Introduction
The intratumor environment is characterized by low
extracellular pH, low nutrition, and chronic hypoxia due to poor
vascular development (1,2). The environment is unsuitable for the
survival of cancer cells; however, a minor population of cancer
cells becomes resistant to the environment and acquires the ability
to survive (1). In prostate cancer,
the characteristics of the intratumor environment have been
demonstrated using in vivo electrode measurements of oxygen
levels (3). The cancer cells that
survive in the environment have been considered potential
therapeutic targets for the treatment of solid tumors, including
prostate cancer.
The human prostate cancer cell line LNCaP is a
widely used model of androgen-sensitive prostate cancer, and
several sublines of LNCaP cells have been established and used to
study the molecular and cellular biology of prostate cancer cells.
The androgen-independent LNCaP subline LNCaP-C4-2 was established
from LNCaP tumors in castrated mice (4). LNCaP-abl and AIDL cells are also
androgen-independent sublines, generated in vitro by
culturing LNCaP cells under androgen-depleted conditions (5,6).
LNCaP-IL6+ and LNCaP-CR cells are resistant to
cytokine-induced apoptosis (7,8).
LNCaP-H1 cells were raised under chronic hypoxia, and exhibited
androgen-independent growth in vitro and in vivo
(9).
We previously established sublines from prostate
cancer LNCaP cells by the limiting dilution method, i.e., LNCaP-E9
and LNCaP-G4, which are slightly and highly androgen-sensitive,
respectively (10). In the present
study, the LNCaP-F10 cell line, which was isolated in a similar
manner, was further characterized by its ability to survive in an
acidic environment under low-nutrient conditions. LNCaP-F10 cells
were examined to determine the mechanisms underlying their
adaptation to a low-pH/low-nutrient environment.
Materials and methods
Materials
2′,7′-Bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein,
acetoxymethyl ester (BCECF-AM) was obtained from Dojin Laboratory
(Kumamoto, Japan). The anti-androgen receptor (AR) rabbit
polyclonal antibody (N-20) and anti-prostate-specific antigen (PSA)
mouse monoclonal antibody (A67-B/E3) used for western blotting were
from Santa Cruz Biotechnology (Santa Cruz, CA, USA). A rabbit
polyclonal anti-PSA antibody for immunohistochemistry was purchased
from DakoCytomation, Inc. (Copenhagen, Denmark). Anti-Bcl-2 and
anti-β-actin mouse monoclonal antibodies were purchased from Sigma
(St. Louis, MO, USA). Anti-caspase-3 rabbit polyclonal and
anti-poly (ADP ribose) polymerase (PARP) monoclonal antibodies were
from BD Biosciences (Franklin Lakes, NJ, USA). Anti-bad and
anti-bax rabbit polyclonal antibodies, and the anti-survivin (6E4)
mouse monoclonal antibody were purchased from Cell Signaling
Technology (Beverly, MA, USA). Mouse monoclonal anti-E-cadherin
(clone 36) antibody was purchased from BD Transduction
Laboratories, Inc. (Lexington, KY, USA). All other chemicals were
of analytical grade.
Cell culture
Human prostatic carcinoma LNCaP cells were obtained
from the American Type Culture Collection (Rockville, MD, USA).
LNCaP-F10 cells were previously isolated as a subline of LNCaP
cells by the limiting dilution method (10). LNCaP and LNCaP-F10 cells were
cultured in RPMI-1640 medium containing 10% fetal bovine serum
(FBS) under a humidified atmosphere with 5% CO2 at
37°C.
Cell growth assay
Cells were seeded onto 6-cm dishes at a density of
7×105 cells/dish with normal medium (RPMI-1640 + 10%
FBS, pH 7.4) or low-pH/low-nutrient medium (RPMI-1640 + 0.5% FBS
and 10 mM PIPES, pH 6.3). The cells were incubated for the
indicated times, and the detached and adherent cells were
collected. The cells were stained with a 0.1% trypan blue solution
and counted under a microscope using a hemocytometer.
Intracellular pH measurement
Cells were incubated in a normal medium or a
low-pH/low-nutrient medium for 24 h. The intracellular pH was then
measured using BCECF-AM, a pH-sensitive probe, as previously
described (11). Cellular pH was
analyzed by FACScan using CellQuest Pro software (BD Biosciences)
with excitation at 488 nm and a ratio of emissions at 520 (pH
sensitive) and 680 nm (pH insensitive). For each experiment,
standard curves were established with a series of cells at pH 6.3,
6.7, 7.0 and 7.3.
DNA fragmentation assay
The DNA fragmentation assay was performed as
previously described (11). Cells
were seeded onto 10-cm dishes at a density of 2×106 in a
normal medium or a low-pH/low-nutrient medium for the indicated
times. Detached cells were collected and lysed in buffer [10 mM
Tris-HCl, 10 mM ethylenediaminetetraacetic acid (EDTA), and 1%
Triton X-100] and treated with Proteinase K and RNase A. The DNA
was separated by electrophoresis on 1.5% gels and visualized by
staining with ethidium bromide.
Real-time reverse
transcriptase-polymerase chain reaction (Real-time RT-PCR)
Real-time RT-PCR was performed with a slightly
modified version of a previously described protocol (10). Total RNA was extracted using the
TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and first-strand
complementary DNA was synthesized from 1 μg of total RNA using
PrimeScript reverse transcriptase (Takara, Otsu, Japan). Real-time
monitoring of PCR reactions was performed using the Thermal Cycler
Dice Real-Time system (Takara) with SYBR Premix Ex Taq (Takara). At
the end of the reaction, a dissociation curve analysis was
performed to examine the specificity of the product. PCR was
performed using the following conditions: 35 cycles of 10 sec at
95°C and 30 sec at 60°C. Gene-specific primers were designed for
each gene and used for RT-PCR (Table
I). B2MG, a housekeeping gene, was used for normalizing the
target mRNA expression.
 | Table IPrimer sequences for real-time
RT-PCR. |
Table I
Primer sequences for real-time
RT-PCR.
| Gene name | Sequence |
|---|
| APAF | Sense:
5′-TCTGCTGATGGTGCAAGGAT-3′ |
| Antisense:
5′-CCGTGTGGATTTCTCCCAAT-3′ |
| BAD | Sense:
5′-CCAGCTGTGCCTTGACTACG-3′ |
| Antisense:
5′-TGCTCACTCGGCTCAAACTC-3′ |
| BAX | Sense:
5′-CAGCAAACTGGTGCTCAAGG-3′ |
| Antisense:
5′-CAACCACCCTGGTCTTGGAT-3′ |
| BCL2 | Sense:
5′-TCCAGATGGCAAATGACCAG-3′ |
| Antisense:
5′-CTGGGTGGGTCTGTGTTGAA-3′ |
| BCL2L1 | Sense:
5′-GGGTTCCCTTTCCTTCCATC-3′ |
| Antisense:
5′-GGAGTCCTGGTCCTTGCATC-3′ |
| BIK | Sense:
5′-GTGCTGGAACACTGCTGAGG-3′ |
| Antisense:
5′-GGCAGGAGTGAATGGCTCTT-3′ |
| BIRC5 | Sense:
5′-CAGCTTCGCTGGAAACCTCT-3′ |
| Antisense:
5′-AGGCTAGGGACGACGATGAA-3′ |
| BIRC6 | Sense:
5′-GCAGTGATAAGCGGGTAGGC-3′ |
| Antisense:
5′-TTGTGGCTCTGCACACCTCT-3′ |
| DAPK1 | Sense:
5′-ATGGCAACATGCCTATCGTG-3′ |
| Antisense:
5′-TGGCTCCCATCAGACAGAGA-3′ |
| MCL1 | Sense:
5′-GGGGCAGGATTGTGACTCTC-3′ |
| Antisense:
5′-AAACCCATCCCAGCCTCTTT-3′ |
| XIAP | Sense:
5′-GACTACCGCCCCAGCATTAG-3′ |
| Antisense:
5′-CCCAGCCCCATTTTATTTCA-3′ |
| B2MG | Sense:
5′-TGTGCTCGCGCTACTCTCTC-3′ |
| Antisense:
5′-CCATTCTCTGCTGGATGACG-3′ |
Preparation of cell lysates and western
blot analysis
Cell lysis and western blotting were performed as
previously described (12). In
brief, cells were lysed in RIPA buffer [10 mM Tris-HCl (pH 7.5), 1%
Nonidet P-40, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate
(SDS), 150 mM NaCl and 1 mM EDTA] containing 200 μM
phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 1 μg/ml
leupeptin, 1 μg/ml pepstatin, 2 mM sodium orthovanadate, 10 mM
sodium fluoride and 2.5 mM β-glycerophosphate. A total of 20 μg of
each cell lysate was subjected to SDS-polyacrylamide gel
electrophoresis (10%), and transferred to a polyvinylidene fluoride
(PVDF) membrane. After blocking with 1% bovine serum albumin, the
membrane was incubated with primary antibodies overnight at 4°C.
After repeated washing, the membrane was incubated with the
indicated secondary horseradish peroxidase-conjugated antibody for
2 h at room temperature. Immunoreactive bands were detected using
the ECL or ECL Plus reagent (GE Healthcare, Buckinghamshire,
UK).
Xenografting
All animals were maintained in a specific
pathogen-free environment. The Mie University’s Committee on Animal
Investigation approved the experimental protocol. As previously
described, 50×104 cells of the LNCaP and LNCaP-F10 lines
were prepared in 50 μl of neutralized type 1 rat tail collagen gel
(13). The cells were grafted into
the sub-renal capsule of 8-week-old adult homozygous athymic
cluster of differentiation-1 male nude mice (CLEA Japan, Inc.,
Tokyo, Japan) (13). The mice were
sacrificed and the grafts were harvested at 4 weeks post-grafting.
The grafts were fixed in a 10% formalin neutral buffer solution
(Wako Pure Chemical Industries, Osaka, Japan) for hematoxylin and
eosin (H&E) and regular immunohistochemical staining.
Immunohistochemistry
Sections (3 μm) were cut from the representative
paraffin-embedded samples. For immunohistochemistry, the sections
were deparaffinized in Histo-Clear (National Diagnostics, Atlanta,
GA, USA) and rehydrated in a graded series of ethanol
concentrations. Endogenous peroxidase activity was blocked by 0.3%
hydrogen peroxide in methanol for 20 min. After extensive washing
in tap water, antigen retrieval was performed using 10 mM sodium
citrate buffer (pH 6.0) for AR and antigen unmasking solution
(Vector Laboratories, Burlingame, CA, USA) for PSA and E-cadherin
immunostaining. After rinsing in phosphate-buffered saline (PBS),
the sections were incubated with the appropriate normal serum for
at least 3 h at room temperature to block nonspecific binding. The
sections were then incubated with anti-AR, anti-PSA, and
anti-E-cadherin antibodies at 4°C overnight. After incubation with
the primary antibody, sections were incubated with the appropriate
biotinylated secondary anti-mouse, rabbit, or rat immunoglobulin
diluted with PBS for 30 min at room temperature. The
antigen-antibody reaction was visualized with the Vectastain
avidin-biotin complex (ABC) kit (Vector Laboratories) using
3,3′-diaminobenzidine tetrahydrochloride (DAB) as a substrate. The
sections were counterstained with hematoxylin and examined by light
microscopy.
Statistical analysis
Statistical significance was assessed by two-way
ANOVA followed by Bonferroni test or Student’s t-test using the
PRISM4 software (Graphpad Software, San Diego, CA, USA). P<0.05
was considered to indicate a statistically significant result.
Results
Growth of LNCaP and LNCaP-F10 cells in a
normal or low-pH/low-nutrient environment
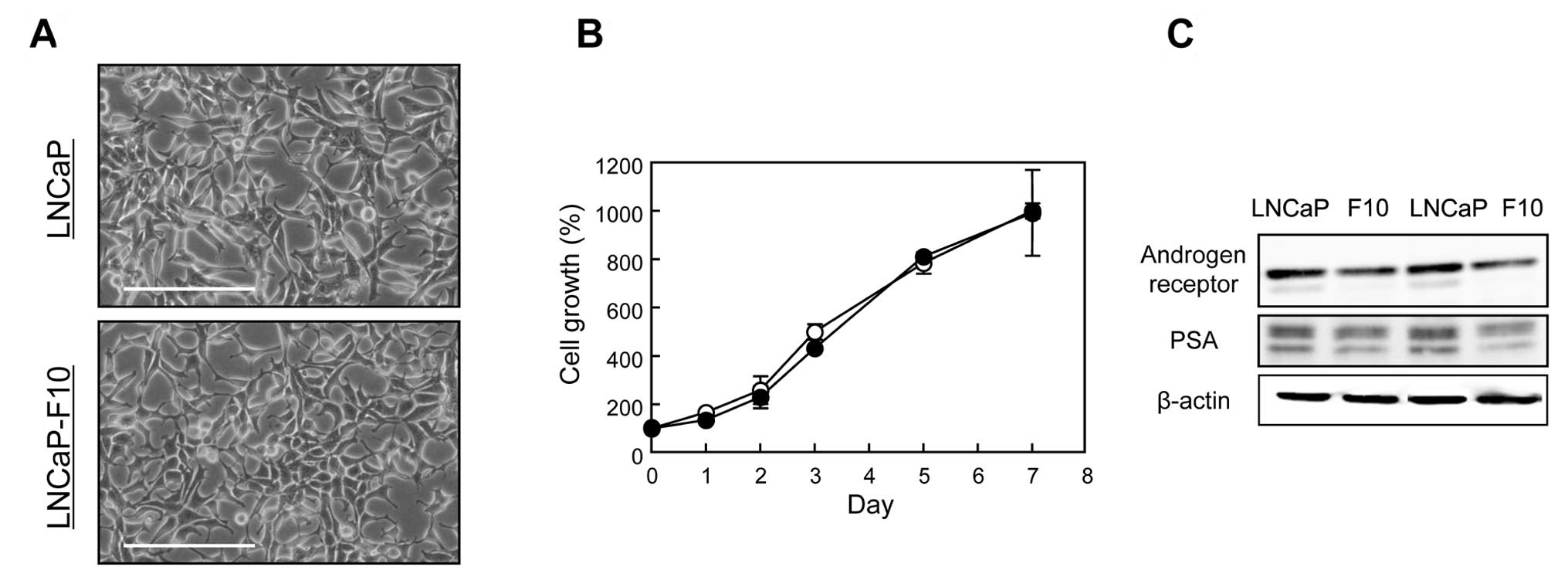
LNCaP-F10 and LNCaP cells grown in a normal medium
(RPMI-1640 + 10% FBS, pH 7.4) showed similar morphology and growth
rates (Fig. 1A and B). AR and PSA
were expressed at lower levels in LNCaP-F10 cells than the levels
in LNCaP cells (Fig. 1C).
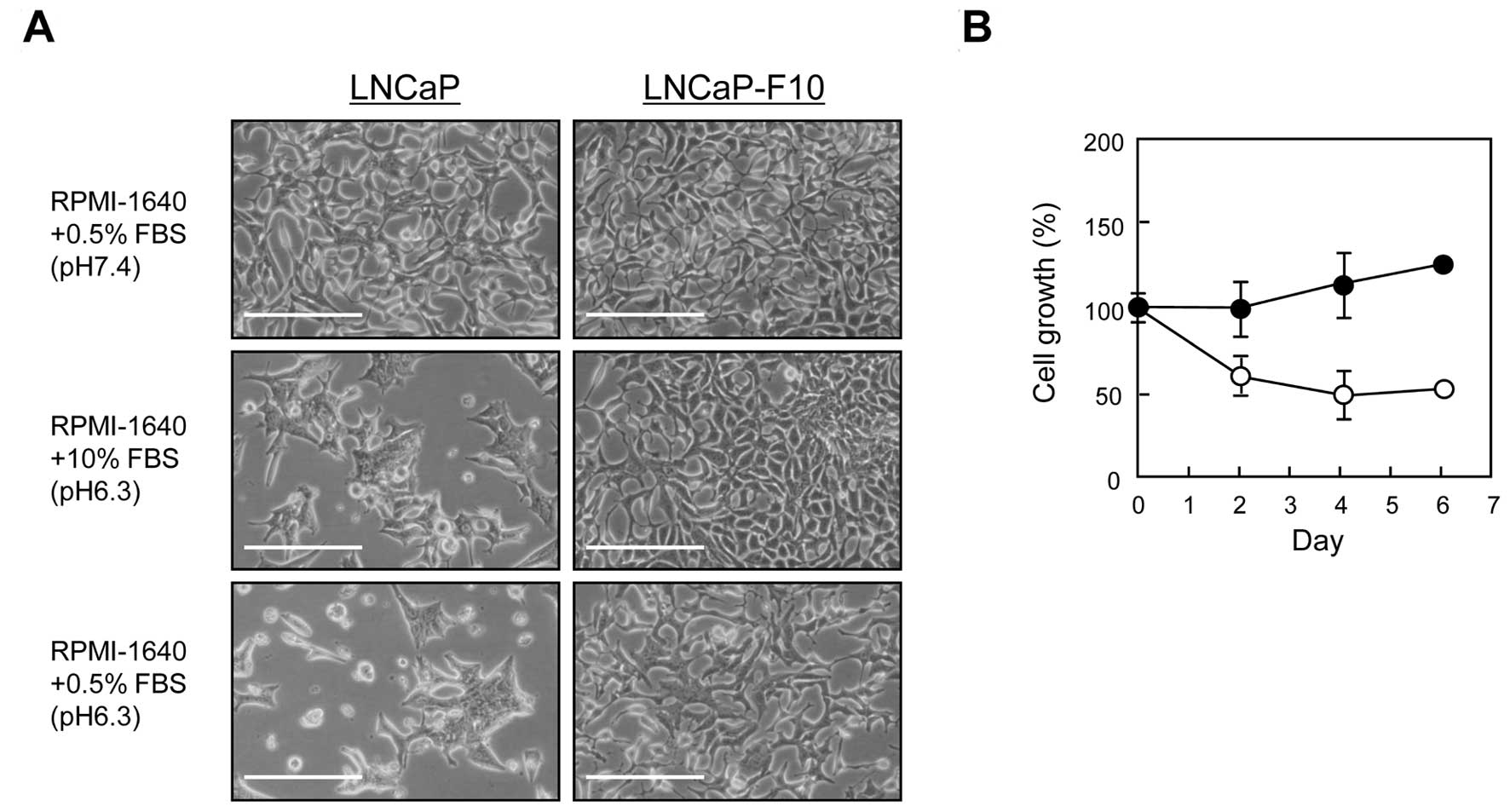
Incubation in a low-pH/low-nutrient medium (RPMI-1640 + 0.5% FBS +
10 mM PIPES, pH 6.3) caused swelling and cell death in LNCaP cells,
whereas LNCaP-F10 cells showed reduced growth rates but no cell
death (Fig. 2A and B).
Intracellular pH of LNCaP and LNCaP-F10
cells in the low-pH/low-nutrient environment
To determine whether LNCaP-F10 cells maintain a
normal intracellular pH in an acidic extracellular environment, the
intracellular pH of cells incubated in the low-pH/low-nutrient
medium was measured for 1 day. No significant differences in the
intracellular pH were found between the 2 cell lines; cells reached
an intracellular pH of 6.6 when incubated in the
low-pH/low-nutrient medium (data not shown).
Death of LNCaP cells in the
low-pH/low-nutrient environment
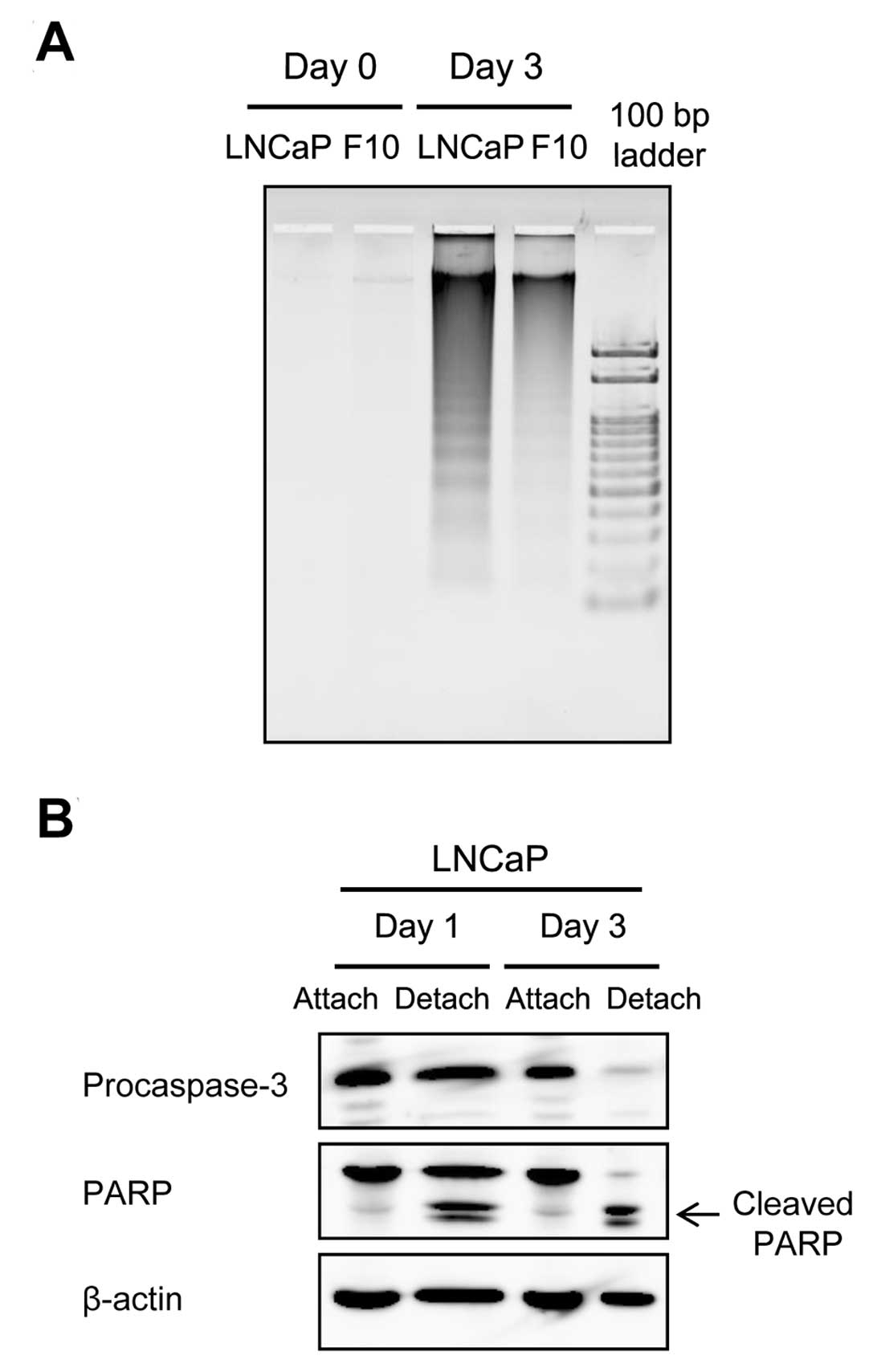
We analyzed DNA fragmentation, which is a hallmark
of apoptosis, to determine whether the death of LNCaP cells in the
low-pH/low-nutrient medium was due to apoptosis. As shown in
Fig. 3A, DNA fragmentation was
detected in LNCaP cells detached after incubation in the
low-pH/low-nutrient medium for 3 days. Furthermore, cleavage of
procaspase-3 and PARP was observed in these cells, confirming that
cell death was caused by apoptosis (Fig. 3B). These results suggested that
LNCaP-F10 cells may be resistant to apoptosis induced by
extracellular environmental stress, i.e., low pH and low
nutrition.
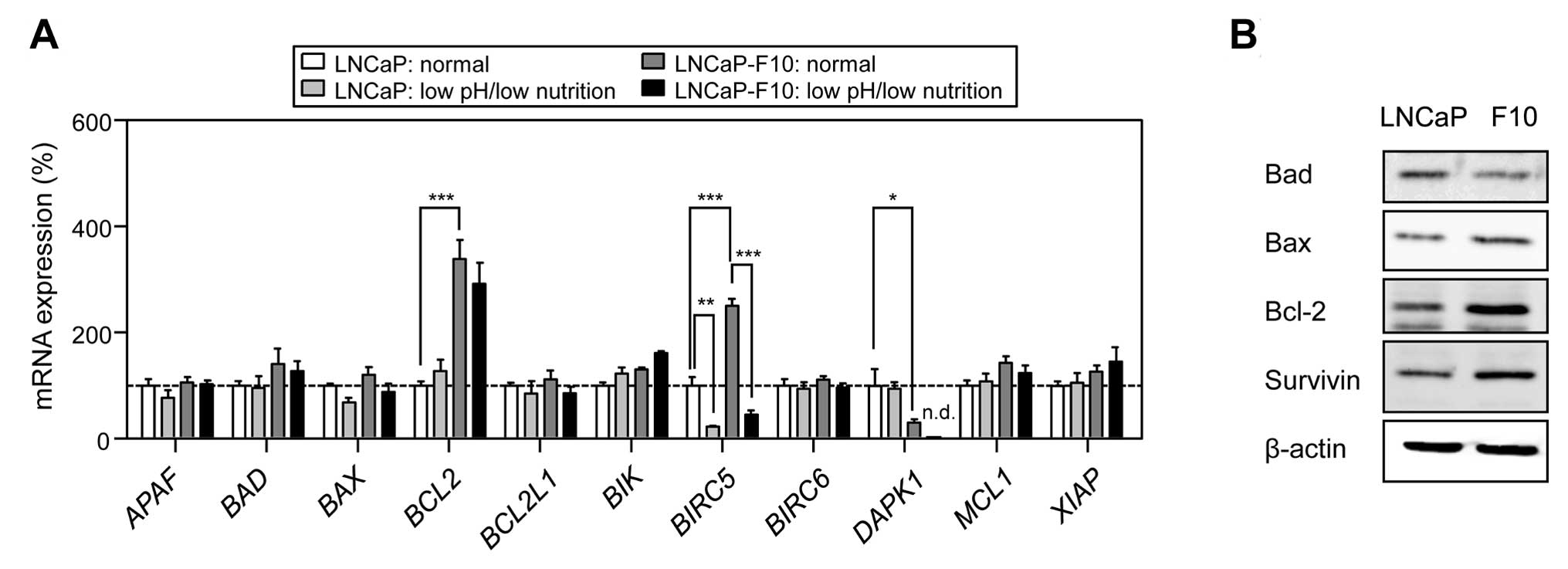
We then determined the expression levels of
apoptosis-related genes in LNCaP and LNCaP-F10 cells incubated in
normal or low-pH/low-nutrient media. When grown in a normal medium,
the expression levels of BCL2 and BIRC5 were
significantly higher in the LNCaP-F10 cells compared to LNCaP cells
(Fig. 4A). DAPK1 expression
in LNCaP-F10 cells was markedly lower compared to LNCaP cells.
BIRC5 expression was decreased in cells incubated in the
low-pH/low-nutrient medium in the 2 cell lines. As shown in
Fig. 4B, the expression of the
bcl-2 and survivin proteins (encoded by BCL2 and
BIRC5, respectively) was also markedly higher in LNCaP-F10
cells compared to that in the LNCaP cells.
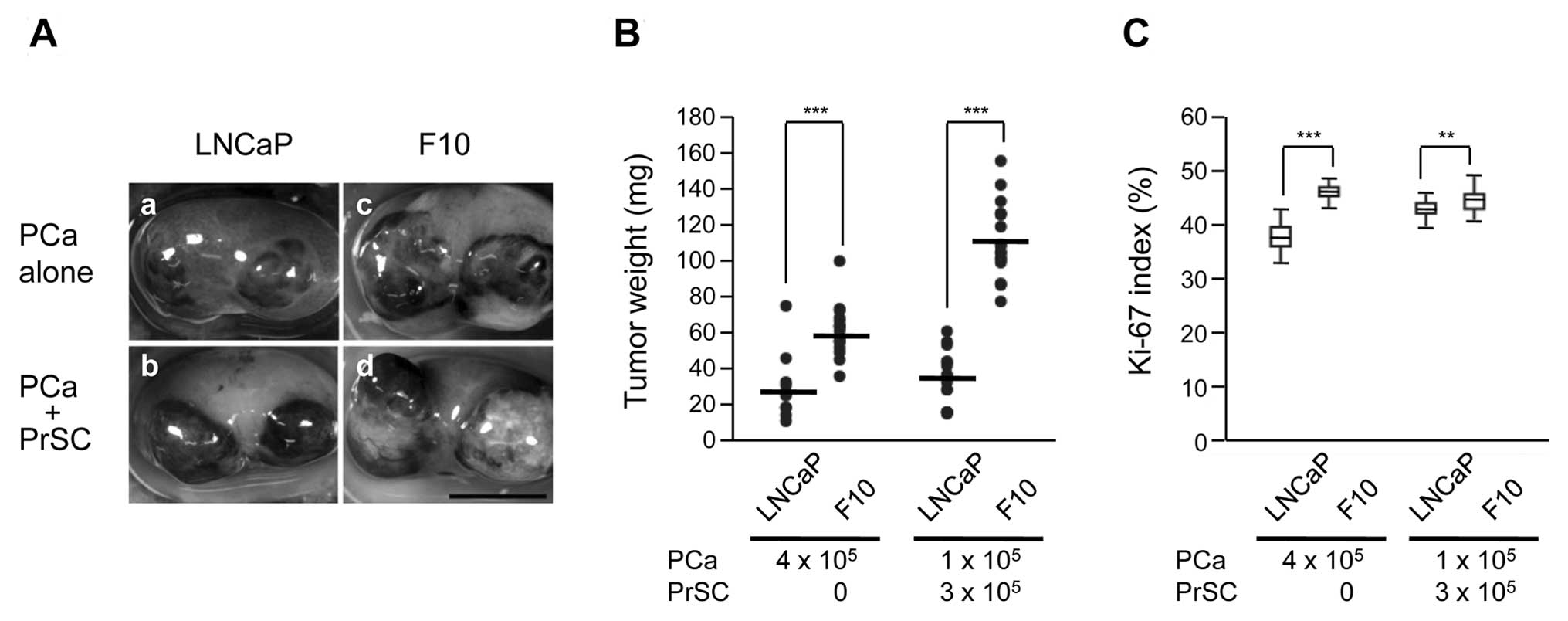
Tumorigenic characteristics of LNCaP and
LNCaP-F10 cells in vivo
The tumorigenicity of LNCaP-F10 cells was compared
to that of the parental LNCaP cell line. When the cells were
implanted into the renal subcapsular space, the weight of LNCaP-F10
tumors with or without prostate stromal cell (PrSC) stimulation was
significantly greater compared to that of the parental LNCaP tumors
(Fig. 5A and B). Cell proliferation
(Ki-67 labeling index) in LNCaP-F10-derived tumors was also
increased compared to that of the parental LNCaP tumors (Fig. 5C).
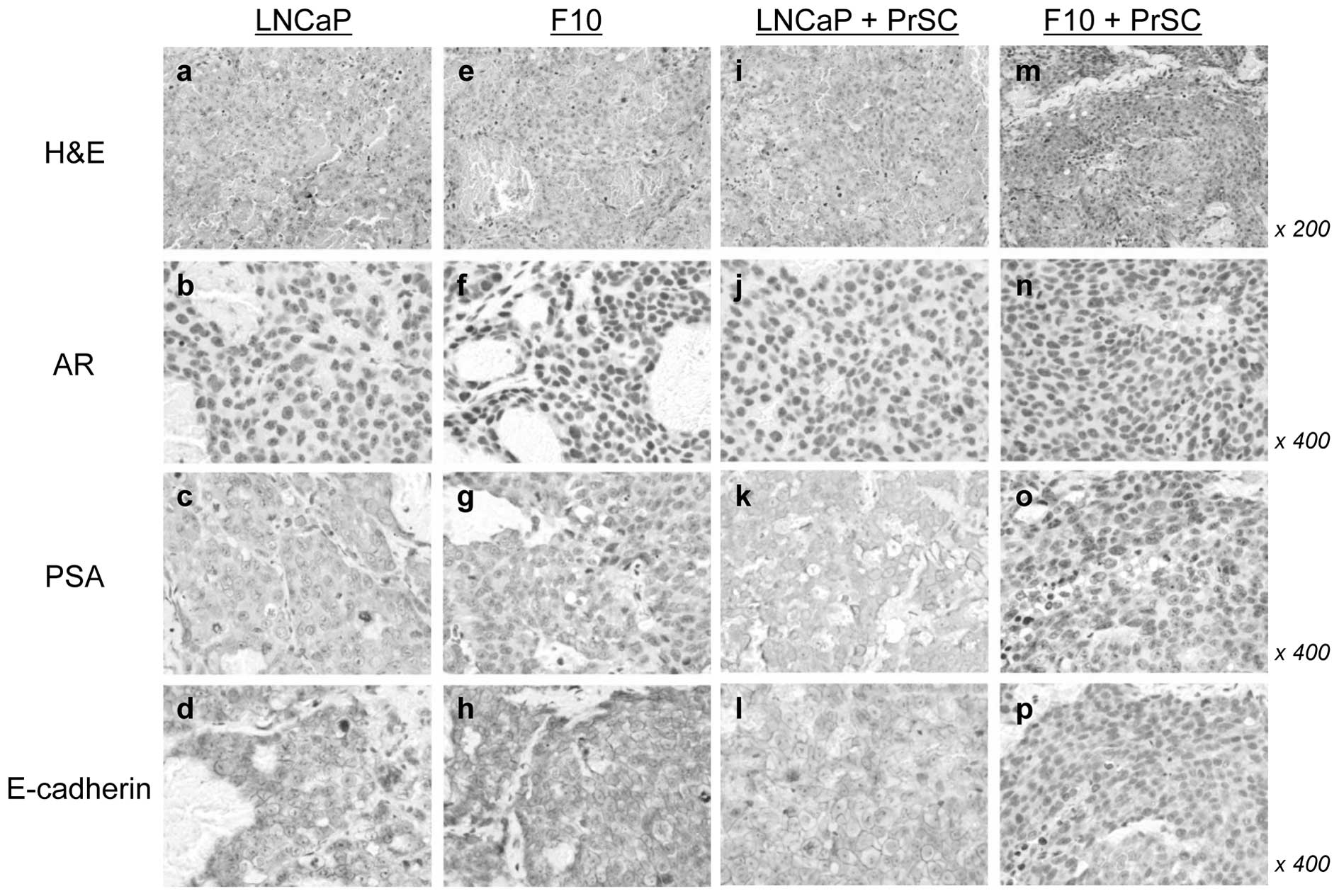
H&E staining showed that LNCaP-F10 + PrSC tumors
had fewer blood spaces than LNCaP + PrSC tumors (Fig. 6i and m). No significant differences
in AR, PSA, and E-cadherin expression levels were found between the
2 tumors without PrSC (Fig. 6b–d and
f–h). Of note, the number of AR-positive cells was
significantly decreased and PSA was undetectable in the LNCaP-F10 +
PrSC tumors (Fig. 6n and o). In
addition, the membrane localization of E-cadherin was decreased in
the LNCaP-F10 + PrSC tumors (Fig.
6p).
Discussion
In the present study, LNCaP-F10 cells, isolated from
prostate cancer LNCaP cells by limiting dilution, were observed to
survive in a low-pH/low-nutrient condition, whereas parental LNCaP
cells demonstrated significant cell death. Cell death of LNCaP
cells occurred in a low-pH/high-nutrient medium (pH 6.3/10% FBS),
but not in a neutral-pH/low-nutrient medium (pH 7.4/0.5% FBS),
suggesting that an acidic extracellular environment was the main
factor inducing cell death (Fig.
2A). Moreover, dead cells were not observed after incubation in
a medium containing 10 mM PIPES at pH 7.4 compared to pH 6.3,
indicating that the cell death was not caused by PIPES toxicity
(data not shown). Previous studies have demonstrated that an acidic
environment leads to p53-dependent and caspase-mediated apoptosis
(14,15). Since LNCaP cells express wild-type
p53, the apoptosis observed in this study may be mediated by p53
activation under an acidic environment. Although the detailed
mechanisms underlying the resistance of LNCaP-F10 cells to
apoptosis remain unclear, the possibility of p53 dysfunction in
LNCaP-F10 cells should be considered. We are currently
investigating the differences in the p53 function between parental
LNCaP cells and the LNCaP-F10 subline.
Our results showed differentially expressed genes
between LNCaP and LNCaP-F10 cells, including BCL2, BIRC5 and
DAPK1, which are involved in apoptosis. Bcl-2 is a key
anti-apoptotic factor and its mRNA and protein expression levels
were significantly higher in the LNCaP-F10 cells compared to that
of the LNCaP cells. The high level of bcl-2 expression in LNCaP-F10
cells suggested that this cell line may be resistant to cell death
induction. However, no significant differences in cell viability
were observed between LNCaP and LNCaP-F10 cells treated with
chemotherapeutic drugs such as etoposide, paclitaxel or docetaxel
(data not shown). These results indicate that LNCaP-F10 cells
exhibit specific resistance to the low pH/low nutrient stress
rather than nonspecific resistance. Survivin (BIRC5), an
anti-apoptosis regulator, was upregulated in the LNCaP-F10 cells
compared to LNCaP cells, and the expression was suppressed by the
acidic environment. The mechanisms underlying the upregulation of
survivin and its physiological implications remain unclear.
However, p53 has been shown to regulate survivin expression
negatively in human cancer cells, suggesting the involvement of p53
in the regulation of apoptosis by the acidic environment (16).
In our xenograft model, LNCaP-F10 tumors were
significantly larger compared to those of the parental LNCaP cell
line even without stromal stimulation. We previously showed that
tumors derived from the LNCaP sublines, i.e., LNCaP-E9 and AIDL,
induced by stromal cells were larger than LNCaP tumors, whereas in
the absence of stromal cell stimulation, LNCaP and LNCaP subline
tumors did not show a significant size difference (13,17).
These results suggest that LNCaP-F10 cells have a more aggressive
potential than LNCaP cells. In the present study, the pro-apoptotic
factor DAPK1 was expressed at significantly lower levels in
LNCaP-F10 cells compared to LNCaP cells. DAPK1 is a known
tumor suppressor; its downregulation is strongly correlated with
tumor recurrence and metastasis, and its expression is often absent
in cancer tissues (18–20). In addition, bcl-2 and survivin have
been reported to be critical for the tumorigenicity of cancer
cells, including prostate cancer cells (21,22).
The differential expression of these genes in LNCaP-F10 cells may
be associated with the differences in the tumorigenicity of these
cells observed in our in vivo experiments, and in the
sensitivity to low-pH/low-nutrient-induced apoptosis determined
in vitro.
In summary, LNCaP-F10 cells were found to be
resistant to a low-pH/low-nutrient environment in vitro and
to exhibit a more malignant phenotype compared to LNCaP cells in
vivo. BCL2, BIRC5 and DAPK1 were differentially
expressed in LNCaP-F10 cells compared to LNCaP cells, suggesting
that these factors may be involved in the unique features of
LNCaP-F10 cells. A tumor-aggressive phenotype has been associated
with the overexpression, mutation or deletion of specific oncogenic
products (23,24); therefore, further identification of
genes differentially expressed in LNCaP and LNCaP-F10 cells would
be beneficial in designing efficient therapeutic strategies.
Acknowledgements
This study was supported by a Grant-in-Aid for Young
Scientists (no. 23791768) from the Ministry of Education, Culture,
Sports, Science, and Technology of Japan.
References
|
1
|
Kizaka-Kondoh S, Inoue M, Harada H and
Hiraoka M: Tumor hypoxia: a target for selective cancer therapy.
Cancer Sci. 94:1021–1028. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stewart GD, Ross JA, McLaren DB, Parker
CC, Habib FK and Riddick AC: The relevance of a hypoxic tumour
microenvironment in prostate cancer. BJU Int. 105:8–13. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Movsas B, Chapman JD, Horwitz EM, et al:
Hypoxic regions exist in human prostate carcinoma. Urology.
53:11–18. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thalmann GN, Anezinis PE, Chang SM, et al:
Androgen-independent cancer progression and bone metastasis in the
LNCaP model of human prostate cancer. Cancer Res. 54:2577–2581.
1994.PubMed/NCBI
|
|
5
|
Culig Z, Hoffmann J, Erdel M, et al:
Switch from antagonist to agonist of the androgen receptor
bicalutamide is associated with prostate tumour progression in a
new model system. Br J Cancer. 81:242–251. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Onishi T, Yamakawa K, Franco OE, et al:
Mitogen-activated protein kinase pathway is involved in alpha6
integrin gene expression in androgen-independent prostate cancer
cells: role of proximal Sp1 consensus sequence. Biochim Biophys
Acta. 1538:218–227. 2001. View Article : Google Scholar
|
|
7
|
Hobisch A, Ramoner R, Fuchs D, et al:
Prostate cancer cells (LNCaP) generated after long-term interleukin
6 (IL-6) treatment express IL-6 and acquire an IL-6 partially
resistant phenotype. Clin Cancer Res. 7:2941–2948. 2001.
|
|
8
|
Kawada M, Inoue H, Usami I, et al:
Establishment of a highly tumorigenic LNCaP cell line having
inflammatory cytokine resistance. Cancer Lett. 242:46–52. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Butterworth KT, McCarthy HO, Devlin A, et
al: Hypoxia selects for androgen independent LNCaP cells with a
more malignant geno- and phenotype. Int J Cancer. 123:760–768.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iguchi K, Ishii K, Nakano T, et al:
Isolation and characterization of LNCaP sublines differing in
hormone sensitivity. J Androl. 28:670–678. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iguchi K, Usui S, Ishida R and Hirano K:
Imidazole-induced cell death, associated with intracellular
acidification, caspase-3 activation, DFF-45 cleavage, but not
oligonucleosomal DNA fragmentation. Apoptosis. 7:519–525. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Otsuka T, Iguchi K, Fukami K, et al:
Androgen receptor W741C and T877A mutations in AIDL cells, an
androgen-independent subline of prostate cancer LNCaP cells. Tumour
Biol. 32:1097–1102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishii K, Imamura T, Iguchi K, et al:
Evidence that androgen-independent stromal growth factor signals
promote androgen-insensitive prostate cancer cell growth in vivo.
Endocr Relat Cancer. 16:415–428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park HJ, Lyons JC, Ohtsubo T and Song CW:
Acidic environment causes apoptosis by increasing caspase activity.
Br J Cancer. 80:1892–1897. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Williams AC, Collard TJ and Paraskeva C:
An acidic environment leads to p53 dependent induction of apoptosis
in human adenoma and carcinoma cell lines: implications for clonal
selection during colorectal carcinogenesis. Oncogene. 18:3199–3204.
1999. View Article : Google Scholar
|
|
16
|
Chang GC, Yu CT, Tsai CH, et al: An
epidermal growth factor inhibitor, gefitinib, induces apoptosis
through a p53-dependent upregulation of pro-apoptotic molecules and
downregulation of anti-apoptotic molecules in human lung
adenocarcinoma A549 cells. Eur J Pharmacol. 600:37–44. 2008.
View Article : Google Scholar
|
|
17
|
Kanai M, Ishii K, Kanda H, et al:
Improvement in predicting tumorigenic phenotype of
androgen-insensitive human LNCaP prostatic cancer cell subline in
recombination with rat urogenital sinus mesenchyme. Cancer Sci.
99:2435–2443. 2008. View Article : Google Scholar
|
|
18
|
Bialik S and Kimchi A: The
death-associated protein kinases: structure, function, and beyond.
Annu Rev Biochem. 75:189–210. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee TH, Chen CH, Suizu F, et al:
Death-associated protein kinase 1 phosphorylates Pin1 and inhibits
its prolyl isomerase activity and cellular function. Mol Cell.
42:147–159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raveh T and Kimchi A: DAP kinase-a
proapoptotic gene that functions as a tumor suppressor. Exp Cell
Res. 264:185–192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kajiwara T, Takeuchi T, Ueki T, et al:
Effect of Bcl-2 overexpression in human prostate cancer cells in
vitro and in vivo. Int J Urol. 6:520–525. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen J, Liu J, Long Y, et al: Knockdown of
survivin expression by siRNAs enhances chemosensitivity of prostate
cancer cells and attenuates its tumorigenicity. Acta Biochim
Biophys Sin. 41:223–230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baldi E, Bonaccorsi L and Forti G:
Androgen receptor: good guy or bad guy in prostate cancer invasion?
Endocrinology. 144:1653–1655. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mimeault M and Batra SK: Novel therapies
against aggressive and recurrent epithelial cancers by molecular
targeting tumor- and metastasis-initiating cells and their
progenies. Anticancer Agents Med Chem. 10:137–151. 2010. View Article : Google Scholar
|