Introduction
Nasopharyngeal carcinoma (NPC) is a malignant cancer
of epithelial cell origin. It has a high incidence in Southeast
Asia and Southern China and is closely associated with infection by
Epstein-Barr virus (EBV) (1–5).
According to the different antigens generated during infection,
latent EBV infections in human can be classified into latency type
I, II and III infections. During latency I infection, only a small
number of gene products are produced [EBV-encoded small RNAs
(EBERs) and EBV nuclear antigen 1 (EBNA1)]. In type II latent
infection, the infected host cell will produce EBERs, EBNA1, latent
membrane protein 1 (LMP1), LMP2A, LMP2B and other transcripts. In
type III latent infection, EBNAs, EBERs and LMPs are also seen.
Latent EBV infection in NPC is usually of type II (1,4).
LMP1 is one of the latent EBV infection-associated
antigens, which is closely related with the carcinogenic effects of
EBV. As an oncoprotein, LMP1 can cross-link by using its C-terminal
cytoplasmic domain and analog CD40 to mediate
intracellular signalling pathways. LMP1 can activate transcription
factor activator protein 1 (AP-1) and nuclear factor
kappa-light-chain-enhancer of activated B cells (NF-κB) signal
transduction through its binding to the tumor necrosis factor
receptor-associated death domain protein (TRADD) or to the tumor
necrosis factor receptor-associated factor (TRAF). Therefore, LMP1
plays a key role in cell proliferation, apoptosis, cell
transformation, invasion and metastasis via regulating the
expression of downstream target genes (6–8). Most
of NPC biopsies are positive for LMP1 expression (3,9).
EBV can integrate into the host cell genome at
chromosomal fragile sites which are often prone to sister chromatid
exchanges, chromosomal translocations, gene deletions, gene
amplifications and oncogenic virus integrations. EBV can integrate
into human chromosomes at 1p, 1q, 2q, 3p, 3q, 4q, 5q, 6q, 7p, 7q,
9q, 11p, 14q and 15q (10,11). EBV infection also can cause a loss
of heterozygosity (LOH) at chromosome 5q11-q14 and 5q31-q33 loci
(10).
microRNAs (miRNAs) are endogenous, 19–25-nucleotide
long, single-stranded small non-coding RNA transcripts. They
interact with the 3′ UTR of target mRNA, resulting in inhibition of
translation of the target gene or target degradation. It has been
found that the expression levels and patterns of miRNAs in human
cancer cells and tissues show significant difference from those of
normal cells and tissues (12).
Human miRNAs have been found frequently (52%) at fragile sites in
the cancer-associated genomic regions or genes, including LOH
region, genomic breakpoints and fragile sites where genetic
abnormality often occurs in the cancer cells (13). Functionally, some miRNAs are similar
to oncogenes or tumor suppressor genes and post-transcriptionally
regulate the expression of cancer-related genes (14–17).
Human miRNA-146a is located within the second exon
of 5q33.3 LOC285628 gene which has two exons and an intermediate
intron of approximate size of 16 kb (18,19).
Overexpression of miRNA-146a has been observed in various malignant
tumors, such as papillary thyroid carcinoma, pediatric acute
leukemia and hepatocellular carcinoma (20–25).
LMP1 can also increase the expression of miR-146a in malignant
lymphoma (21,22).
Recent studies have shown that miRNA encoded by
oncogenic viruses or by the human genome may play an important
regulatory role in virus-host interactions (26–29).
Clarifying the mechanism of action of miRNAs not only helps to
understand the virus-host interaction but also helps to decipher
the development of virus-induced tumors. Our present study aims to
explore the relationship between miRNA-146a expression and
EBV-associated antigen LMP1 in the context of NPC and its possible
mechanism.
Materials and methods
Specimens and cell lines
Specimens of non-keratinizing undifferentiated
nasopharyngeal carcinoma, and chronic nasopharyngitis were obtained
from the diagnosed outpatients at the Affiliated Hospital of
Guangdong Medical College. Informed consent was provided by all
patients. All NPC patients had not been treated with radiotherapy
or chemotherapy.
Of the 40 cases of NPC patients whose specimens were
used for immunohistochemical detection, 12 cases were at TNM stage
I, 12 cases were at stage II, 10 cases were at stage III and 6
cases were at stage IV. Twenty-seven were males and 13 were
females, with age ranging from 30 to 75 years, median age of 46
years and mean age of 46.5 years. Of the 28 cases of chronic
nasopharyngitis patients, 17 were males and 11 were females, with
age ranging from 17 to 80 years, median age of 40 years and mean
age of 40.6 years. Of the 16 NPC patients (5 cases of TNM stage I,
4 cases of stage II, 4 cases of stage III and 3 cases of stage IV)
whose specimens were detected by qRT-PCR, 11 were male and 5 were
females, with age ranging from 28 to 78 years, median age of 46
years and mean age of 46.6 years. Of the 13 patients with chronic
nasopharyngitis, 7 were males and 6 were females, with age ranging
from 24 to 73 years, median age of 44 years and mean age of 44.1
years.
Nasopharyngeal carcinoma cell line CNE1 and CNE1-GL
were from the Department of Pathology, Guangdong Medical College.
CNE1-GL was transfected with a eukaryotic expression plasmid
pAT-GFP-LMP containing LMP1 gene and green fluorescent protein
(GFP) as reporter gene, which was a gift from Dr J.R. Arrand from
the Paterson Institute for Cancer Research, Christie CRC Research
Center, Manchester, UK (30).
Immunohistochemical staining
Slides of paraffin-embedded specimens and cells were
incubated overnight with mouse anti-human LMP1 monoclonal antibody
(Dako) (1:100) in a humidified box at 4°C, washed thrice with PBS,
incubated with streptavidin-peroxidase (SP) for 1 h at room
temperature, washed for three times and stained by diaminobenzidine
(DAB). Sections with known high LMP1 expression were used as
positive controls and sections incubated with PBS instead of
primary antibody served as negative controls.
For each section, 5 low magnification fields (x100)
were randomly selected under the microscope, and the proportions of
positively stained cells per field was counted under higher
magnification (x400). The results are presented as the mean
percentage of the 5 fields. The rating criteria were as follows: no
positive cells in the field was ranked as 0; positive cells ≤10% as
1; 11–50% as 2; 51–75% as 3; >75% as 4. The criteria for
staining intensity was: no staining was ranked as 0; light yellow
staining as 1; brown staining as 2; dark brown staining as 3. The
LMP1 staining was evaluated according to the number of positive
cells and staining intensity. The sections with a number of
positive cells of 0 were recognized as negative (−), while sections
with a number ≥1 were positive (+). The results of all the sections
were subjected to review by three pathologists who were blinded to
the study design.
Quantitative reverse transcription
polymerase chain reaction (qRT-PCR)
The total RNAs of NPC tissues and chronic
nasopharyngitis tissues were extracted with TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) and were reversely transcribed into
complementary DNA (cDNA) using miScript Reverse Transcription kit
(Qiagen, Germany). The cDNAs were diluted 1:10, 1:100 and 1:1000
and then amplified by miScript SYBR-Green PCR kit (Qiagen) in the
Applied Biosystems 7300 Real-time PCR System (Applied Biosystems,
Foster City, CA, USA). The primers for miRNA-146a and reference
gene were as follows: upstream primer of miRNA-146a
5′-TGAGAACTGAATTCCATGGGTT-3′, downstream primer
5′-ATCTACTCTCTCCAGGTCCTCA-3′ and upstream primer for the reference
gene U6 small nuclear RNA (snRNA) 5′-CTCGCTTCGGCAGCACA-3′,
downstream primer 5′-AA CGCTTCACGAATTTGCGT-3′. The relative changes
in the expression of miRNA-146a were calculated in accordance with
2−ΔΔCt method (31).
Construction of pGL3-miR146a-pri-pro
plasmid
Primers for the miRNA-146a promoter were synthesized
as previously described (22):
upstream primer 5′-GCAGCTAGCTTTCGG TCCATGAGCACGT-3′
(NheI restriction site is underlined); downstream primer
5′-GCAAAGCTTAGCGGTCAAGCGT CTTGG-3′
(HindIII restriction site is underlined). The sequence from
−1153 to +21 of miRNA-146a gene promoter was amplified by PCR. The
PCR products were purified, digested with NheI and
HindIII and ligated with the expression vector pGL3-Basic
containing firefly (Photinus pyralis) luciferase gene
(Promega, Madison, WI, USA) with the aid of T4 DNA ligase. After
transformation, positive clones were screened by restriction enzyme
digestion. The correct construct was verified by DNA sequencing and
named as pGL3-miR146a-pri-pro.
Transfection and detection of luciferase
activity
CNE1 or CNE1-GL cells (2×104) were seeded
into 24-well plates and incubated in a humidified 5% CO2
incubator at 37°C for 24 h. When the cells reached 60–70%
confluence, 1 μg of pGL3-Basic or pGL3-miR146a-pri-pro with 10 ng
pRL-TK, a plasmid containing Renilla luciferase reporter gene and
100 ng pRL-SV40 plasmid were co-transfected using Lipofectamine
2000 (Invitrogen, Carlsbad, CA, USA). After 48 h, the firefly and
Renilla luciferase enzyme assays were conducted by using the dual
luciferase reporter assay system (Promega) and fluorescence
intensity was detected by GloMax® 96 Microplate
Luminometer (Promega) in the transfected cells. The expression of
firefly luciferase was calculated using the formula ΔCT =
(F/R)sample/(F/R)control, where F refers to
the intensity of firefly luciferase and R is the intensity of
Renilla luciferase. Each treatment was performed in triplicate.
Statistical analysis
Data are presented as means ± standard deviation
(SD). Statistical comparisons for percentages were performed using
χ2 analysis in each case. For continuous variables,
Mann-Whitney U test was used. All statistical analyses were carried
out by using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). P-values
<0.05 were considered statistically significant (2-tailed).
Results
Different expression of LMP1 in chronic
nasopharyngitis, NPC specimens and NPC cell lines
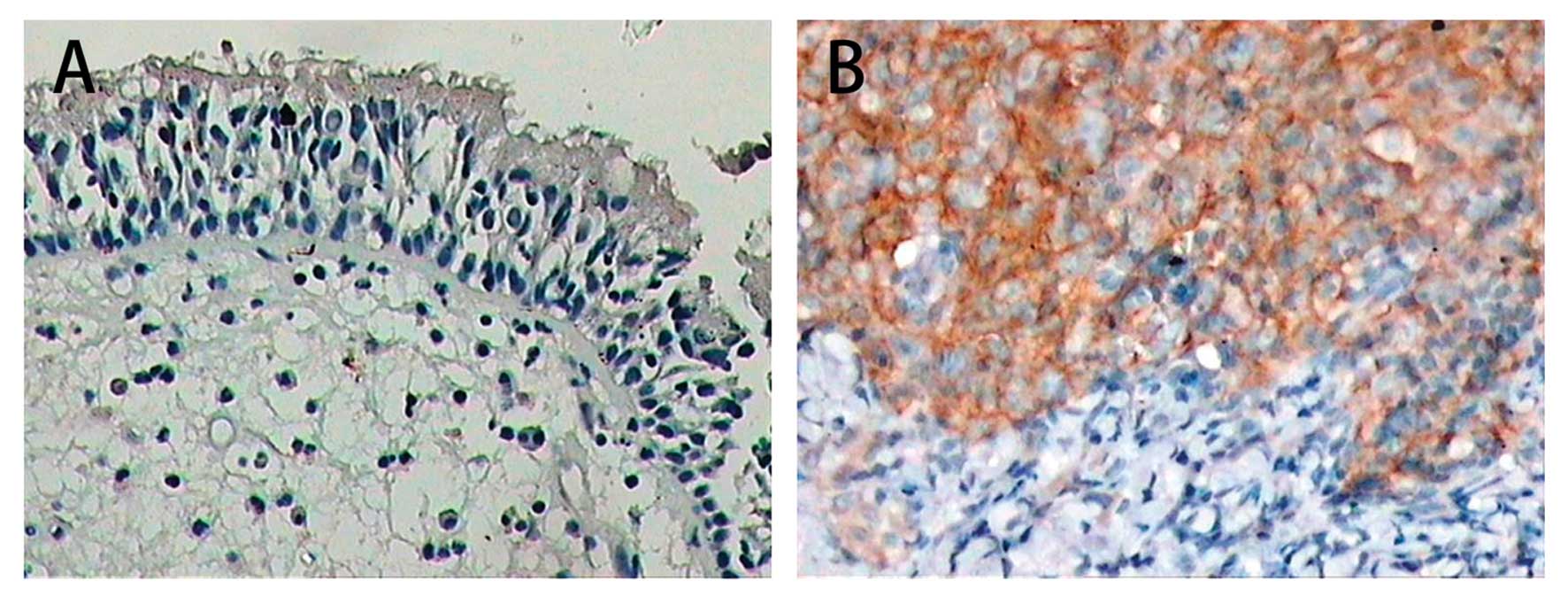
LMP1 expression in NPC and chronic nasopharyngitis
specimens was detected by immunohistochemical staining. It was
observed that LMP1 protein was located on the cell membrane
(Fig. 1). LMP1 expression was found
in 5 out of 28 (17.9%) cases of chronic nasopharyngitis, while the
percentage of positive staining of LMP1 was 62.5% (25/40) in the
NPC specimens. The difference of LMP1 expression between the two
groups was statistically significant (P<0.01) (Table I).
 | Table IExpression of LMP1 in NPC and chronic
nasopharyngitis specimens. |
Table I
Expression of LMP1 in NPC and chronic
nasopharyngitis specimens.
| Conditions | n | Positive | Negative | Positive (%) |
|---|
| Chronic
nasopharyngitis | 28 | 5 | 23 | 17.9 |
| NPC | 40 | 25 | 15 | 62.5a |
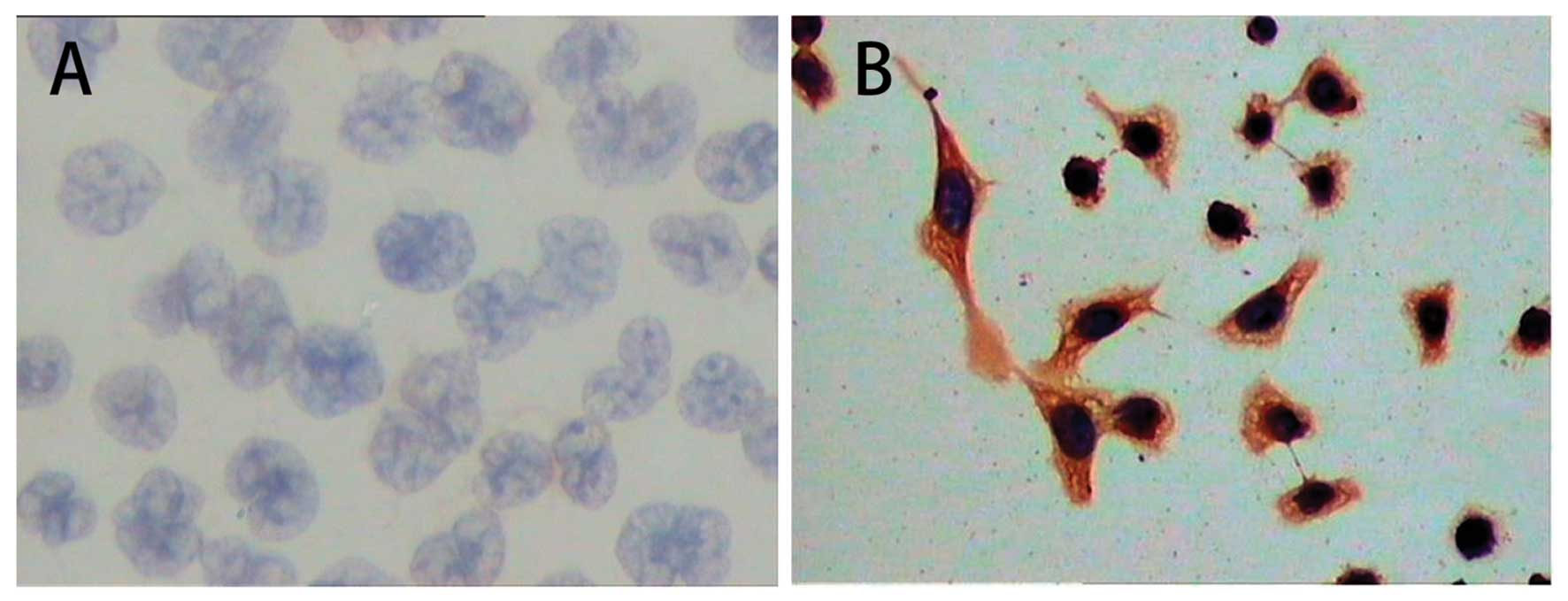
The results of immunocytochemical staining showed
that LMP1 protein was abundantly expressed and distributed in the
cell membrane and cytoplasm of CNE1-GL cells. However, expression
of LMP1 protein was not detected in CNE1 cells (Fig. 2 and Table II).
 | Table IIExpression of LMP1 in different NPC
cell lines. |
Table II
Expression of LMP1 in different NPC
cell lines.
| LMP1 |
|---|
|
|
|---|
| Cell lines | Positive (n) | Positive (%) |
|---|
| CNE1 | 0 | 0 |
| CNE1-GL | 492±11 | 98.4±2.2 |
Expression of miRNA-146a in chronic
nasopharyngitis, NPC specimens and NPC cell lines
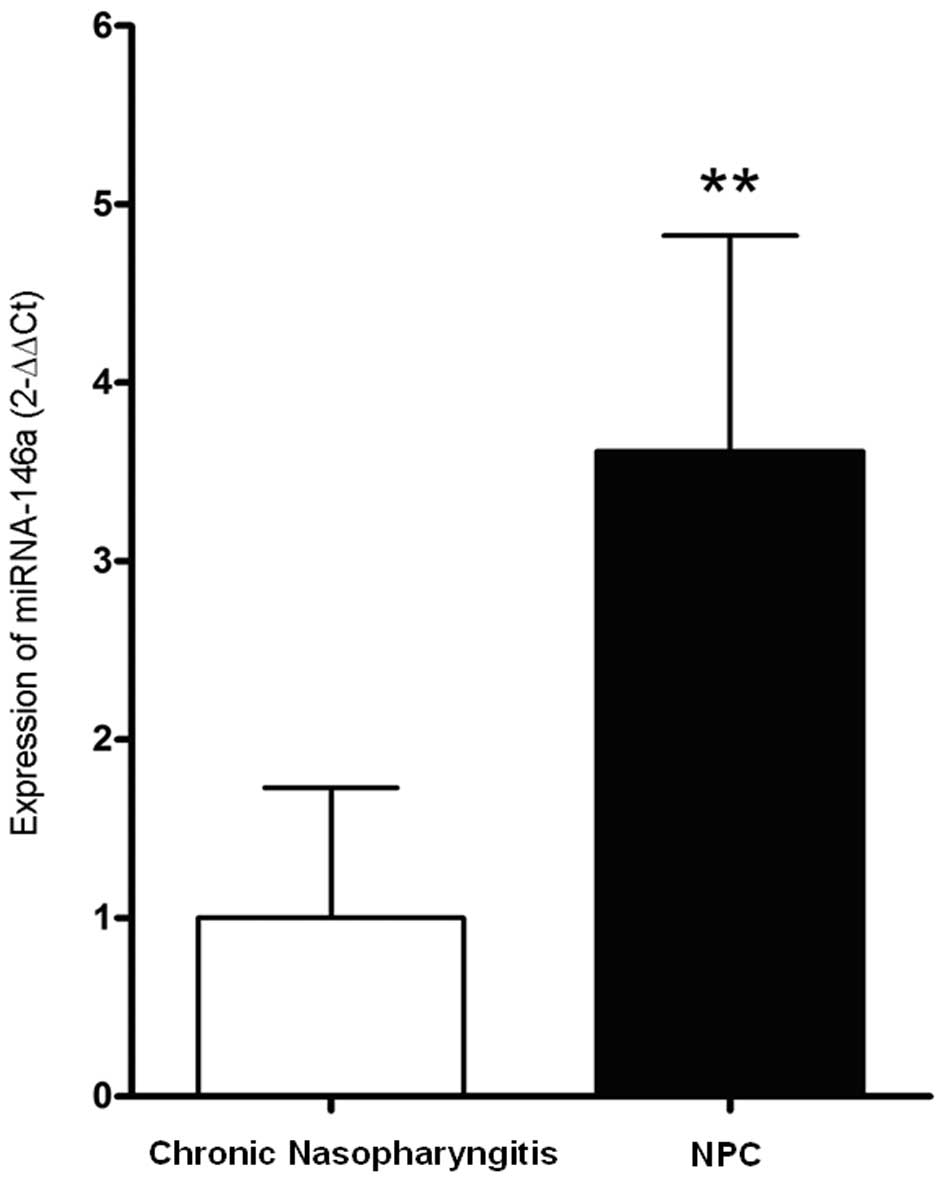
The result of qRT-PCR revealed that the expression
level of miRNA-146a in NPC was 3.62±1.20 times higher than that in
chronic nasopharyngitis (P<0.01) (Fig. 3).
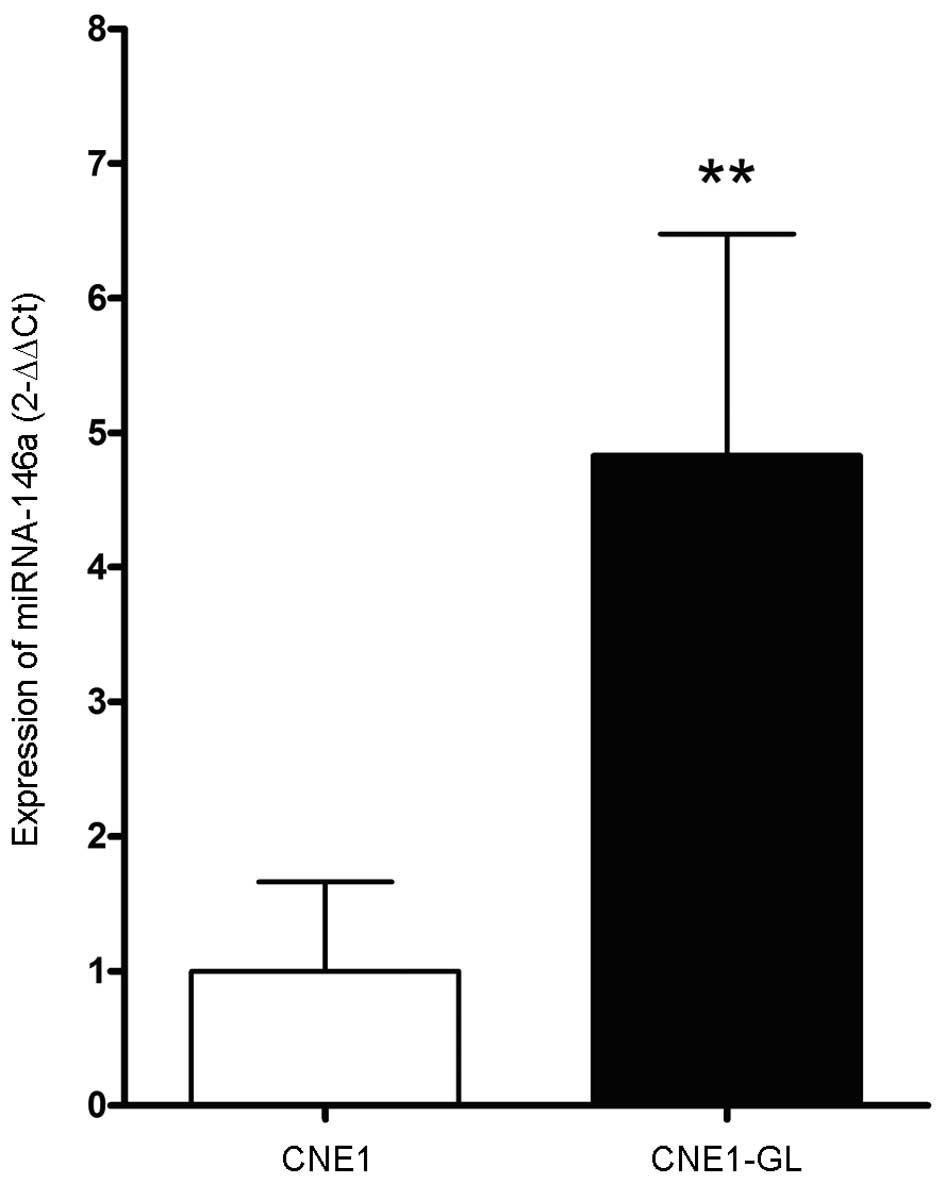
Similarly, as shown in Fig. 4, the miRNA-146a level in CNE1-GL
cells was 4.83±1.64 times higher than that in CNE-1 cells
(P<0.01).
The relationship between LMP1 and
miRNA-146a
To explore the relationship between LMP1 and
miRNA-146a, we constructed the plasmid pGL3-miR146a-pri-pro to
transfect NPC cells (CNE1 and CNE1-GL) with different LMP1 status.
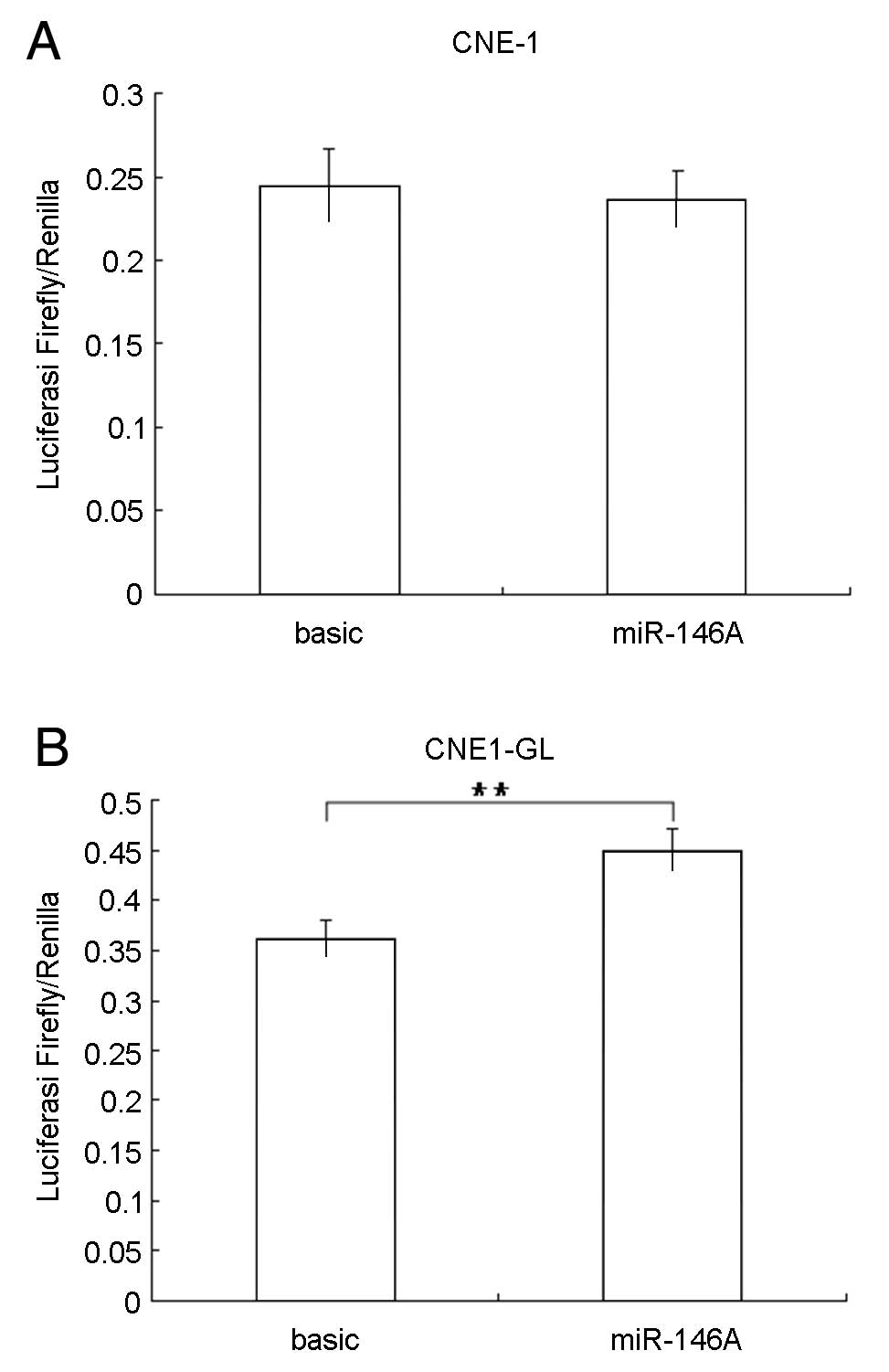
It was shown that ΔCT values of CNE1 cells transfected with
pGL3-Basic or the recombinant plasmid pGL3-miR146a-pri-pro did not
make significant difference (P=0.207) (Fig. 5A). The expression level of firefly
luciferase in pGL3-miR146a-pri-pro transfected CNE1 cells was 0.967
that of pGL3-Basic transfected cells, suggesting that the activity
of miR146a promoter was not effective in CNE1 cells.
In contrast, the ΔCT value of CNE1-GL cells
transfected with plasmid pGL3-miR146a-pri-pro was significantly
higher than that of CNE1-GL cells transfected with pGL3-Basic
(P=0.0042) (Fig. 5B). The
expression level of firefly luciferase in CNE1-GL cells was 1.240
higher than that of pGL3-Basic transfected cells, suggesting that
activity of miR146a promoter may be upregulated by LMP1.
Discussion
In the present study, we found that the miRNA-146a
expression level in 16 cases of human non-keratinizing
undifferentiated NPC was significantly elevated (P<0.01),
compared to that in 13 cases of chronic nasopharyngitis, indicating
that the higher expression of miRNA-146a could be a potential
indicator for NPC development. EBV infection is an important
stimulus for oncogenesis of NPC (1–5), and
EBV is detectable in almost all of the non-keratinizing
undifferentiated nasopharyngeal carcinoma. The latent EBV infection
involved in NPC is type II (1,4). In
general, in the state of latent infection EBV genome persists in
the host cell as an episome, but sometimes the EBV genome also
integrates into the chromosomes of the host cell (32). Many integration sites in human
chromosomes have been confirmed, such as 1p, 1q, 2q, 3p, 3q, 4q,
5q, 6q, 7p, 7q, 9q, 11p, 14q and 15q (10,11).
miRNA-146a is in the second exon of LOC285628 gene which is located
coincidentally, on chromosome 5q33.3 (18,19).
This suggests that the increased expression of miRNA-146a may be
the result of the enhanced promoter activity of miRNA-146a, due to
integration of the EBV genome.
On the other hand, LMP1 as an EBV latent
infection-associated antigen that can mimic
CD40-mediated signal transduction pathways, which
participate in regulation of cell proliferation, apoptosis,
malignant transformation, invasion and metastasis. To further
determine whether the elevated expression level of miRNA-146a in
human NPC is directly related to LMP1, we used
immunohistochemical/immunocytochemical methods to detect LMP1
protein expression in paraffin-embedded sections of the NPC cell
lines with/without stably transfected LMP1 gene. The expression of
miRNA-146a was quantified by qRT-PCR in parallel. The results
showed that in both the specimens and cell lines, the expression
levels of miRNA-146a were positively correlated with that of LMP1
protein expression. It was confirmed that the LMP1 can upregulate
the expression level of miRNA-146a. To investigate the mechanism
involved in regulation of miRNA-146a by LMP1, we constructed a dual
luciferase reporter gene vector pGL3-mir146a-pri-pro which was
transfected into CNE1 and CNE1-GL cells, using pGL3-Basic as a
control. The results showed that when compared to CNE1 cells (no
LMP1 expression), the pri-miRNA-146a level in CNE1-GL cells (LMP1
expressing) was significantly upregulated. The dual luciferase
reporter assay indicated that in NPC cells LMP1 protein
significantly enhanced the expression of miRNA-146a via promoter
activation. As one of the main regulators of EB virus, LMP1 can
simulate the CD40 receptor and activate multiple cell signaling
pathways such as NF-κB, AP-1, inhibitor of differentiation 1 (Id)1,
Id3, signal transducers and activators of transcription (STAT) and
TNF receptor-associated factors (TRAFs), thus enlarging the
downstream regulation of target gene expression. CD40
activates cells by a signaling pathway that begins with the
association of adapter proteins known as TRAFs. TRAFs are thought
to interact with CD40 to nuclear factor (NF)-κB and
c-Jun kinase (JNK) activation. Similar to CD40, LMP1
also binds TRAFs which are then thought to interact with kinases
such as NF-κB-inducing kinase that ultimately promote activation of
NF-κB (33–38). However, further investigation on the
exact interaction between LMP1 and miRNA-146a is still needed.
In conclusion, this study confirmed that miR-146a
expression was correlated with the expression of EBV-associated
antigen LMP1 in NPC in vitro and in vivo. LMP1 may
elevate the expression of miRNA-146a via promoter activation, which
adds new evidence to EBV-host interactions.
Acknowledgements
This study is supported by the National Basic
Research Program of China (973 Program), 2011CB504800.
References
|
1
|
Brooks L, Yao QY, Rickinson AB and Young
LS: Epstein-Barr virus latent gene transcription in nasopharyngeal
carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts.
J Virol. 66:2689–2697. 1992.PubMed/NCBI
|
|
2
|
Thompson MP and Kurzrock R: Epstein-Barr
virus and cancer. Clin Cancer Res. 10:803–821. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang M, Zong YS, He JH, Lin SX, Zhong BL
and Liang YJ: Comparison of Epstein-Barr virus infection and 30
bp-deleted LMP1 gene among four histological types of
nasopharyngeal carcinoma. Chin Med J. 117:608–611. 2004.PubMed/NCBI
|
|
4
|
Tao Q, Young LS, Woodman CB and Murray PG:
Epstein-Barr virus (EBV) and its associated human cancers -
genetics, epigenetics, pathobiology and novel therapeutics. Front
Biosci. 11:2672–2713. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng H, Li LL, Hu DS, Deng XY and Cao Y:
Role of Epstein-Barr virus encoded latent membrane protein 1 in the
carcinogenesis of nasopharyngeal carcinoma. Cell Mol Immunol.
4:185–196. 2007.PubMed/NCBI
|
|
6
|
Akiba H, Nakano H, Nishinaka S, et al:
CD27, a member of the tumor necrosis factor receptor superfamily,
activates NF-kappaB and stress-activated protein kinase/c-Jun
N-terminal kinase via TRAF2, TRAF5, and NF-kappaB-inducing kinase.
J Biol Chem. 273:13353–13358. 1998. View Article : Google Scholar
|
|
7
|
Deng L, Yang J, Zhao XR, et al: Cells in
G2/M phase increased in human nasopharyngeal carcinoma cell line by
EBV-LMP1 through activation of NF-kappaB and AP-1. Cell Res.
13:187–194. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thornburg NJ, Kulwichit W, Edwards RH,
Shair KH, Bendt KM and Raab-Traub N: LMP1 signaling and activation
of NF-kappaB in LMP1 transgenic mice. Oncogene. 25:288–297.
2006.PubMed/NCBI
|
|
9
|
Young LS and Rickinson AB: Epstein-Barr
virus: 40 years on. Nat Rev Cancer. 4:757–768. 2004.PubMed/NCBI
|
|
10
|
Shao JY, Huang XM, Yu XJ, et al: Loss of
heterozygosity and its correlation with clinical outcome and
Epstein-Barr virus infection in nasopharyngeal carcinoma.
Anticancer Res. 21:3021–3029. 2001.PubMed/NCBI
|
|
11
|
Gao J, Luo X, Tang K, Li X and Li G:
Epstein-Barr virus integrates frequently into chromosome 4q, 2q, 1q
and 7q of Burkitt’s lymphoma cell line (Raji). J Virol Methods.
136:193–199. 2006.PubMed/NCBI
|
|
12
|
Bandres E, Cubedo E, Agirre X, et al:
Identification by Real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Tang S, Le SY, et al: Aberrant
expression of oncogenic and tumor-suppressive microRNAs in cervical
cancer is required for cancer cell growth. PLoS One. 3:e25572008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hurst DR, Edmonds MD, Scott GK, Benz CC,
Vaidya KS and Welch DR: Breast cancer metastasis suppressor 1
up-regulates miR-146, which suppresses breast cancer metastasis.
Cancer Res. 69:1279–1283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo X, Yang W, Ye DQ, et al: A functional
variant in microRNA-146a promoter modulates its expression and
confers disease risk for systemic lupus erythematosus. PLoS Genet.
7:e10021282011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He H, Jazdzewski K, Li W, et al: The role
of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad
Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Motsch N, Pfuhl T, Mrazek J, Barth S and
Grasser FA: Epstein-Barr virus-encoded latent membrane protein 1
(LMP1) induces the expression of the cellular microRNA miR-146a.
RNA Biol. 4:131–137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cameron JE, Yin Q, Fewell C, et al:
Epstein-Barr virus latent membrane protein 1 induces cellular
MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J
Virol. 82:1946–1958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jazdzewski K, Murray EL, Franssila K,
Jarzab B, Schoenberg DR and de la Chapelle A: Common SNP in
pre-miR-146a decreases mature miR expression and predisposes to
papillary thyroid carcinoma. Proc Natl Acad Sci USA. 105:7269–7274.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu T, Zhu Y, Wei QK, et al: A functional
polymorphism in the miR-146a gene is associated with the risk for
hepatocellular carcinoma. Carcinogenesis. 29:2126–2131. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang H, Luo XQ, Zhang P, et al: MicroRNA
patterns associated with clinical prognostic parameters and CNS
relapse prediction in pediatric acute leukemia. PLoS One.
4:e78262009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Couturier JP and Root-Bernstein RS: HIV
may produce inhibitory microRNAs (miRNAs) that block production of
CD28, CD4 and some interleukins. J Theor Biol. 235:169–184. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sullivan CS, Grundhoff AT, Tevethia S,
Pipas JM and Ganem D: SV40-encoded microRNAs regulate viral gene
expression and reduce susceptibility to cytotoxic T cells. Nature.
435:682–686. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gupta A, Gartner JJ, Sethupathy P,
Hatzigeorgiou AG and Fraser NW: Anti-apoptotic function of a
microRNA encoded by the HSV-1 latency-associated transcript.
Nature. 442:82–85. 2006.PubMed/NCBI
|
|
29
|
Rana TM: Illuminating the silence:
understanding the structure and function of small RNAs. Nat Rev Mol
Cell Biol. 8:23–36. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Y and Chen XY: Effect of Epstein-Barr
virus latent membrane protein 1 (LMP1) on apoptosis of
nasopharyngeal carcinoma cell line CNE1. Ai Zheng. 21:498–503.
2002.(In Chinese).
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
32
|
Henderson A, Ripley S, Heller M and Kieff
E: Chromosome site for Epstein-Barr virus DNA in a Burkitt tumor
cell line and in lymphocytes growth-transformed in vitro. Proc Natl
Acad Sci USA. 80:1987–1991. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kieser A, Kilger E, Gires O, Ueffing M,
Kolch W and Hammerschmidt W: Epstein-Barr virus latent membrane
protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase
cascade. EMBO J. 16:6478–6485. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gires O, Kohlhuber F, Kilger E, et al:
Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3
and activates STAT proteins. EMBO J. 18:3064–3073. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Everly DN Jr, Mainou BA and Raab-Traub N:
Induction of Id1 and Id3 by latent membrane protein 1 of
Epstein-Barr virus and regulation of p27/Kip and cyclin-dependent
kinase 2 in rodent fibroblast transformation. J Virol.
78:13470–13478. 2004. View Article : Google Scholar
|
|
36
|
Li HM, Zhuang ZH, Wang Q, et al:
Epstein-Barr virus latent membrane protein 1 (LMP1) upregulates Id1
expression in nasopharyngeal epithelial cells. Oncogene.
23:4488–4494. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hau PM, Tsang CM, Yip YL, Huen MS and Tsao
SW: Id1 interacts and stabilizes the Epstein-Barr virus latent
membrane protein 1 (LMP1) in nasopharyngeal epithelial cells. PLoS
One. 6:e211762011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Talaty P, Emery A and Everly DN Jr:
Characterization of the latent membrane protein 1 signaling complex
of Epstein-Barr virus in the membrane of mammalian cells with
bimolecular fluorescence complementation. Virol J. 8:4142011.
View Article : Google Scholar
|