Introduction
Small cell lung cancer (SCLC) is an aggressive
malignancy representing ~13% of 220,000 new lung cancer cases
projected for 2009 (1–3). Despite moderate progress achieved in
the past two decades, the survival rate of SCLC patients is still
very poor (3,4). The poor prognosis of SCLC patients is
due to its high metastatic potential and chemoresistance upon
relapse. Cancer stem cells (CSCs) are thought to be responsible for
tumor initiation, therapy resistance, progression, relapse, and
metastasis. Therefore, targeting of CSCs may be one of approaches
to eradicate cancerous tumors early. However, lack of understanding
the regulatory mechanisms underlying CSCs of SCLC is an obstacle in
the progress of targeting of CSCs to improve SCLC therapy.
microRNAs (miRNAs) are small noncoding regulatory
RNAs that regulate the translation of mRNAs by inhibiting ribosome
function, decapping the 5′Cap structure, deadenylating the poly(A)
tail, and degrading the target mRNA (5). They can regulate a variety of cell
functions, including stem cell maintenance and differentiation, and
play roles in controlling cancer initiation and progression
(6,7). Previous studies have shown an aberrant
miRNA expression in a various types of cancer stem cells, and
changes in specific miRNAs have been associated with stem cell
self-renewal and differentiation (6,8,9). For
example, the expression of let-7 reduces tumor sphere formation in
breast cancer cell lines and inhibits tumorigenicity in an in
vivo xenograft tumor assay (8).
miR-130b promotes CD133+ liver tumor-initiating cell
growth and self-renewal (9). The
downregulation of miR-200 family miRNAs suggests that breast cancer
stem cells and normal stem cells share common molecular mechanisms
that regulate stem cell functions, such as self-renewal,
proliferation, and the epithelial-mesenchymal transition (6). These findings demonstrate that miRNAs
are critical regulators of self-renewal and differentiation.
Currently, miRNA dysregulation in many CSCs,
including breast (6,8), prostate (10), non-small cell lung cancer (11), liver (9), pancreas (12), and glioblastoma (13) stem cells have been identified.
However, the dysregulation of miRNAs in CSCs of SCLC remains
unclear. Understanding the miRNA expression of CSCs may provide
insight into the origin of and new therapeutics for SCLC.
Therefore, we looked at whether differences in miRNA expression
might distinguish CSCs from their more differentiated progeny.
Previous work from our laboratory showed that tumor
spheres from SCLC cell lines were characterized by stem-like
properties, and stem-like cells were enriched after consecutively
passaging in a defined serum-free medium. Furthermore, the
stem-like cell population may be enriched in cells expressing
urokinase plasminogen activator receptor (uPAR) cell surface marker
(14). In this study, we undertook
a systematic comparison of the miRNAs in SCLC stem/progenitor cell
populations and their differentiated progeny. Our results showed
that SCLC stem/progenitor cell populations expressed a limited set
of miRNAs compared to differentiated cells. Moreover, miR-27a was
consistently downregulated in stem/progenitor cells of all 3 SCLC
cell lines. By antagonizing miR-27a in parental cells, we found
that miR-27a regulates the key feature of SCLC stem cells - self
renewal in vitro. Therefore, lack of miR-27a may be critical
to maintaining a stem cell function in SCLC.
Materials and methods
Cell lines and cell culture
SCLC cell lines H446, H209, and H69 were obtained
from the American Type Culture Collection (ATCC). Cells were
cultured in RPMI-1640 (Gibco-BRL, Grand Island, NY, USA) medium
supplemented with 10% fetal bovine serum, 100 U/ml penicillin and
100 μg/ml streptomycin in humidified air at 37°C with 5%
CO2. The procedure for obtaining tumor sphere cell
culture has been described in our recent study (14).
microRNA microarray
Microarray assay was performed using a service
provider (LC Sciences). The assay started from 5 μg total RNA,
which was size-fractionated using a YM-100 Microcon centrifugal
filter (from Millipore). The small RNAs (<300 nt) were isolated
and labeled with Cy5 fluorescent dyes. Hybridization was performed
overnight on a μParaflo microfluidic chip containing 1212 mature
human microRNA (miRNA) probes (Sanger miRBase, release 16.0). Each
detection probe consisted of a chemically modified nucleotide
coding segment complementary to target miRNA (from miRBase,
http://microrna.sanger.ac.uk/sequences/). After
hybridization detection used fluorescence labeling using
tag-specific Cy5 dyes. Hybridization images were collected using a
laser scanner (GenePix 4000B, Molecular Device) and digitized using
Array-Pro image analysis software (Media Cybernetics). Raw data
were normalized the signals using a LOESS filter (15) (locally-weighted regression) after
first subtracting the background. A Student’s t-test was performed
to analyze the statistical significance of the signal differences
between the fourth passage sphere cells and parental cells, and
differentially detected signals were those with P<0.01.
qRT-PCR analysis of miRNAs
expression
The total RNA (2 μg) extracted separately from cell
samples was used to generate cDNA by using SuperScript II reverse
transcriptase (Invitrogen) with special stem-loop primer for miRNA.
The primers for the analysis of miRNAs expression were designed
according to Chen et al(16). Human small nuclear U6 RNA was
amplified as an internal control. Real-time qPCR was performed on a
Bio-Rad iQ5 real-time PCR detection system using SYBR®
Premix Ex Taq™ (Takara, RR041A). The PCR reaction contained 1 μl RT
product, 10 μl 2× SYBR Premix Ex Taq, 1 μl forward primer and 1 μl
reverse primer (5 μmol/l each), and nuclease-free water in a final
volume of 20 μl. Standard PCR samples were analyzed with a Bio-Rad
iQ5 thermal cycler. Melting curves were generated for each
real-time RT-PCR to verify the specificity of each PCR reaction.
All quantitative PCR reactions were performed in triplicate.
Expression levels of each miRNA were evaluated using the
comparative threshold cycle (Ct) method as normalized to that of U6
(2−ΔCt). Relative fold changes were calculated by the
equation 2−ΔΔCt.
Cell transfection
Transfection was performed with Lipofectamine 2000
Reagent (Invitrogen) following the manufacturer’s protocol.
Briefly, cells were trypsinized, counted and seeded in plates the
day before transfection to ensure a suitable cell confluence on the
day of transfection. miR-27a inhibitor or scrambled
oligonucleotides as the negative control (GenePharma, Shanghai,
China) were transfected into H446 cells at a final concentration of
40 nM in antibiotic free Opti-MEM medium (Invitrogen). Transfection
efficiency was monitored by FAM-oligonucleotides.
MTT assay
H446 cells were seeded in 96-well plate at 3000
cells per well in 100 μl cell culture medium and incubated at 37°C
for 24 h. Then, they were transfected with miR-27a inhibitor or
scrambled oligonucleotides performed as mentioned above. After
incubation for 72 h, the cells were incubated with 10 μl MTT (at a
final concentration of 0.5 mg/ml) at 37°C for 4 h. The medium was
removed and the precipitated formazan was dissolved in 100 μl DMSO.
After shaking for 10 min, the absorbance at 570 nm was detected
using TECAN GENios Pro (BioTek Instruments). This procedure was
repeated at 24, 48 and 72 h after transfection.
Tumor sphere formation assay and colony
formation assay
After transfection with 40 nM of miR-27a inhibitor,
40 nM of scrambled oligonucleotides, cells were trypsinized,
counted, and seeded for tumor sphere formation assay and colony
formation assay in 24-well ultra low attachment plates at 500 cells
per well. The tumor sphere culture performed as mentioned above.
The total number of tumor spheres was counted after 5–14 days of
culture. During colony growth, the culture medium was replaced
every 3 days. The colony was counted only if it contained >50
cells, and the number of colonies was counted the 5th day after
seeding. Each experiment was carried out in triplicate.
Immunofluorescence
For immunofluorescence studies, H446 cells were
treated with miR-27a inhibitor or scrambled oligonucleotides for 72
h in 96-well plate and then fixed with 4% paraformaldehyde for 15
min at 37°C, permeabilized with 0.1% Triton X-100/PBS for 15 min at
room temperature and then incubated with the following monoclonal
antibodies: uPAR (American Diagnostica), pan-cytokeratin (CK,
Zymed). FITC-conjugated rabbit anti-mouse IgG (Dako) was used as
the secondary antibody. Cell nuclei were counterstained with DAPI.
Images were recorded on a Nikon Eclipse Ti.
Statistical analysis
All data were expressed as the mean ± standard
deviation (SD). Statistical analysis was performed with Student’s
t-test and P<0.05 was considered statistically significant.
Results
miRNA expression profiles of
sphere-forming cells and parental cells
Previous work from our laboratory showed that tumor
spheres from SCLC cell lines were characterized by stem-like
properties, and stem-like cells were enriched after consecutively
passaging (14). We further studied
the underlying mechanism that regulates these cells. As miRNAs are
critical regulators of self-renewal and differentiation in both
normal and cancer stem cells, we compared the miRNA expression
profiles of the fourth passage sphere cells and parental cells from
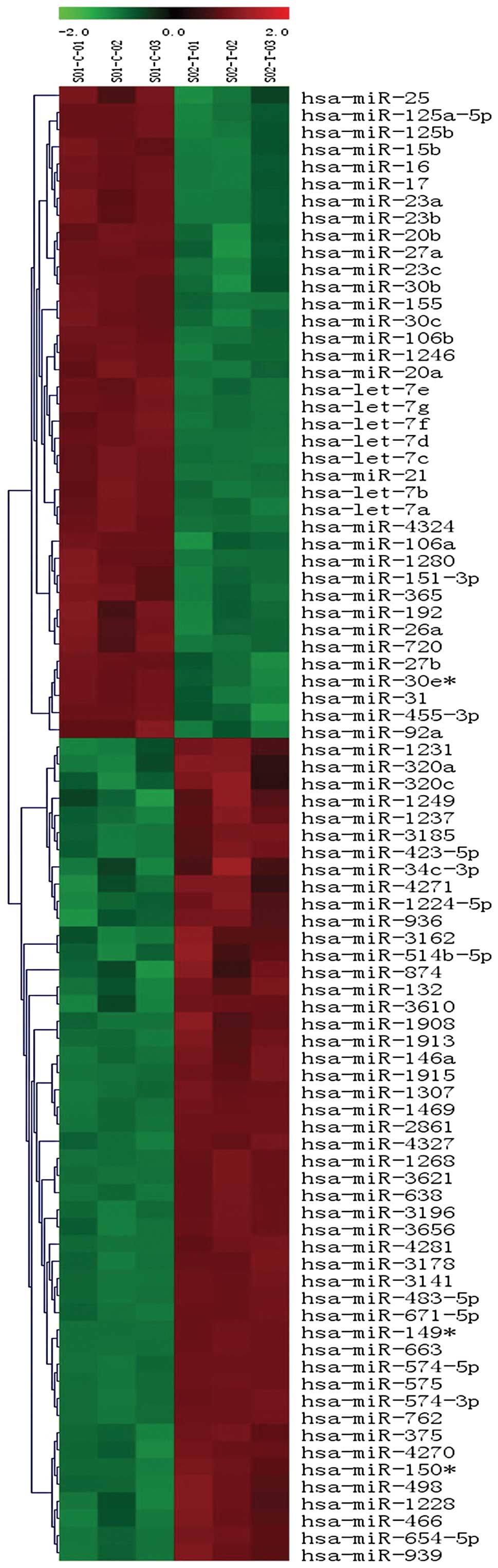
H446 using miRNA array containing 1212 mature miRNAs. In the array
86 miRNAs were found to be differentially expressed (P<0.01),
including 48 upregulated miRNAs and 38 downregulated miRNAs in the
fourth passage sphere cells (Fig.
1). Among 86 differentially expressed miRNAs, 18 of the 48
upregulated miRNAs and 20 of the 38 downregulated miRNAs (Table I) showed at least a 4-fold changes
in the fourth passage sphere cells relative to parental cells. Of
the upregulated miRNAs, several miRNAs were not previously
implicated in lung cancer, such as miR-1469, miR-2861, miR-3141,
miR-3621, miR-663, miR-762, miR-763 and miR-4281.
 | Table ISignificantly differentially expressed
miRNAs in the fourth passage sphere cells relative to parental
cells (P<0.01, fold changes ≥4). |
Table I
Significantly differentially expressed
miRNAs in the fourth passage sphere cells relative to parental
cells (P<0.01, fold changes ≥4).
| Downregulated
miRNAs | Upregulated
miRNAs |
|---|
|
|
|---|
| miRNA name | Fold changes | Location | miRNA name | Fold changes | Location |
|---|
| hsa-miR-125a-5p | 4.2 | 19q13.41 | hsa-miR-3185 | 8.49 | 17 |
| hsa-miR-125b | 6.88 | 11q24.1 | hsa-miR-146a | 7.07 | 5q34 |
| | 21q21.1 | hsa-miR-1915 | 12.23 | 10 |
| hsa-miR-15b | 5.43 | 3q25.33 | hsa-miR-1469 | 76.24 | 15q26.2 |
| hsa-miR-16 | 8.85 | 13q14.2 | hsa-miR-2861 | 54.21 | 9 |
| | 3q25.33 | hsa-miR-3621 | 31.91 | 9 |
| hsa-miR-23b | 4.39 | 9q22.32 | hsa-miR-4281 | 20.16 | 5 |
| hsa-miR-20b | 28.77 | Xq26.2 | hsa-miR-3656 | 4.09 | 11 |
| hsa-miR-27a | 12.48 | 19p13.13 | hsa-miR-3178 | 17.28 | 16 |
| hsa-miR-23c | 14.08 | X | hsa-miR-3141 | 41.71 | 5 |
| hsa-miR-30b | 25.21 | 8q24.22 | hsa-miR-483-5p | 15.11 | 11p15.5 |
| hsa-miR-30c | 12.5 | 1p34.2 |
hsa-miR-149* | 112.56 | 2q37.3 |
| hsa-miR-1246 | 6.86 | 2q31.1 | hsa-miR-663 | 31.53 | 20p11.1 |
| hsa-miR-20a | 4.77 | 13q31.3 | hsa-miR-574-5p | 16.78 | 4 |
| hsa-let-7g | 7.17 | 3p21.1 | hsa-miR-575 | 531.92 | 4q21.22 |
| hsa-let-7f | 8.96 | 9q22.32 | hsa-miR-574-3p | 13.31 | 4 |
| | Xp11.22 | hsa-miR-762 | 28.68 | 16 |
| hsa-let-7d | 6.49 | 9q22.32 | hsa-miR-763a | 28.68 | - |
| hsa-let-7c | 6.36 | 21q21.1 | | | |
| hsa-miR-21 | 8.97 | 17q23.1 | | | |
| hsa-let-7a | 7.1 | 9q22.32 | | | |
| | 11q24.1 | | | |
| | 22q13.31 | | | |
| hsa-miR-1280 | 6.04 | 3 | | | |
| hsa-miR-27b | 10.29 | 9q22.32 | | | |
Validation of miRNA expression profiles
with qRT-PCR
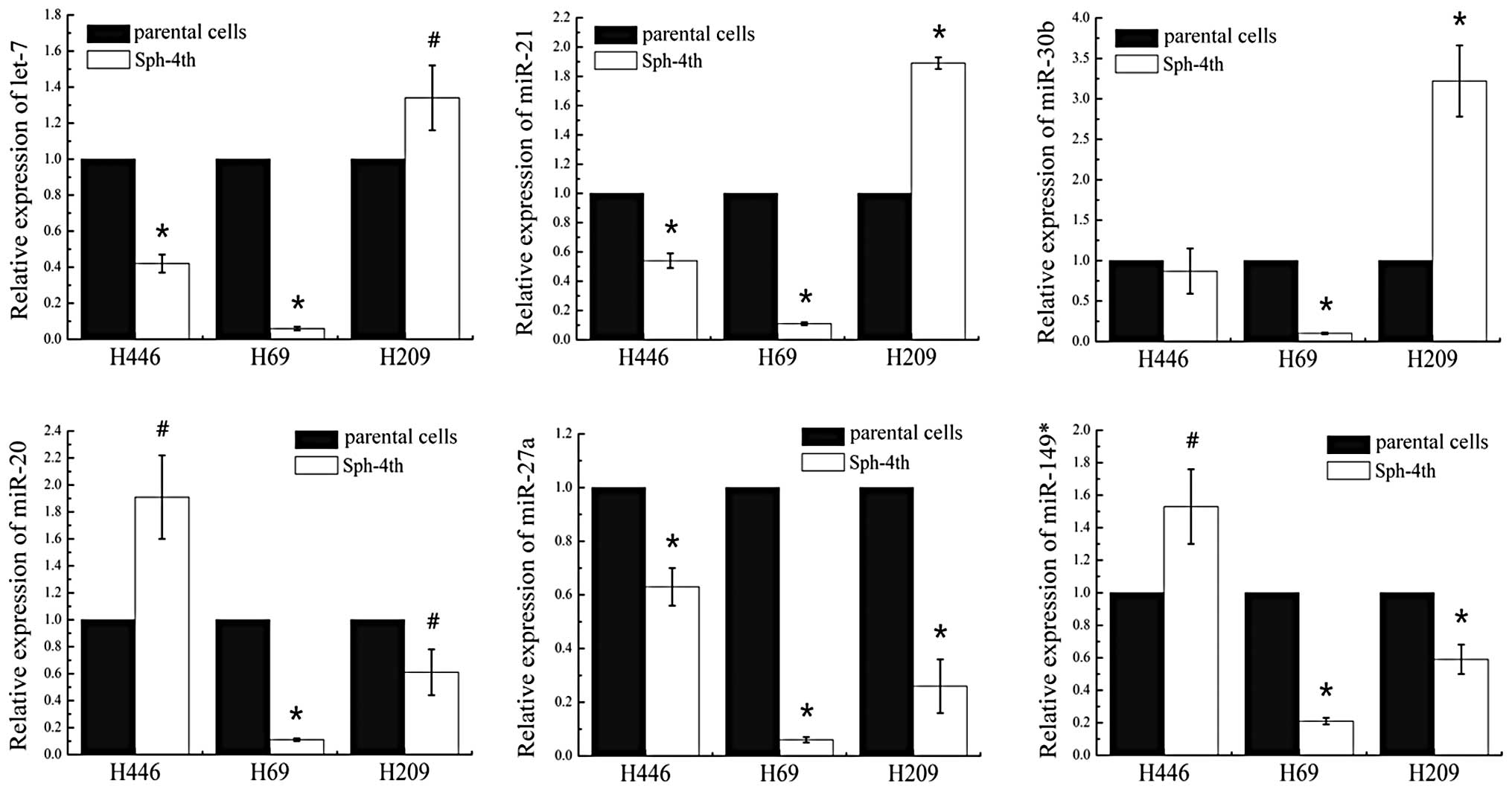
To confirm the miRNA microarray data and filter out
the common miRNAs, 6 tumor-related miRNAs with same seed sequence
and significant fold changes (Table
I; downregulated: let-7, miR-20, 21, 27a and 30b; upregulated:
miR-149*) were selected for validation in 3 sets of human SCLC
fourth passage sphere cells and parental cells by qRT-PCR. For each
sample, the expression values were normalized to the U6 gene. As
Fig. 2 shown, qRT-PCR analysis
confirmed that these 6 miRNAs were indeed differentially expressed.
The trend of expression change of five miRNAs (let-7, miR-20, 21,
30b and 149*) were aberrant in the fourth passage sphere cells of 3
cell lines. let-7, miR-21 and miR-30b were downregulated in H446
and H69, but upregulated in H209. Similarly, miR-149* and miR-20
were downregulated in H69 and H209, but upregulated in H446.
However, miR-27a was consistently downregulated in the fourth
passage sphere cells of all 3 cell lines (Fig. 2). Therefore, we focused on miR-27a
in the following studies.
Reduced miR-27a increases tumor sphere
formation and colony formation
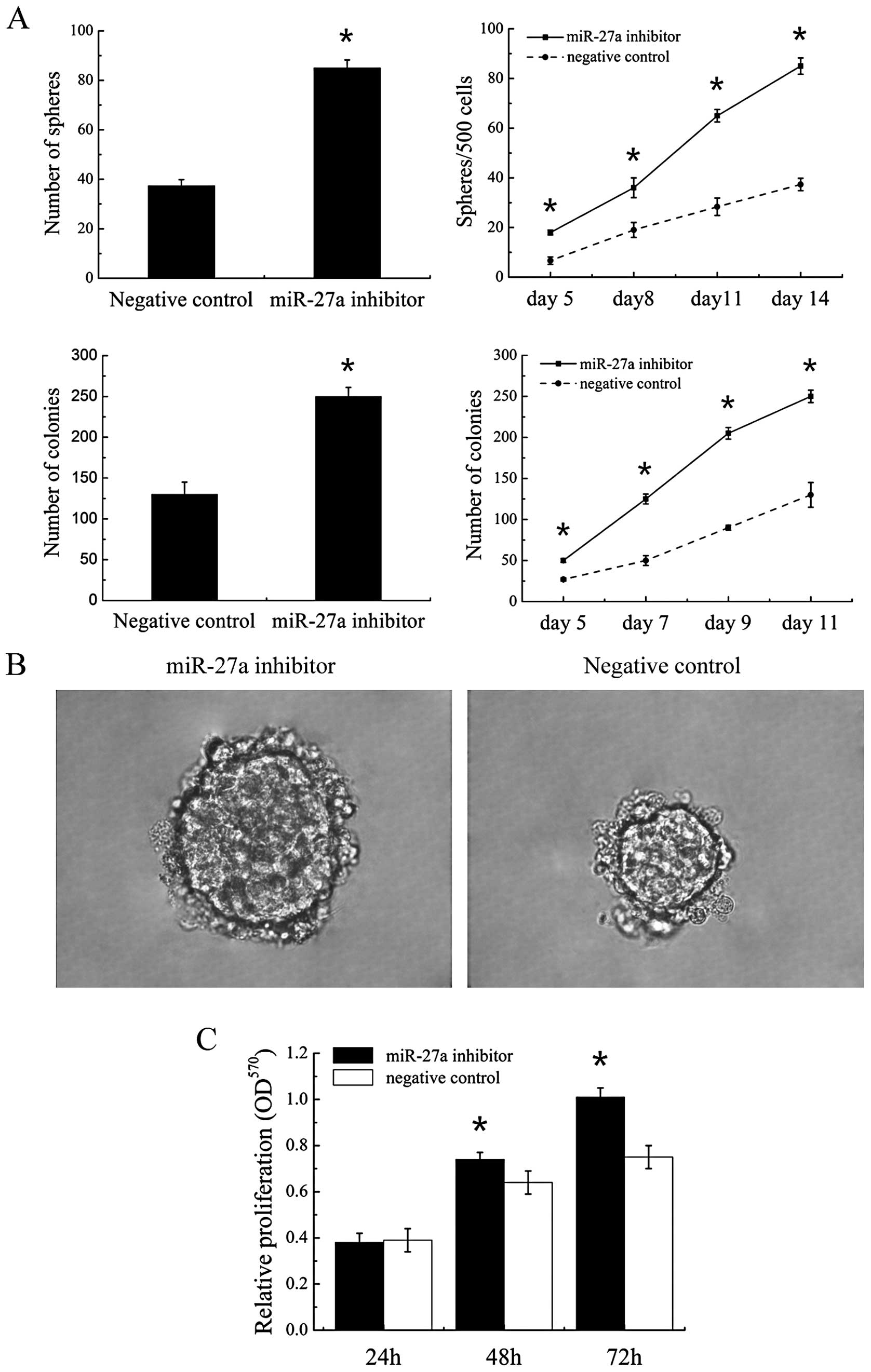
To evaluate miR-27a function, we used a
‘loss-of-function’ approach by transfecting H446 with a specific
inhibitor to miR-27a. Tumor sphere formation assay and colony
formation assay was performed to investigate the effect of miR-27a
on self renewal. The cells transfected with miR-27a inhibitor
formed significantly more tumor spheres and more clones than those
transfected with negative control (Fig.
3A). Meanwhile, the tumor spheres that formed were larger in
transfected with miR-27a-inhibitor cells than negative control
cells (Fig. 3B). Therefore miR-27a
inhibitor enhanced self-renewal capacity of parental cells in
vitro.
Reduced miR-27a increases cell
proliferation but inhibits differentiation
The effect of miR-27a on the proliferation and
differentiation was investigated in cell line. At different time
points post-transfection, the vitality of cells was tested by the
MTT assay. The cells transfected with miR-27a inhibitor showed
enhanced proliferation at 48 and 72 h compared with cells
transfected with negative control (Fig.
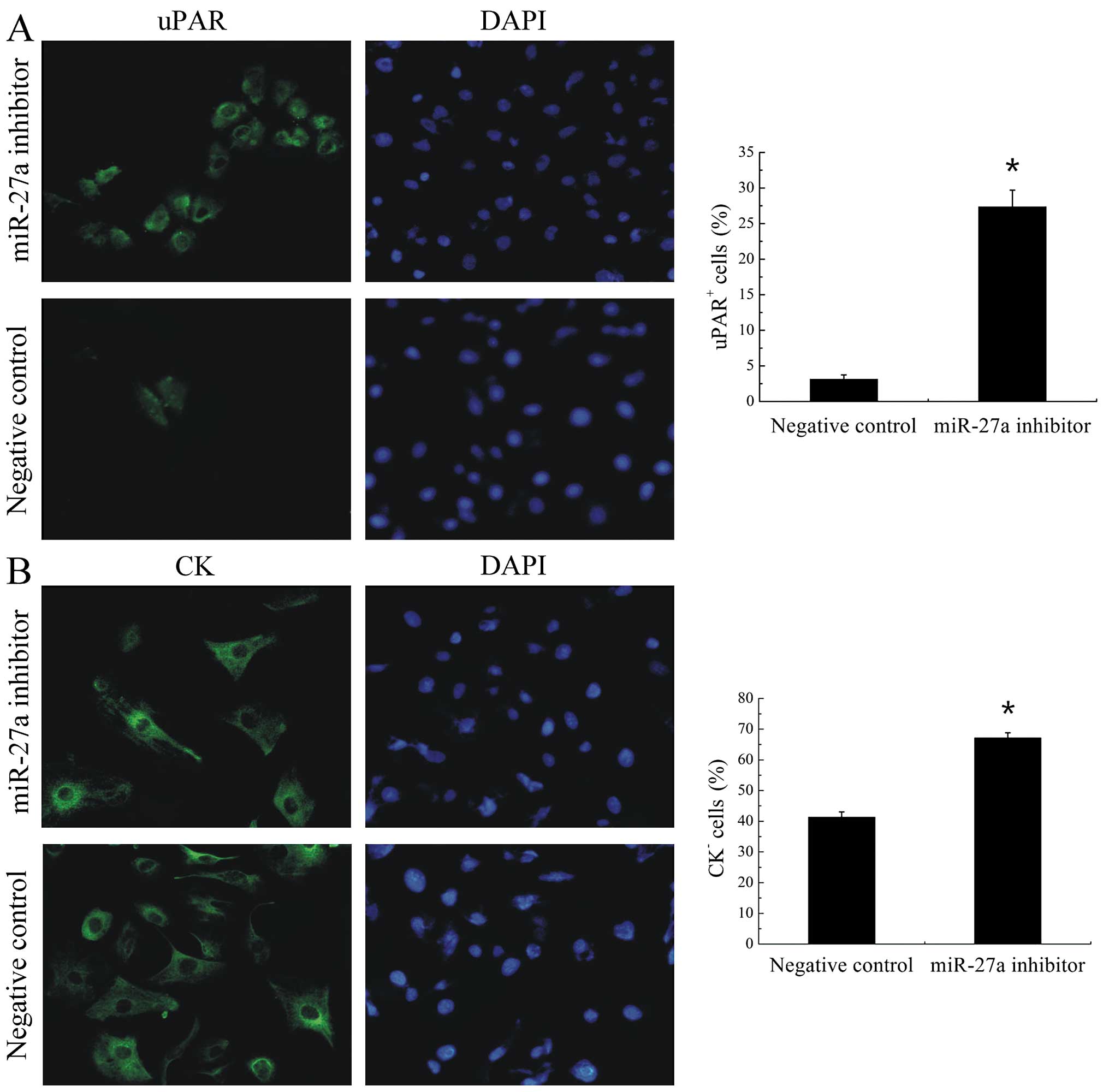
3C). Another hallmarker of stem-like cells is their
undifferentiated state. Previous work from our laboratory showed
that stem-like cell population may be enriched in the
uPAR+ fraction of tumor cells in the H446 cell line. To
test the effect of miR-27a on the differentiation, we further
detected the expression of uPAR and cell differentiation marker CK
in cell line. miR-27a inhibitor significantly (P<0.001)
increased the proportion of uPAR+ cells from 3.13±0.6%
to 27.37±2.32% (Fig. 4A) and the
proportion of CK− cells from 41.33±1.72% to 67.17±1.66%
(Fig. 4B) compared with negative
control inhibitor in H446 cells. Therefore, low miR-27a enhanced
the proliferative potential and increased the population of
undifferentiated cells.
Prediction of miRNA targets
miRNAs participate in various physiological and
pathological processes by regulating the expression of their target
genes. Identification of miRNAs target genes is very important to
understand the mechanisms involved in the malignant biological
behavior of SCLC. So the candidate miRNA and some miRNAs with same
seed sequence were picked for target prediction and analyzed with
TargetScan 5.1 (http://www.targetscan.org/). By analysis of the
database, human genes which were known to be involved in cell
proliferation, apoptosis and cell cycle were selected from the
TargetScan 5.1. Then, those genes predicted to be targets of the
miRNAs were chosen and listed in Table
II. All data might provide the foundation for further analysis
of mechanisms of self-renewal and tumor growth in SCLC.
 | Table IITarget prediction of miRNAs. |
Table II
Target prediction of miRNAs.
| Putative targets of
miRNAs and their functions |
|---|
|
|
|---|
| miRNA name | Apoptosis and
proliferation | Cell cycle |
|---|
| miR-27a | BCLAF1, BAK1 | CCNG1 |
| miR-27b | BAK1 | CCNG1 |
| let-7ga | DAPK | - |
| let-7fa | - | CCND2 |
| let-7d | TP53, FASLG,
TNFSF9, BCL2L11, TP53INP1 | CCNF, E2F2,
TP53 |
| miR-30b | BCLAF1, KIPK5,
CASP3, TANK | CCNE2, CCNK, TFDP1,
E2F7 |
| miR-30ca | - | CCNT2 |
| miR-23a | APAF1, SATB1, FAS,
BNIP2, CASP7 | CCNH, CCND1,
RBL2 |
| miR-23ba | - | CCNT2 |
Discussion
There is growing evidence that cancers are initiated
and maintained by cancer stem cells. miRNAs regulate both normal
stem cells and CSCs (6,8,17–19),
and alterations in the expression of miRNA genes contribute to the
pathogenesis of most human malignancies (10,11,20).
miRNA dysregulation in many CSCs has been identified, but how
miRNAs regulate CSCs in SCLC remains unclear. In this study, we
obtained a large number of self-renewing cells to study changes in
miRNA expression compared to parental cells. The microarray data
showed that 86 miRNAs were differentially expressed in the fourth
passage sphere cells versus parental cells of H446 cell line. The
upregulation and downregulation miRNAs in sphere-forming cells
suggest a role for small RNA molecules in the maintenance of stem
cell properties in SCLC cells. Among them, we selected 6
tumor-related miRNAs for validation and further screened the miRNAs
of steady expression trend in 3 SCLC cell lines by qRT-PCR.
However, only miR-27a was consistently downregulated in the fourth
passage sphere cells of all 3 cell lines and others were
downregulated in two cell lines.
Furthermore, we used the ‘loss-of-funtion’ approach
to study the role of miR-27a in the H446 SCLC cell line. Our
results showed that antagonization of miR-27a in parental cells
enhanced proliferation, self renewal, and increased the proportion
of undifferentiated cells. Downregulation of miR-27a enhanced the
stem-like properties of SCLC cells in vitro, and converted
less malignant cells into highly malignant cells. These results
suggest that reduced miR-27a may be critical to maintaining a stem
cell function in SCLC. This statement is also supported by previous
studies. For example, miR-27a plays important roles in normal stem
cells and poorly differentiated cells. In the hematopoietic cells,
reduced miR-27a can antagonize the colony-stimulating
factor-mediated granulocyte differentiation (21). In muscle cells, miR-27 can modify
muscle stem cell behavior by regulating the Pax3, which
downregulation ensures rapid and robust entry into the myogenic
differentiation program (22).
miR-27a was also reported to be downregulated in SP cells of human
lung adenocarcinoma A-549 cell (11). Therefore, we speculate that
correction of miR-27a expression represents a good candidate
strategy for targeted therapy.
Among the other five miRNAs, let-7 and miR-21 are
well studied in other cancers. In breast cancer, there are absent
expression of let-7 in cancer stem cells. Moreover, lack of let-7
is required for self-renewal in vitro and tumorigenicity
in vivo(8). This family is
also downregulated in SP cells of human lung adenocarcinoma A-549
cell(11) and associated with poor
lung cancer prognosis (23).
However, in our study, let-7 shows different expression patterns in
SCLC cell lines, which is downregulated in the fourth passage
sphere cells of H446 and H69 cell lines, yet upregulated in H209
cell line. These results indicate that the expression level of
let-7 is different in different classes of cancer, and the
underlying association mechanisms also might be different.
miR-21 was reported by many studies and was
overexpressed in a wide variety of cancers, such as breast
(24,25), colorectal (26), gastric (27), and non-small cell lung cancer
(28), which causally linked to
poor prognosis, invasion and metastases (25,26,28–32).
But in mouse embryonic stem cells, miR-21 suppressed the
self-renewal (33) and increased
dramatically upon differentiation (34). In our study, miR-21 was
downregulated in tumor sphere cells of two cell lines. The same
miRNA exhibiting diverse expression and functions in different
cellular contexts, may depend on the specific microenvironment and
the combination of its direct target genes, but the specific
function of miRNAs need to be further verified through function
studies.
In summary, these results established global
expression profiles for SCLC stem/progenitor cells and
differentiated cells, which provide new knowledge on gene
regulation during the SCLC developmental progress, and are valuable
in further studies of SCLC tumorigenic mechanism. Moreover, reduced
miR-27a is a potential intrinsic property of SCLC stem/progenitor
cells. Downregulation of miR-27a enhanced the cell stem-like
properties of SCLC cells in vitro and may be critical in
maintaining the stem cell function in SCLC.
Acknowledgements
The current work was supported by the Research
Programme of the Applied Basic and Cutting-edge Technologies of
Tianjin under contract No. 09JCZDJC20300.
References
|
1
|
Jackman DM and Johnson BE: Small-cell lung
cancer. Lancet. 366:1385–1396. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
3
|
Govindan R, Page N, Morgensztern D, et al:
Changing epidemiology of small-cell lung cancer in the United
States over the last 30 years: analysis of the surveillance,
epidemiologic, and end results database. J Clin Oncol.
24:4539–4544. 2006.PubMed/NCBI
|
|
4
|
Lally BE, Urbanic JJ, Blackstock AW,
Miller AA and Perry MC: Small cell lung cancer: have we made any
progress over the last 25 years? Oncologist. 12:1096–1104.
2007.PubMed/NCBI
|
|
5
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimono Y, Zabala M, Cho RW, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
8
|
Yu F, Yao H, Zhu P, et al: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma S, Tang KH, Chan YP, et al: miR-130b
promotes CD133(+) liver tumor-initiating cell growth and
self-renewal via tumor protein 53-induced nuclear protein 1. Cell
Stem Cell. 7:694–707. 2010.PubMed/NCBI
|
|
10
|
Liu C, Kelnar K, Liu B, et al: The
microRNA miR-34a inhibits prostate cancer stem cells and metastasis
by directly repressing CD44. Nat Med. 17:211–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hua S, Xiaotao X, Renhua G, et al: Reduced
miR-31 and let-7 maintain the balance between differentiation and
quiescence in lung cancer stem-like side population cells. Biomed
Pharmacother. 66:89–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji Q, Hao X, Zhang M, et al: MicroRNA
miR-34 inhibits human pancreatic cancer tumor-initiating cells.
PLoS One. 4:e68162009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gal H, Pandi G, Kanner AA, et al: MIR-451
and Imatinib mesylate inhibit tumor growth of glioblastoma stem
cells. Biochem Biophys Res Commun. 376:86–90. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu X, Wang Z, Li Y, Miao Y, Ren Y and
Luan Y: Characterization of sphere-forming cells with stem-like
properties from the small cell lung cancer cell line H446. Cancer
Lett. 323:161–170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Melton C, Judson RL and Blelloch R:
Opposing microRNA families regulate self-renewal in mouse embryonic
stem cells. Nature. 463:621–626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu C and Tang DG: MicroRNA regulation of
cancer stem cells. Cancer Res. 71:5950–5954. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
DeSano JT and Xu L: MicroRNA regulation of
cancer stem cells and therapeutic implications. AAPS J. 11:682–692.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kohu K, Feng J, Iwama A and Satake M:
MicroRNA-27 enhances differentiation of myeloblasts into
granulocytes by post-transcriptionally downregulating Runx1. Br J
Haematol. 145:412–423. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Buckingham M, Crist CG, Montarras D, et
al: Muscle stem cell behavior is modified by microRNA-27 regulation
of Pax3 expression. Proc Natl Acad Sci USA. 106:13383–13387. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takamizawa J, Konishi H, Yanagisawa K, et
al: Reduced expression of the let-7 microRNAs in human lung cancers
in association with shortened postoperative survival. Cancer Res.
64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang GL, Zhang XH, Guo GL, et al:
Clinical significance of miR-21 expression in breast cancer:
SYBR-Green I-based real-time RT-PCR study of invasive ductal
carcinoma. Oncol Rep. 21:673–679. 2009.PubMed/NCBI
|
|
25
|
Shao JY, Yan LX, Huang XF, et al: MicroRNA
miR-21 overexpression in human breast cancer is associated with
advanced clinical stage, lymph node metastasis and patient poor
prognosis. RNA. 14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY and
Yan M: microRNA-21 promotes tumor proliferation and invasion in
gastric cancer by targeting PTEN. Oncol Rep. 27:1019–1026.
2012.PubMed/NCBI
|
|
28
|
Gao W, Shen H, Liu L, Xu J and Shu Y:
MiR-21 overexpression in human primary squamous cell lung carcinoma
is associated with poor patient prognosis. J Cancer Res Clin Oncol.
137:557–566. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hiyoshi Y, Kamohara H, Karashima R, et al:
MicroRNA-21 regulates the proliferation and invasion in esophageal
squamous cell carcinoma. Clin Cancer Res. 15:1915–1922. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Horiuchi A, Iinuma H, Akahane T, Shimada R
and Watanabe T: Prognostic significance of PDCD4 expression and
association with microRNA-21 in each Dukes’ stage of colorectal
cancer patients. Oncol Rep. 27:1384–1392. 2012.PubMed/NCBI
|
|
32
|
Zhu Q, Wang Z, Hu Y, et al: miR-21
promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback
loop in human hepatocellular carcinoma. Oncol Rep. 27:1660–1668.
2012.PubMed/NCBI
|
|
33
|
Singh SK, Kagalwala MN, Parker-Thornburg
J, Adams H and Majumder S: REST maintains self-renewal and
pluripotency of embryonic stem cells. Nature. 453:223–227. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Houbaviy HB, Murray MF and Sharp PA:
Embryonic stem cell-specific MicroRNAs. Dev Cell. 5:351–358. 2003.
View Article : Google Scholar : PubMed/NCBI
|