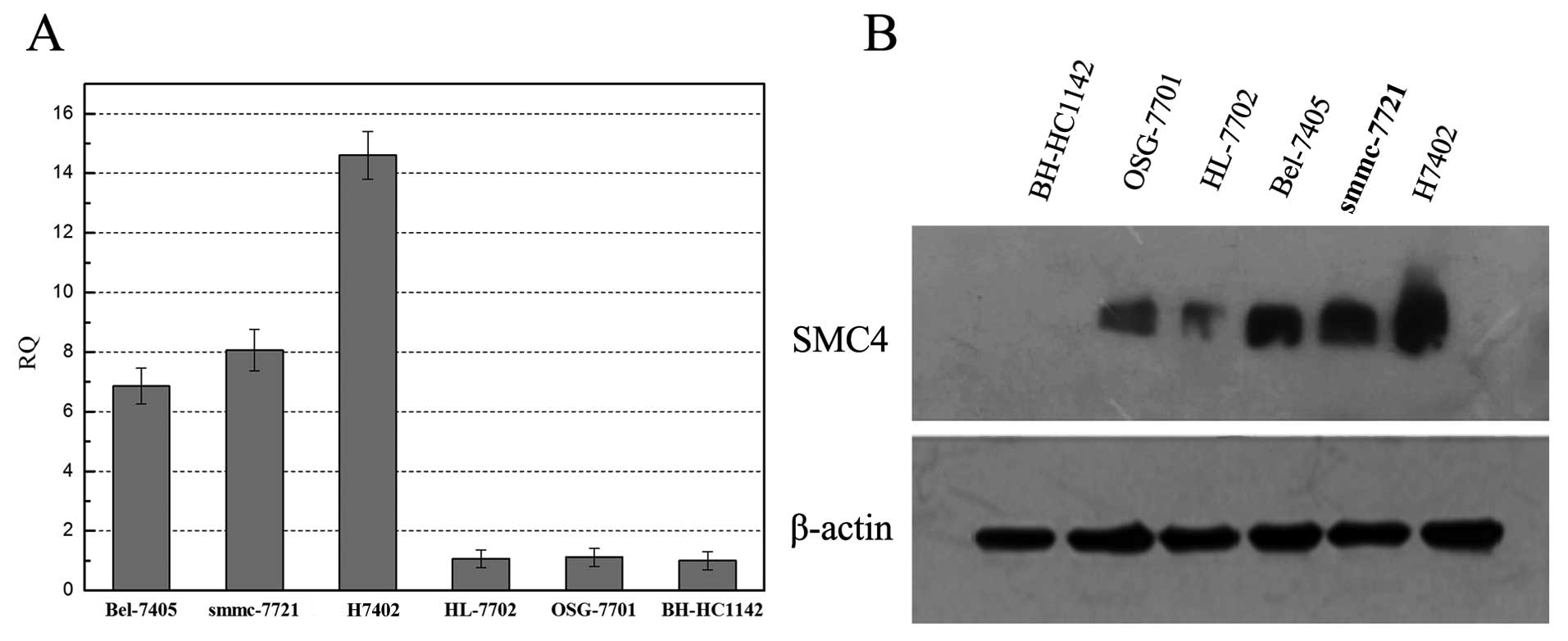
In this study, we first detected expression of SMC4
mRNA and protein in HCC cell lines using qRT-PCR and western blot
analyses, respectively, and found that SMC4 mRNA was highly
expressed in HCC cell lines (i.e., Bel-7405, smmc-7721, and H7402)
compared to normal cells (i.e., HL-7702, OSG-7701, and BH-HC1142;
Fig. 1A). Similarly, the western
blot data showed high expression of SMC4 in HCC cell lines
(Fig. 1B).
Overexpression of SMC4 mRNA and protein
in primary liver cancer tissues
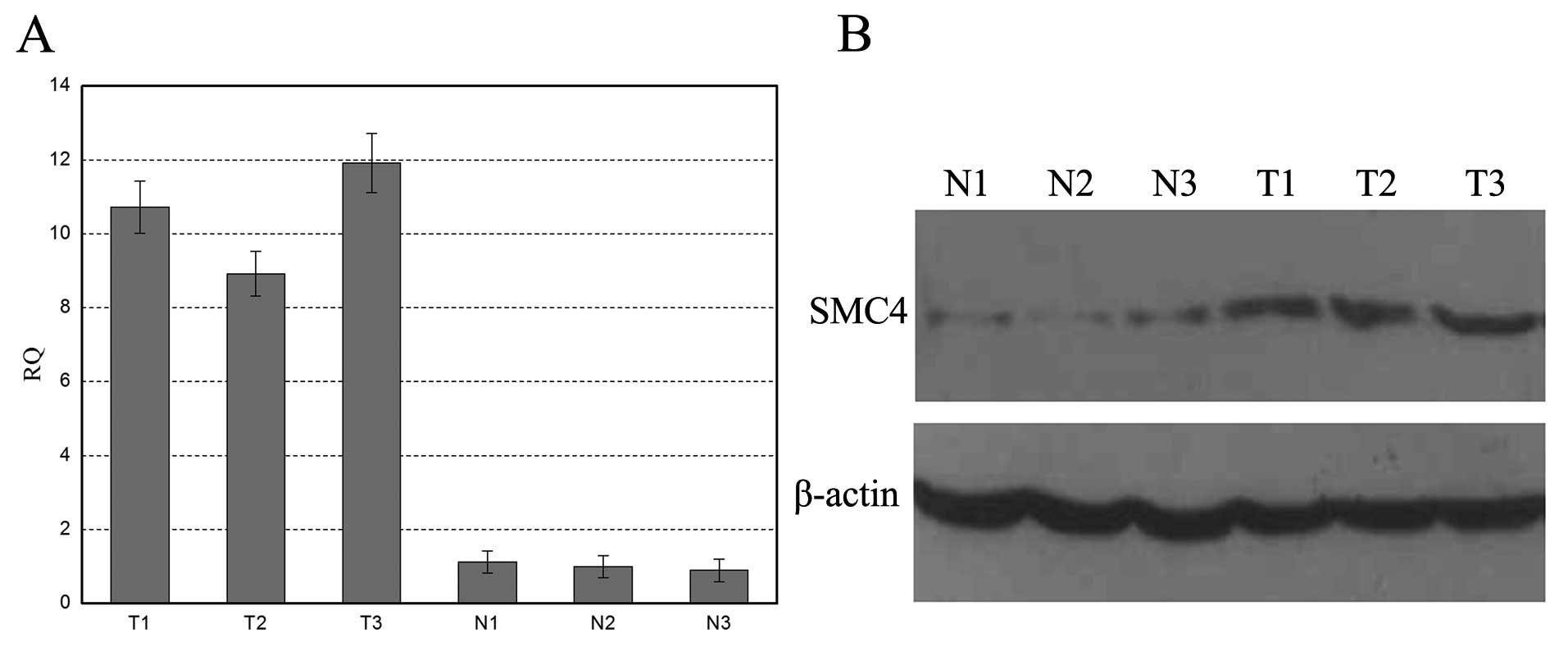
Next, we detect SMC4 expression in primary liver
cancer tissue samples using qRT-PCR and western blotting. The
primary liver cancer tissues showed significantly higher levels of
SMC4 mRNA than that of the paired normal liver tissues (Fig. 2A). Western blot analysis also showed
that SMC4 protein was highly expressed in primary liver cancer
tissues, whereas it was weakly detected in normal liver tissues
(Fig. 2B). Furthermore, we
performed immunohistochemistry, and the data shown in Fig. 2C revealed that strong SMC4
immunoreactions were detected in 52 (72.2%) primary liver cancer
tissue samples, weak immunoreactions detected in 16 (22.2%) cases,
and only 4 (5.9%) cases with negative immunoreactions. In contrast,
paired normal liver tissues only showed 6 (8.3%) positive cases.
The difference in SMC4 protein expressions between normal and
cancer tissues was statistically significant (P=0.000 by Student’s
t-test).
Next, we associated SMC4 expression with
clinicopathological data from primary liver cancer patients. The
data are shown in Table III. We
found a statistically significant association between SMC4 protein
expression with tumor size (P=0.027), tumor differentiation
(P=0.025), TNM stage (P=0.001), and vascular invasion (P=0.002);
however, there was no association between SMC4 expression and the
age and sex of patients with primary liver cancer (P>0.05) or
tumor type (P=0.170), indicating that SMC4 expression might
associate with the poor survival of the patients with primary liver
cancer.
 | Table IIIAssociation between SMC4 expression
and clinicopathological data. |
Table III
Association between SMC4 expression
and clinicopathological data.
| SMC4 expression | |
|---|
|
| |
|---|
| Strong (%)
(n=52) | Weak (%) (n=20) | P-value |
|---|
| Gender |
| Male | 41 (75.9) | 13 (24.1) | 0.228 |
| Female | 11 (61.1) | 7 (38.9) | |
| Age (years) |
| <45 | 14 (60.1) | 9 (39.1) | 0.143 |
| ≥45 | 38 (77.6) | 11 (22.4) | |
| Tumor size (cm) |
| <5 | 10 (52.6) | 9 (47.4) | 0.027 |
| ≥5 | 42 (79.2) | 11 (20.8) | |
| Tumor type |
| Cholangiocellular
carcinoma | 13 (56.5) | 10 (43.5) | 0.170 |
| Hepatocellular
carcinoma | 34 (82.9) | 7 (17.1) | |
| Mixed type | 5 (62.5) | 3 (37.5) | |
| Cell
differentiationa |
| I+II | 8 (50) | 8 (50) | 0.025 |
| III+IV | 44 (78.6) | 12 (21.4) | |
| TNM stageb |
| I+II | 7 (41.2) | 10 (58.8) | 0.001 |
| III+IV | 45 (81.8) | 10 (18.2) | |
| Vascular
invasion |
| Negative | 18 (54.5) | 15 (45.4) | 0.002 |
| Positive | 34 (87.2) | 5 (12.8) | |
Effects of SMC4 knockdown in HCC
cells
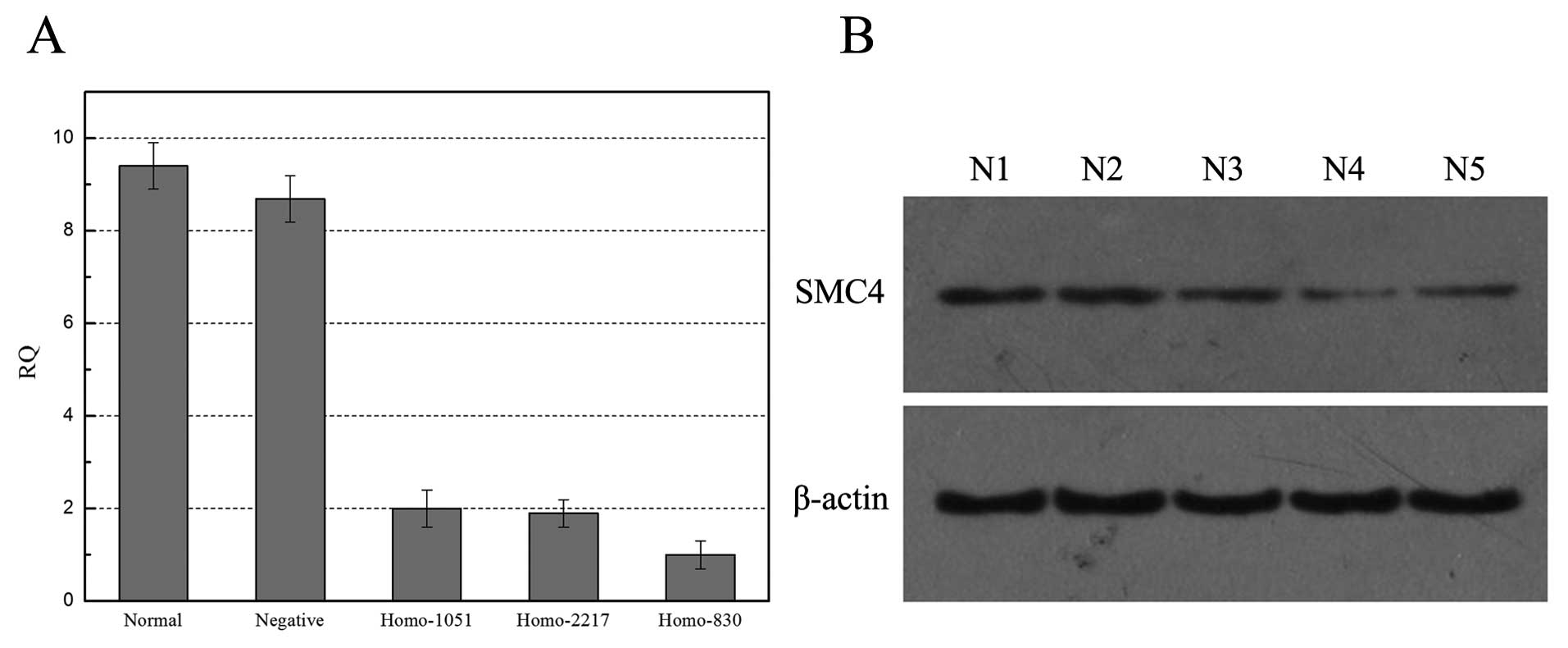
Next, we designed SMC4 siRNAs and knocked down SMC4
expression in HCC H-7402 cell line. qRT-PCR and western blot data
showed that three siRNA fragments were all able to knockdown SMC4
expression in HCCH-7402 cells compared to the negative control and
parental cells (Fig. 3). We then
chose Homo-830 SMC4 siRNA, the most effective fragment to knockdown
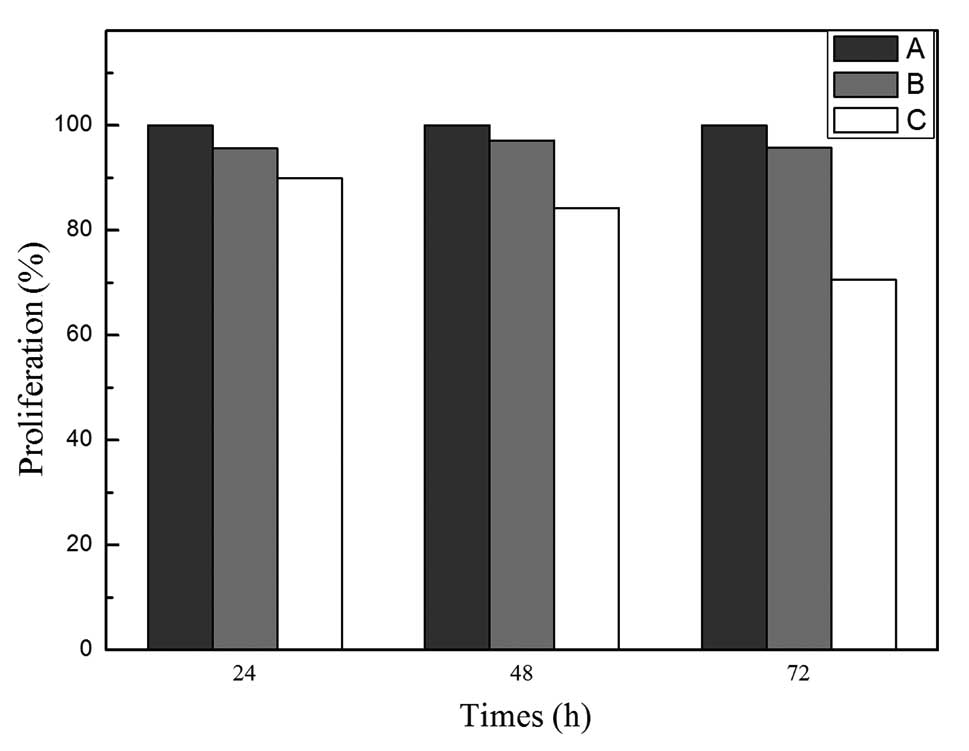
SMC4 expression in H-7402 cells and found that SMC4 siRNA
transfection was able to reduce tumor cell viability. The survival
rates of negative control siRNA and SMC4 siRNA were 95.6 and 89.9%,
97.1 and 84.2%, 95.7 and 70.6%, respectively, at 24, 48, and 72 h
(Fig. 4). These data indicate that
knockdown of SMC4 expression reduced HCC cell proliferation.
Discussion
In this study, we first detected SMC4 expression in
HCC cell lines and cancer tissue specimens and found that
expression of SMC4 mRNA and protein was upregulated in HCC tissues.
Expression of SMC4 protein was significantly associated with tumor
size, differentiation, TNM stage, and vascular invasion of primary
liver cancer. Furthermore, SMC4 expression was associated with poor
prognosis of patients with primary liver cancer. Next we
investigated the effects of SMC4 knockdown in an HCC cell line and
found that three SMC siRNA fragments were all able to knockdown
SMC4 protein expression, and that use of one the best SMC4 siRNA
fragment was able to reduce HCC cell viability. This study
indicates the usefulness of using SMC4 protein as a tumor marker in
the early detection of primary liver cancer, although further
investigation is needed to verify the present data.
In the present study, we detected SMC4 expression in
primary liver cancer tissue samples and found that 52 of 72 (72.2%)
paraffin-embedded cancer tissues displayed strong cytoplasm
staining of SMC4 protein, but only 6 (8.3%) cases had
immunostaining in normal liver tissues. The difference was
statistically significant, indicating the potential use of SMC4
protein as biomarker for detecting early primary liver cancer in
the clinic. Furthermore, we also associated SMC4 expression with
clinicopathological data from the patients, and found a significant
association between SMC4 expression with tumor size,
differentiation, TNM stage, and vascular invasion, indicating that
overexpression of SMC4 protein is associated with a poor prognosis
of the primary liver cancer patients. These findings provide
evidence indicating that increased SMC4 expression is associated
with development and progression of primary liver cancer, and thus
could be used as an independent predictor for prognosis in
patients. Our data are novel and have not been previously reported.
This study provides the first link between alterations in SMC4
expression and primary liver cancer. Further studies are needed to
determine the underlying mechanisms responsible for alterations in
SMC4 expression in primary liver cancer, and the possible role of
SMC4 in liver carcinogenesis.
We also investigated the effects of SMC4 knockdown
in HCC. Our data showed that three SMC4 siRNA fragment were all
able to significantly reduce SMC4 protein expression, and the
Homo-830 siRNA was able to reduce HCC cell viability. These
findings provide evidence that SMC4 has an obvious growth-promoting
effect on HCC cells. Future studies will be needed to further
investigate the role of SMC4 alterations in primary liver cancer,
and the underlying molecular mechanisms.
Indeed, SMC4 gene encodes a structural maintenance
of chromosomes protein, is a subunit of condensin, and plays a
central role in chromosome assembly and segregation in eukaryotic
cells (32). During synchronous
progression from G1 into S phase, SMC4 protein is required for
normal S phase progression (33).
SMC4 has been reported to play an important role in DNA replication
and recombination in breast cancer (34). Altered SMC4 expression could affect
chromosomal stability, and increase DNA double strand breaks and
unique chromosomal rearrangements in breast epithelial cells
(30).
In this study, we were for the first time able to
evaluate SMC4 overexpression in primary liver cancer tissue
specimens and to associate the expression with tumor size,
differentiation, TNM stage, and vascular invasion in primary liver
cancer tissues. Our data demonstrated that altered expression of
SMC4 protein could be a novel tumor marker in the early detection
of primary liver cancer and in predicting primary liver cancer
progression. However, there are limitations to our present study,
such as lack of Kaplan-Meier analysis of survival data and lack of
data showing the mechanism responsible for SMC4 overexpression.
Future study will investigate the role of SMC4 in HCC growth,
invasion, and apoptosis.
Acknowledgements
We would like to thank Professor Kai-Tai Zhang of
Cancer Institute and Hospital, Chinese Academy of Medical Sciences
for valuable advice, Medjaden Bioscience Ltd. for editing and
proofreading of the manuscript. We also specially thank Yan Li of
the Department of Gastroenterology, Daping Hospital and Research
Institute of Surgery for consultation.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ozenne V, Bouattour M, Goutté N, et al:
Prospective evaluation of the management of hepatocellular
carcinoma in the elderly. Dig Liver Dis. 43:1001–1005. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lau WY and Lai EC: Hepatocellular
carcinoma: current management and recent advances. Hepatobiliary
Pancreat Dis Int. 7:237–257. 2008.PubMed/NCBI
|
|
4
|
Hawkins MA and Dawson LA: Radiation
therapy for hepatocellular carcinoma from palliation to cure. Clin
Cancer Res. 106:1653–1661. 2006.PubMed/NCBI
|
|
5
|
Takayama T: Surgical treatment for
hepatocellular carcinoma. Jpn J Clin Oncol. 41:447–454. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cha CH, Saif MW, Yamane BH and Weber SM:
Hepatocellular carcinoma: current management. Curr Probl Surg.
47:10–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma S, Jiao B, Liu X, et al: Approach to
radiation therapy in hepatocellular carcinoma. Cancer Treat Rev.
36:157–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wertheim JA, Petrowsky H and Saab S: Major
challenges limiting liver transplantation in the United States. Am
J Transplant. 11:1773–1784. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wiggermann P, Jung EM and Stroszczynski C:
Radiofrequency ablation is a technique finished? Radiologe.
52:9–14. 2012.PubMed/NCBI
|
|
10
|
Liapi E and Geschwind JF: Medium-sized
HCC: achieving effective local tumor control with combined
chemoebolization and radiofrequency ablation. Ann Surg Oncol.
18:1527–1528. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smerdou C, Menne S, Hernandez-Alcoceba R
and Gonzalez-Aseguinolaza G: Gene therapy for HCV/HBV-induced
hepatocellular carcinoma. Curr Opin Investig Drugs. 11:1368–1377.
2010.PubMed/NCBI
|
|
12
|
Truty MJ and Vauthey JN: Surgical
resection of high-risk hepatocellular carcinoma: patient selection,
preoperative considerations, and operative technique. Ann Surg
Oncol. 17:1219–1225. 2010. View Article : Google Scholar
|
|
13
|
Abelev GI, Perova SD, Khramkova NI,
Postnikova ZA and Irlin IS: Production of embryonal alpha-globulin
by transplantable mouse hepatomas. Transplantation. 1:174–180.
1963. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lok AS, Sterling RK, Everhart JE, et al:
Des-γ-carboxy prothrombin and α-fetoprotein as biomarkers for the
early detection of hepatocellular carcinoma. Gastroenterology.
138:493–502. 2010.
|
|
15
|
Giardina MG, Matarazzo M, Varriale A,
Morante R, Napoli A and Martino R: Serum alpha-L-fucosidase: a
useful marker in the diagnosis of hepatocellular carcinoma. Cancer.
70:1044–1048. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishizuka H, Nakayama T, Matsuoka S, et al:
Prediction of the development of hepato-cellular carcinoma in
patients with liver cirrhosis by the serial determinations of serum
alpha-L-fucosidase activity. Intern Med. 38:927–931. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu ZW, Friess H, Wang L, et al: Enhanced
glypican-3 expression differentiates the majority of hepatocellular
carcinomas from benign hepatic disorders. Gut. 48:558–564. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Allegretta M and Filmus J: Therapeutic
potential of targeting glypican-3 in hepatocellular carcinoma.
Anticancer Agents Med Chem. 11:543–548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oka H, Tamori A, Kuroki T, Kobayashi K and
Yamamoto S: Prospective study of alpha-fetoprotein in cirrhotic
patients monitored for development of hepatocellular carcinoma.
Hepatology. 19:61–66. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishii M, Gama H, Chida N, et al:
Simultaneous measurements of serum alpha-fetoprotein and protein
induced by vitamin K absence for detecting hepatocellular
carcinoma. Am J Gastroenterol. 95:1036–1040. 2000.
|
|
21
|
Marrero JA, Su GL, Wei W, Emick D,
Conjeevaram HS, Fontana RJ and Lok AS: Des-gamma carboxyprothrombin
can differentiate hepatocellular carcinoma from nonmalignant
chronic liver disease in American patients. Hepatology.
37:1114–1121. 2003. View Article : Google Scholar
|
|
22
|
Gambarin-Gelwan M, Wolf DC, Shapiro R,
Schwartz ME and Min AD: Sensitivity of commonly available screening
tests in detecting hepatocellular carcinoma in cirrhotic patients
undergoing liver transplantation. Am J Gastroenterol. 95:1535–1538.
2000. View Article : Google Scholar
|
|
23
|
Trevisani F, D’Intino PE, Morselli-Labate
AM, et al: Serum alpha-fetoprotein for diagnosis of hepatocellular
carcinoma in patients with chronic liver disease: influence of
HBsAg and anti-HCV status. J Hepatol. 34:570–575. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Toyoda H, Kumada T, Kiriyama S, et al:
Prognostic significance of simultaneous measurement of three tumor
markers in patients with hepatocellular carcinoma. Clin
Gastroenterol Hepatol. 4:111–117. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakamura S, Nouso K, Sakaguchi K, et al:
Sensitivity and specificity of des-gamma-carboxy prothrombin for
diagnosis of patients with hepatocellular carcinomas varies
according to tumor size. Am J Gastroenterol. 101:2038–2043. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Uto H, Kanmura S, Takami Y and Tsubouchi
H: Clinical proteomics for liver disease: a promising approach for
discovery of novel biomarkers. Proteome Sci. 8:702010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sherman M: Alphafetoprotein: an obituary.
J Hepatol. 34:603–605. 2001. View Article : Google Scholar
|
|
28
|
Losada A and Hirano T: Dynamic molecular
linkers of the genome: the first decade of SMC proteins. Genes Dev.
19:1269–1287. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu KH, Patterson AP and Wang L: Selection
of potential markers for epithelial ovarian cancer with gene
expression arrays and recursive descent partition analysis. Clin
Cancer Res. 10:3291–3300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kulawiec M, Safina A, Desouki MM, Still I,
Matsui S, Bakin A and Singh KK: Tumorigenic transformation of human
breast epithelial cells induced by mitochondrial DNA depletion.
Cancer Biol Ther. 7:1732–1743. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Zhang N, Song LB, et al: Astrocyte
elevated gene-1 is a novel prognostic marker for breast cancer
progression and overall patient survival. Clin Cancer Res.
14:3319–3326. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Freeman L, Aragon-Alcaide L and Strunnikov
A: The condensin complex governs chromosome condensation and
mitotic transmission of rDNA. J Cell Biol1. 49:811–824. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu L, Peña Castillo L, Mnaimneh S, Hughes
TR and Brown GW: A survey of essential gene function in the yeast
cell division cycle. Mol Biol Cell1. 7:4736–4747. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Klijn JG, Zhang Y, et al:
Gene-expression profiles to predict distant metastasis of
lymph-node-negative primary breast cancer. Lancet. 365:671–679.
2005. View Article : Google Scholar : PubMed/NCBI
|