Introduction
The oxidored-nitro domain-containing protein 1
(NOR1) gene (also known as OSCP1) is a novel tumor suppressor gene
that was first isolated from a nasopharyngeal carcinoma (NPC)
(1). Human NOR1 expression is
decreased in the HNE1 NPC cell line (1) and tissues when compared with normal
nasopharyngeal epithelial cells (2). Furthermore, mutations in the NOR1
coding region were found in NPC biopsies (1), and a hypermethylated promoter region
of NOR1 was found in NPC biopsies (3), leukemia cell lines, and acute myeloid
leukemia patients (4). Our previous
study has shown that the functional NOR1 promoter is regulated by
heat shock factor 1 (HSF1) and nuclear respiratory factor 1 (NRF1)
and the promoter is located within a CpG island (3). Hypermethylation of this CpG island was
found in NPC tissue samples and cancer cell lines, whereas no
aberrant promoter methylation was detected in non-cancerous
nasopharyngeal tissue samples or normal nasopharyngeal epithelial
cells. Promoter hypermethylation may occur during transcriptional
inactivation of the NOR1 gene in cancer cells and NOR1 may be a
critical tumor suppressor involved in the development of various
cancers.
The human NOR1 gene is located on 1p34.3 and shares
40% homology with nitroreductases from Escherichia coli.
This homology implies that NOR1 may be a novel member of
nitroreductases, a group of FMN- or FAD- and NAD(P)H-dependent
enzymes, which catalyze the reduction of nitro groups in a wide
range of substrates to produce the corresponding hydroxylamine
(5). Since much evidence has shown
that exposure to nitroso compounds such as nitrosamines is a risk
factor for NPC (6–9), NOR1 may play an important role in
chemical carcinogen formation and NPC carcinogenesis due to its
nitrosation function. With a highly specific NOR1 antibody, Xiang
et al demonstrated that NOR1 is predominantly expressed in
the nasopharynx and trachea and is weakly expressed in the central
nervous system (2). These data
explain the selectivity of the potential NOR1 physiological
functions and provide an indispensable marker for the NPC
carcinogenesis process, and the identification or validation of
tissue-specific drug targets may help in the further study of the
selectivity of the organ targets that respond to chemical
carcinogens such as nitroso compounds. However, to date, the tumor
suppression functions of NOR1 have not been completely
elucidated.
In this study, we performed NOR1 expression analysis
in various human cancer and benign tissue specimens and analysis of
the antitumor effects of NOR1 using in vitro cell functional
studies. We demonstrate that NOR1 expression was significantly
decreased in nine types of cancer tissues: ovary, lung, kidney,
vulva, prostate, uterus, cervix, thyroid gland, and testis. The
exogenous NOR1 overexpression results in inhibited proliferation
and colony-forming ability of NPC 6-10B and CCA HeLa cells and
leads to S phase cell cycle arrest.
In addition, NOR1 upregulation enhanced apoptosis
under the stable transfection of pCDNA3.1-myc-his-NOR1. NOR1
overexpression activated caspase 9, released cytochrome c from
mitochondria to cytoplasm, increased Bax and p53 expression, and
decreased Bcl-2 expression, which is involved in the
mitochondria-dependent apoptotic pathway. Furthermore, we also
found that NOR1 is a cytoplasmic protein that is partially
localized to the mitochondria and endoplasmic reticulum. Therefore,
NOR1 is an important tumor suppressor gene that is related to NPC
and CCA and may play antitumor roles by inhibiting proliferation,
preventing colony formation, and promoting the apoptosis of tumor
cells via the mitochondria-dependent apoptotic pathway.
Materials and methods
Reagents
XhoI, BamHI, and primers and probes
were purchased from Takara (Dalian, China). The Cancer Profiling
Array II was purchased from BD Biosciences Clontech (Palo Alto, CA,
USA). The DIG Olignucleotide 3′-tailing labeling kit and
anti-DIG-POD were purchased from Roche (Germany). The RevertAid™
First Strand cDNA synthesis kit and PCR Master mix (2X) were
purchased from Fermentas (Glen Burnie, MD). The RPMI-1640 medium,
fetal bovine serum (FBS), pcDNA3.1-myc-his plasmid, Lipofectamine
2000, TRIzol, MitoTracker Red CMXRos, ER-Tracker Red, and Opti-Mem
I were purchased from Invitrogen (Carlsbad, CA, USA).
Gentamicin-G418, the endoplasmic reticulum isolation kit, and all
other chemicals were obtained from Sigma (St. Louis, MO, USA). The
BCA protein assay and Mitochondria isolation kit were purchased
from Pierce (Rockford, IL).
The ECL system was purchased from Amersham Pharmacia
Biotech (Piscataway, NJ). The antibodies used, obtained from Abcam
(Cambridge, UK), were as follows: anti-His (clone HIS.H8, dilution
1:1500); anti-β-actin (clone mAbcam 8226, dilution 1:2000);
anti-Bax (clone 6A7, dilution 1:1000); anti-Bcl-2 (clone 100/D5,
dilution 1:1000); anti-p53 (clone pAb 240, dilution 1:1000);
anti-NF-κB p65 (dilution 1:1000); anti-NF-κB p50 (dilution 1:1000);
anti-cytochrome c (clone EP1326Y, dilution 1:1000); anti-caspase 9
(dilution 1:1000); HRP-conjugated goat anti-rabbit IgG (dilution
1:3000); and HRP-conjugated goat anti-mouse IgG (dilution
1:3000).
Cell culture and transfection
The HNE1 Human NPC and the HeLa CCA cell lines were
maintained in RPMI-1640 supplemented with 10% FBS in a humidified
incubator with 5% CO2 at 37°C.
For stable transfection, a mammalian expression
system using the pCDNA3.1-myc-his-NOR1 plasmid was created by
inserting the full-length human NOR1 cDNA, which was obtained from
IMAGE Consortium, between the XhoI and BamHI sites of
the pcDNA3.1-myc-his plasmid. The transfection experiment was
performed with a pcDNA3.1-myc-his plasmid containing the
full-length NOR1 cDNA insert or with a pcDNA3.1-myc-his plasmid
containing no insert using Lipofectamine 2000 according to the
protocol of the manufacturer. Gentamicin-G418 (500 μg/ml) was added
for transfectant selection. The stable transfectants were isolated
after 3 weeks using a pooled-cloning technique. Cells that grew to
80% confluence were harvested for experiments.
The transient transfection experiment was performed
with a pGFP-N1-NOR1 vector (a gift from Dr Xinming Nie) containing
the full-length human NOR1 cDNA and a control vector using
Lipofectamine 2000 according to the manufacturer’s protocol. After
transfection for 48 h, the cells were used in subsequent
experiments.
Cancer profiling array
The differential expression of NOR1 mRNA in normal
and cancer tissues from 19 different tissues was determined using
the Cancer Profiling Array II using cDNA from paired cancer and
normal human tissues. This array includes 154 pairs of cDNA blots
prepared from the full thickness of cancer specimens from 19
different tissues, each paired with cDNA from normal tissue from
the same patient. Pre-hybridization, hybridization, and
post-hybridization washings were performed according to the
protocol of manufacturer. A 32P-labeled NOR1-specific
cDNA probe was used for hybridization. The array was then exposed
to a storage phosphor screen (Molecular Dynamics, Amersham
Biosciences) for 36 h at room temperature. A Storm 840
phosphorimager system (Molecular Dynamics, Amersham Biosciences)
and ImageQuant software version 4.1 (Molecular Dynamics, Amersham
Biosciences) were used to quantify the hybridization signals.
Tissue microarrays and in situ
hybridization
The tissue microarrays (TMAs), which were prepared
from NPC and normal tissue specimens collected at the Second
Xiangya Hospital (Changsha, Hunan, China), were constructed in our
laboratory. The antisense sequence of three in situ
hybridization (ISH) nucleotide probes from different regions of the
NOR1 cDNA used were: 5′-ccttcttggagtagagctcttgaggcttga-3′ (325–354
bp); 5′-gaactcccctgcagagagaccaccatata-3′ (594–623 bp);
5′-cataggataactcttctggcctggttagcg-3′ (1126–1155 bp).
The probes were labeled with 11-DIG-dUTP at their 3′
tails. The ISH detection method was performed according to protocol
of the manufacturer. Briefly, the sections were deparaffinized,
rehydrated, digested with 2 μg/ml proteinase K at 37°C for 15 min,
and dehydrated. The slides were incubated with prehybridization
solution at 37°C for two hours and hybridized with the NOR1 probe
at 37°C overnight. Hybridization was detected by incubation with
anti-digoxigenin horseradish peroxidase (anti-DIG-POD) fab
fragments. The color reaction was performed with
3.3′-diaminobenzidine tetrahydrochloride (DAB), and the slides were
counterstained with hematoxylin. A poly d(T) oligo was used as a
control for total RNA preservation. The GAPDH housekeeping gene and
prehybridization solution containing no probe were used as positive
and negative control, respectively. The NOR1 positive hybridization
signals were microscopically scored at ×400 magnification. The
scoring was recorded in accordance with the following staining
proportion and intensity: 0 (negative), 1 (<10% nuclei being
EBER-1 positive), 2 (10–50% positive), or 3 (>50% positive). The
final scores were regarded as negative (0 score) and positive (1–3
score).
RNA extraction and reverse
transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was isolated using TRIzol according to the
manufacturer’s protocol. Total RNA (5 μg) was reverse transcribed
with reverse transcriptase (RevertAid™ First Strand cDNA synthesis
kit) and used for PCR using PCR Master Mix (2X). The primers for
NOR1, GAPDH, and β-actin were: NOR1 5′-ACCTGCACATCCGAGTATCC-3′
(forward) and 5′-CTGGCCAAGAAATTCAGCTC-3′ (reverse); GAPDH
5′-ATGTTCGTCATGGGTGTGAA-3′ (forward) and 5′-TGCTGTAGCCAAATTCGTTG-3′
(reverse); β-actin 5′-CCTCGCCTTTGCTGATCC-3′ (forward) and
5′-GGATCTTCATGAGGTAGTCAGTC-3′ (reverse).
Protein isolation and western
blotting
Cells at 80% confluence were harvested for western
blot analysis. The harvested cells were lysed, and their protein
concentrations were determined using a BCA protein assay. The cell
lysates (50 μg of protein each lane) were separated by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to nitrocellulose membranes. The membranes were blocked
with 5% (v/v) skim milk and probed with primary antibody at 4°C
overnight. After washing, the membranes were incubated with
HRP-conjugated secondary antibody at room temperature for 1 h. The
primary antibodies used were specific for human His, HIF-1α,
β-actin, Bax, Bcl-2, p53, NF-κB p65, NF-κB p50, cytochrome c, and
caspase 9. The bound antibodies were visualized using the ECL
system.
Cell growth assay
Cell proliferation was characterized by a
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT)
assay. Cells (104 cells/well) were incubated with
RPMI-1640 containing 10% FBS in 96-well plates for seven days. MTT
(20 μl of 5 mg/ml) was added daily to each well and incubated at
37°C for 4 h, after which the MTT solution in the medium was
aspirated off. To achieve solubilization of the formazan crystals
formed in viable cells, 150 μl of DMSO was added to each well. Cell
proliferation was determined by measuring the converted formazan at
570 nm using an ELX-800 ELISA plate reader (Bio-Tek Instruments,
Winooski, VT).
Soft agar colony forming assay
For soft agar assays, a bottom layer of 1 ml of the
corresponding culture media containing 0.6% agar and 10% fetal calf
serum was prepared, placed in 35-mm culture dishes, and allowed to
solidify. Cells (2×104) were suspending in 50 μl of
RPMI-1640 containing 10% FBS and G418 (500 μg/ml). Culture medium
(1 ml) containing 0.33% agarose was added to the cell suspension
before seeding the dishes. Triplicates were performed for each cell
type. Cells were incubated at 37°C in a 5% CO2
atmosphere. The dishes were examined twice per week, and the
colonies were manually counted after 2 weeks.
Cell cycle and apoptosis assay
Cell cultures were plated in 10-cm dishes at ~40%
confluence and allowed to grow exponentially. The adherent cells
were collected by trypsinization, pooled with the non-adherent
cells, and washed with phosphate-buffered saline (PBS). The cells
were then fixed in 70% cold ethanol overnight at 4°C. Before
analysis, the cells were adjusted to a final density of
1×106 cells/ml in PBS containing RNase (1 μg/ml) and
stained with 10 μg/ml propidium iodide for 30 min at room
temperature. The multiparameter analysis of 5000 cells was
performed on a FACScan flow cytometer using CellQuest software (BD
Biosciences, San Jose, CA, USA).
Acridine orange (AO)/ethidium bromide
(EB) staining
An AO/EB cocktail (80 μl) containing 1 ml of
RPMI-1640 was added to the culture plate. Fields of stained cells
were selected and focused on using fluorescence microscopy (Nikon
Eclipse E800; Nikon, Tokyo, Japan). Viable cells, which were
stained with only AO, were bright green with intact structure;
early apoptotic cells, which were stained with AO and EB, were
bright green in the nucleus with red-orange chromatin. Late
apoptotic cells, which were stained with both AO and EB, were
red-orange with chromatin condensation (10).
RNA interference (RNAi) treatment
The HIF-1α siRNA oligonucleotides used were:
5′-ATGGAGGGCGCCGGC-3′ (sense) and 5′-GCCGGCGCCCTCCAT-3′
(antisense). On the day of RNAi transfection, the media were
removed from 50% confluent cells cultured in a 12-well plate, and
800 μl of Opti-Mem I was added. Then, HIF-1α siRNA was diluted in
Opti-Mem I to a final volume of 200 μl containing 2 μl of
Lipofectamine 2000 reagent (Invitrogen) and incubated at room
temperature for 25 min before addition to each well. The final
working concentration of HIF-1α siRNA was 100 nM. Twenty-four hours
later, the culture medium was replaced with RPMI-1640 without FBS.
The cells were used for the following assays.
Mitochondria and endoplasmic reticulum
(ER) isolation
Cells were grown to 80–90% confluence and collected
by trypsinization into tubes (4×107 cells per sample).
The mitochondria were isolated using the mitochondria isolation
kit, and the ERs were separated using the Endoplasmic Reticulum
Isolation kit according to the manufacturer’s protocol.
Mitochondria and ER staining
Cells were grown to 70% confluence on a coverslip.
Mitochondria were stained with 50 nM MitoTracker Red CMXRos at 37°C
for 20 min, and the cells were washed twice with PBS and fixed with
freshly prepared 4% formaldehyde at 37°C for another 20 min. The ER
was stained with 1 μM ER-Tracker Red for 30 min at 37°C, and the
cells were washed twice with PBS without fixation. Cell images were
captured with an LSM 510 Zeiss confocal microscope.
Northern blotting
Cells at 80% culture confluence were harvested, the
total RNA was extracted, and 40 μg per sample was dotted onto a
nitrocellulose membrane. The membranes were then prehybridized with
100 μg/ml salmon sperm DNA in a solution containing 6X SSC, 0.5%
SDS, 5X Denhardt’s at 68°C for 3 h. The membranes were hybridized
with 32P-labeled PCR-amplified NOR1- or 28S-specific
probes at 68°C for 24 h. The membranes were then washed twice with
2X SSC/0.1% SDS at room temperature for 15 min followed by a 30 min
wash with 0.1X SSC/0.1% SDS. Subsequently, the filters were exposed
to X-ray film (Kodak BioMax) for 48 h at −80°C. The relative
abundance of the individual mRNAs in each clone was normalized to
the density of the 28S rRNA dot.
Immune electron microscopy
Immune electron microscopy was carried out as
described previously (10). The
cells were fixed with 2% paraformaldehyde, subsequently dehydrated,
embedded, and polymerized under UV light at room temperature for
48–72 h. Ultrathin sections were cut and collected on
pioloform-coated nickel grids to continue the immunolabeling
experiments. The primary antibody was anti-His antibody and the
diameter of colloidal gold immune complexes (1:10, Boster
Bio-Engineering Ltd. Wuhan, P.R. China) was 10 nm. The dried
sections were then stained with sodium acetate and observed under a
transmission electron microscope.
Statistical analysis
The data are presented as the means ± SE.
Differences of the variables between the groups were analyzed by
the Student’s t-test or a one-way ANOVA test. Differences were
considered significant when P<0.05.
Results
Underexpression of NOR1 mRNA in various
cancer types
To examine NOR1 expression in various types of
cancer, the NOR1 probe was labeled and hybridized to a commercial
array containing 19 different human cancer and normal tissues. The
NOR1 expression levels were consistently high in nontumorous
tissues but were underexpressed in the majority of paired tumor
samples (Fig. 1). Notably, NOR1
expression was significantly decreased in 9 types of cancer
tissues: ovary, lung, kidney, vulva, prostate, uterus, cervix,
thyroid gland and testis. The other cancers represented on the
array (i.e., breast, colon, stomach, bladder, rectum, skin, small
intestine, pancreas, trachea and liver) did not show consistent
upregulation or downregulation of NOR1 expression.
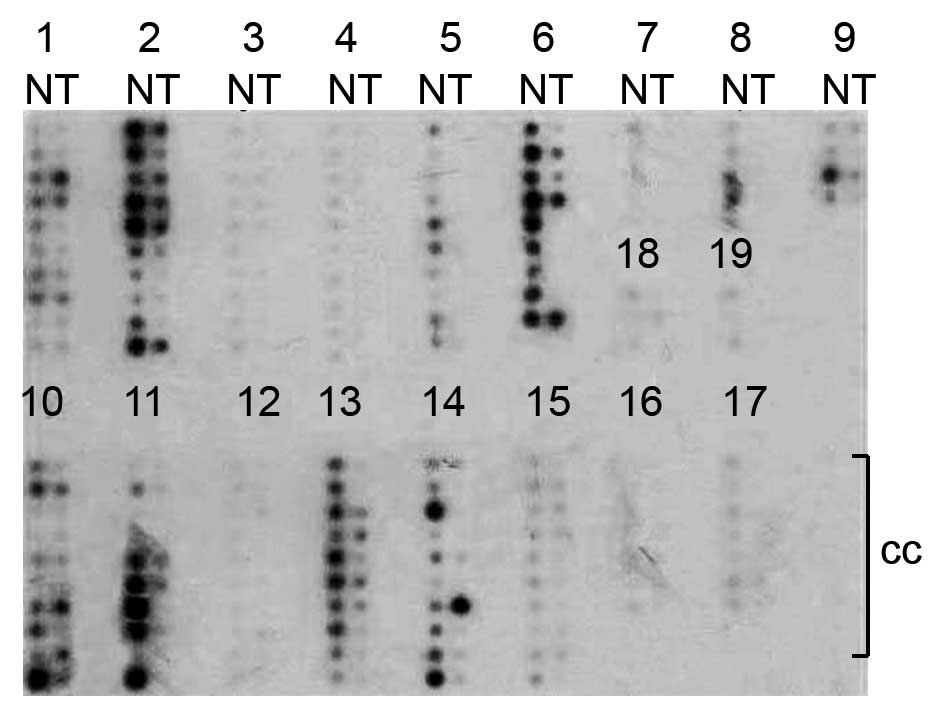 | Figure 1Expression of NOR1 was demonstrated by
the cancer profiling array II. The Cancer Profiling Array II was
hybridized with a NOR1-radiolabeled probe. Hybridization signals
were detected by phosphorimager. The numbers indicate the tissue
types in lanes: 1) breast, 2) ovary, 3) colon, 4) stomach, 5) lung,
6) kidney, 7) bladder, 8) vulva, 9) prostate, 10) uterus, 11)
cervix, 12) rectum, 13) thyroid gland, 14) testis, 15) skin, 16)
small intestine, 17) pancreas, 18) trachea, and 19) liver. N,
normal; T, tumor; cc, cancer cell line cDNA. |
To systematically investigate the epidemiology of
NOR1 expression in normal and neoplastic tissues, we used the TMAs
to analyze the expression of NOR1 in 608 samples from five
different malignant and normal tissue types (nasopharynx, lung,
liver, gastric, colon and rectum) containing 348 cancer tissues,
172 adjacent normal tissues and 88 inflammatory tissues. ISH
revealed a significantly lower rate of NOR1 mRNA expression in all
five types of cancer tissues compared with the benign lesions
(P<0.05) (Table I). However,
there was no significant correlation between NOR1 mRNA expression
and the clinical cancer stage. Therefore, these data clearly
indicate NOR1 mRNA underexpression in NPC, lung, liver, gastric,
colon and rectum cancers and suggest that its low expression may
contribute to the pathogenesis of these cancers.
 | Table INOR1 mRNA expression in five malignant
and normal tissues analyzed by in situ hybridization. |
Table I
NOR1 mRNA expression in five malignant
and normal tissues analyzed by in situ hybridization.
| NOR1 mRNA
expression |
|---|
|
|
|---|
| Tissue | Positive (%) | Negative (%) |
|---|
| Nasopharyngeal |
| Cancer (n=148) | 49 (33.1)a | 99 (66.9) |
| Benign lesion
(n=164) | 103 (62.8) | 61 (37.2) |
| Colon |
| Cancer (n=61) | 29 (47.5)a | 32 (52.5) |
| Benign lesion
(n=32) | 28 (87.5) | 4 (12.5) |
| Lung |
| Cancer (n=89) | 48 (53.9)a | 41 (46.1) |
| Benign lesion
(n=35) | 30 (85.7) | 5 (14.3) |
| Liver |
| Cancer (n=20) | 10 (50.0) | 10 (50.0) |
| Benign lesion
(n=18) | 16 (88.9) | 2 (11.1) |
| Stomach |
| Cancer (n=30) | 15 (50.0)a | 15 (50.0) |
| Benign lesion
(n=11) | 10 (90.9) | 1 (9.1) |
| Total |
| Cancer (n=348) | 151 (43.4)a | 197 (56.6) |
| Benign lesion
(n=260) | 187 (71.9) | 73 (28.1) |
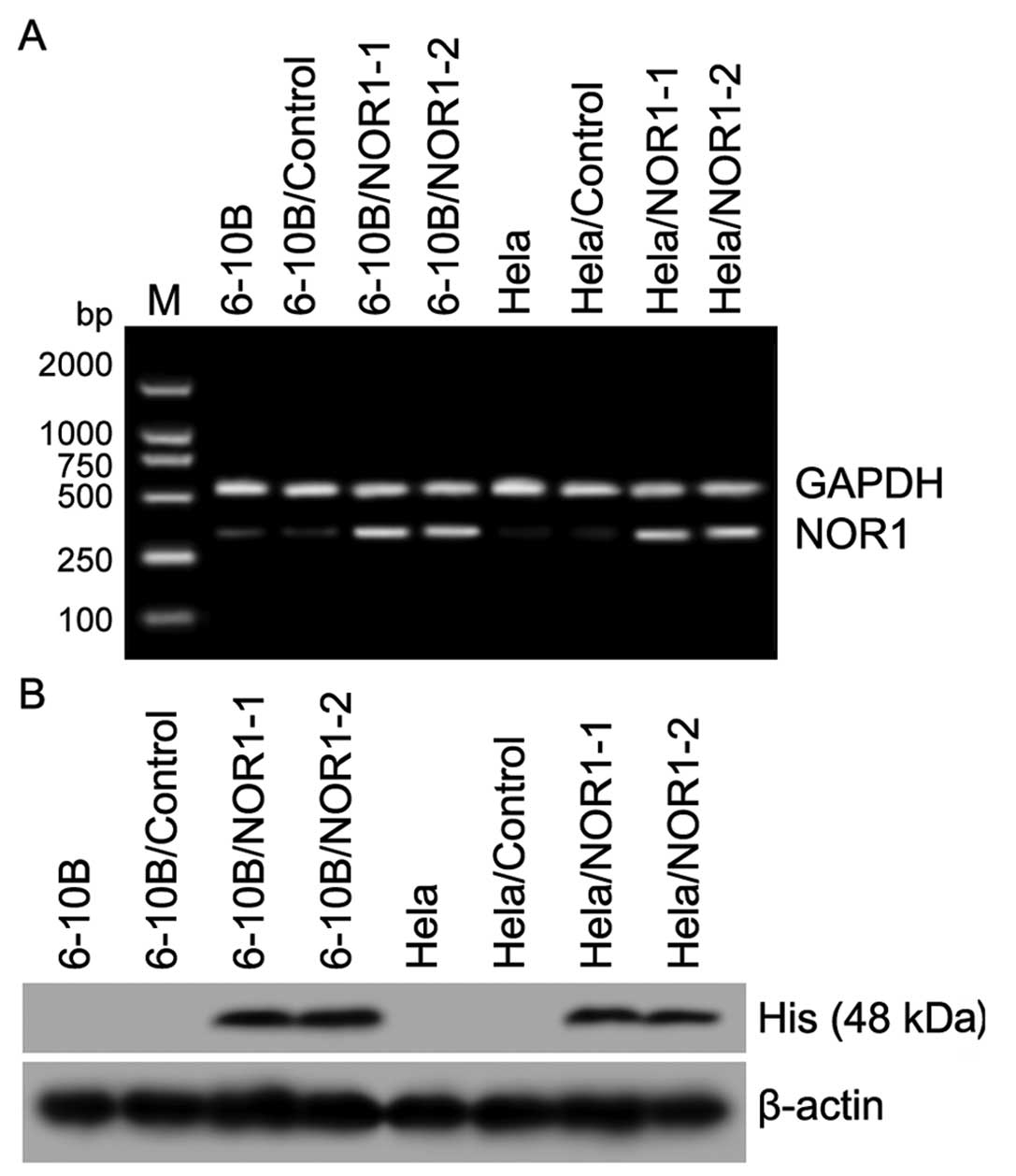
Stable expression of NOR1 in NPC 6-10B
and CCA HeLa cells after transfection with the
pCDNA3.1-myc-his-NOR1 plasmid
Studies in our lab have shown that NOR1 expression
is decreased in NPC and CCA tissues. In this study, we constructed
the recombinant eukaryotic expression vector pCDNA3.1-myc-his-NOR1
and transfected it into NPC 6-10B and CCA HeLa cells. NOR1 mRNA and
protein were stably overexpressed in pCDNA3.1-myc-his-NOR1
transformants (pool clones) as confirmed by RT-PCR and western
blotting (Fig. 2), and this
provided an applicable cell model for further NOR1 functional
analysis.
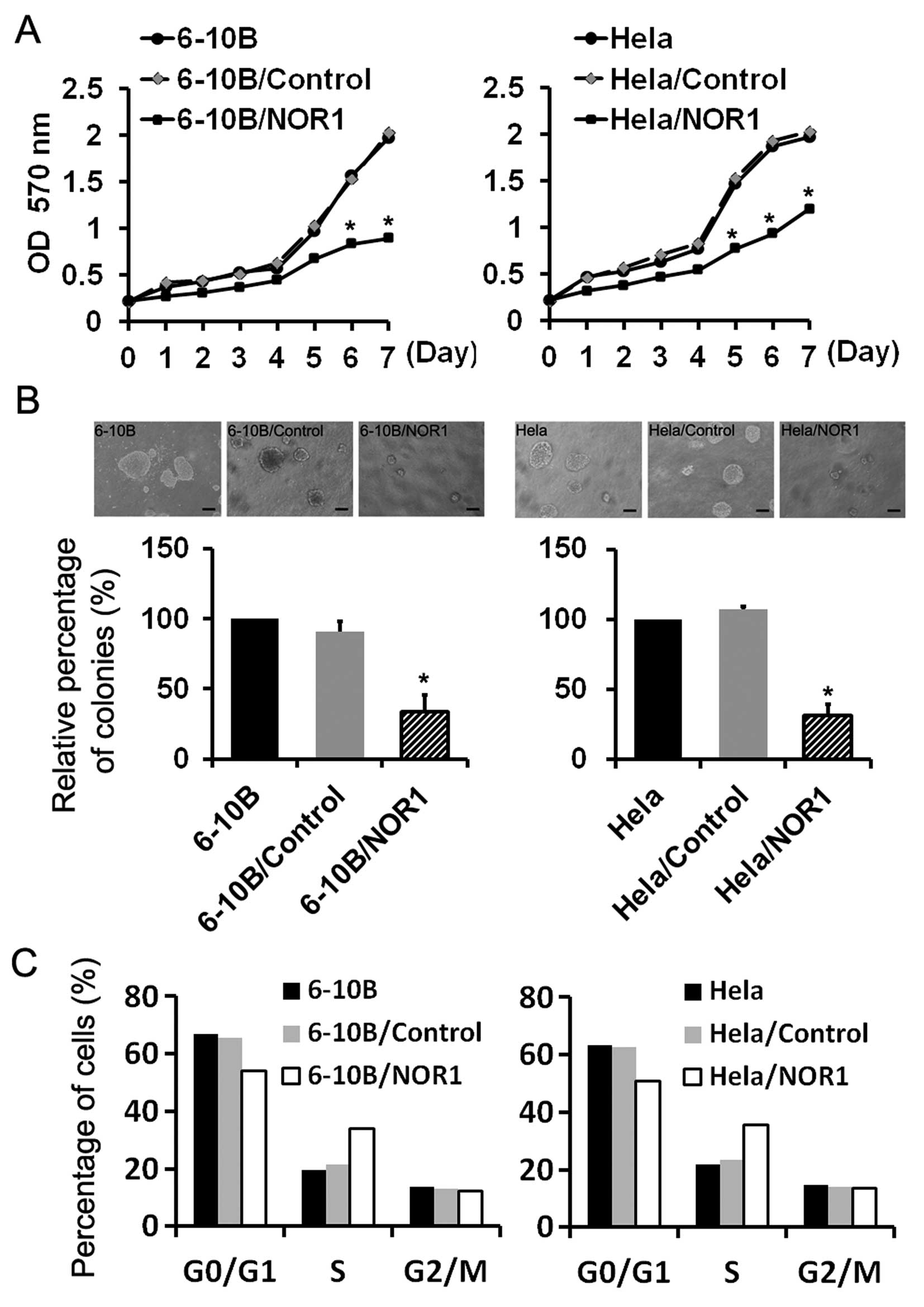
NOR1 overexpression in NPC 6-10B and CCA
HeLa cells inhibits proliferation
An MTT assay demonstrated that the exogenous NOR1
significantly inhibited the proliferative capability of 6-10B and
HeLa cells in NOR1-transfected cells as compared with untransfected
negative controls and empty vector transfected groups (P<0.05)
(Fig. 3A). Anchorage-independent
growth in soft agar semisolid medium was a strong indicator of a
transformed phenotype and a more stringent test of the mitogenic
cancer capacity. In this study, NOR1 overexpression significantly
weakened the potential of NPC and CCA cells to form colonies
(Fig. 3B). Compared with the 6-10B
and 6-10B/Control groups, NOR1 overexpression led to an approximate
65.5% reduction in the number of colonies formed in 6-10B/NOR1
cells. The colony size and number was also consistently reduced
when HeLa cells overexpressed NOR1 as compared with the two control
groups.
To elucidate the impact of upregulated NOR1
expression on the cell cycle, FCM analysis was performed. As shown
in Fig. 3C, the cells arrested in S
phase when NOR1 was overexpressed. In 6-10B, 6-10B/Control, HeLa,
and HeLa/Control groups, the cells in S phase accounted for 19.5,
21.4, 21.9 and 23.4%, respectively. When NOR1 was overexpressed in
6-10B and HeLa cells, the cells in S phase accounted for 33.9 and
35.7%, respectively. This suggested that NOR1 overexpression
arrested cells in S phase.
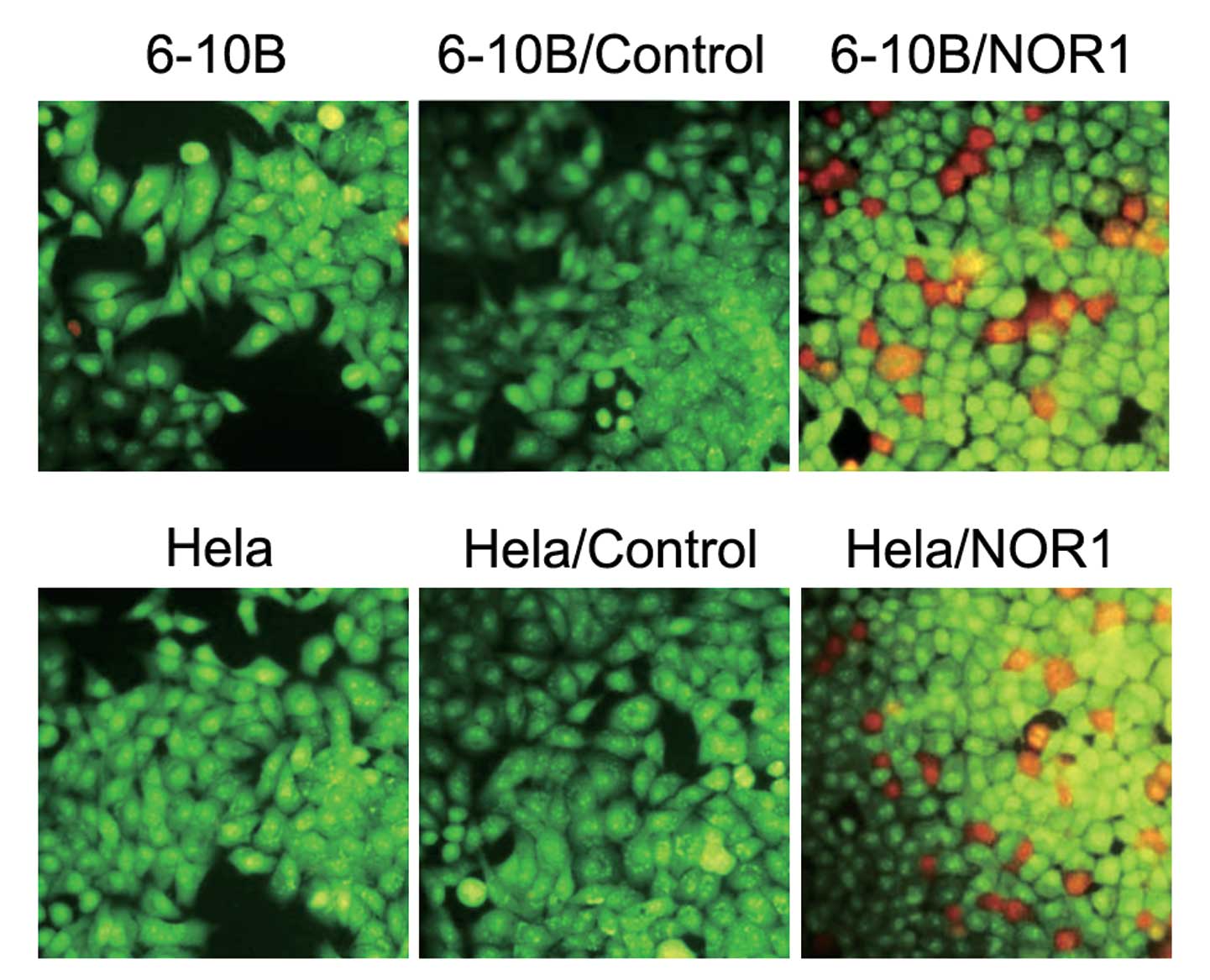
NOR1 overexpression in NPC 6-10B and CCA
HeLa cells promotes apoptosis
We next explored the effect of NOR1 on apoptosis.
Two methods were used to assess apoptosis: AO/EB staining and FCM
analysis. AO/EB staining showed the characteristic changes of
apoptotic morphology in 6-10B and HeLa cells overexpressing NOR1
(Fig. 4). Uniformly green live
cells with normal morphology were observed in the control groups,
whereas orange apoptotic cells with fragmented chromatin and
apoptotic bodies were observed in 6-10B/NOR1 and HeLa/NOR1 cells.
These results suggested that NOR1 overexpression was able to induce
a marked apoptotic morphology in NPC and CCA cells. FCM analysis
results also revealed that 6-10B and HeLa cells transfected with
NOR1 contain a certain level of apoptotic cells (1.97±0.22% and
2.39±0.45%, respectively) which is more than that found in 6-10B,
6-10B/Control, HeLa and HeLa/Control cells (0.27±0.11%, 0.21±0.09%,
0.16±0.13% and 0.17±0.11%, respectively). There was no difference
in the number of apoptotic cells between the two control
groups.
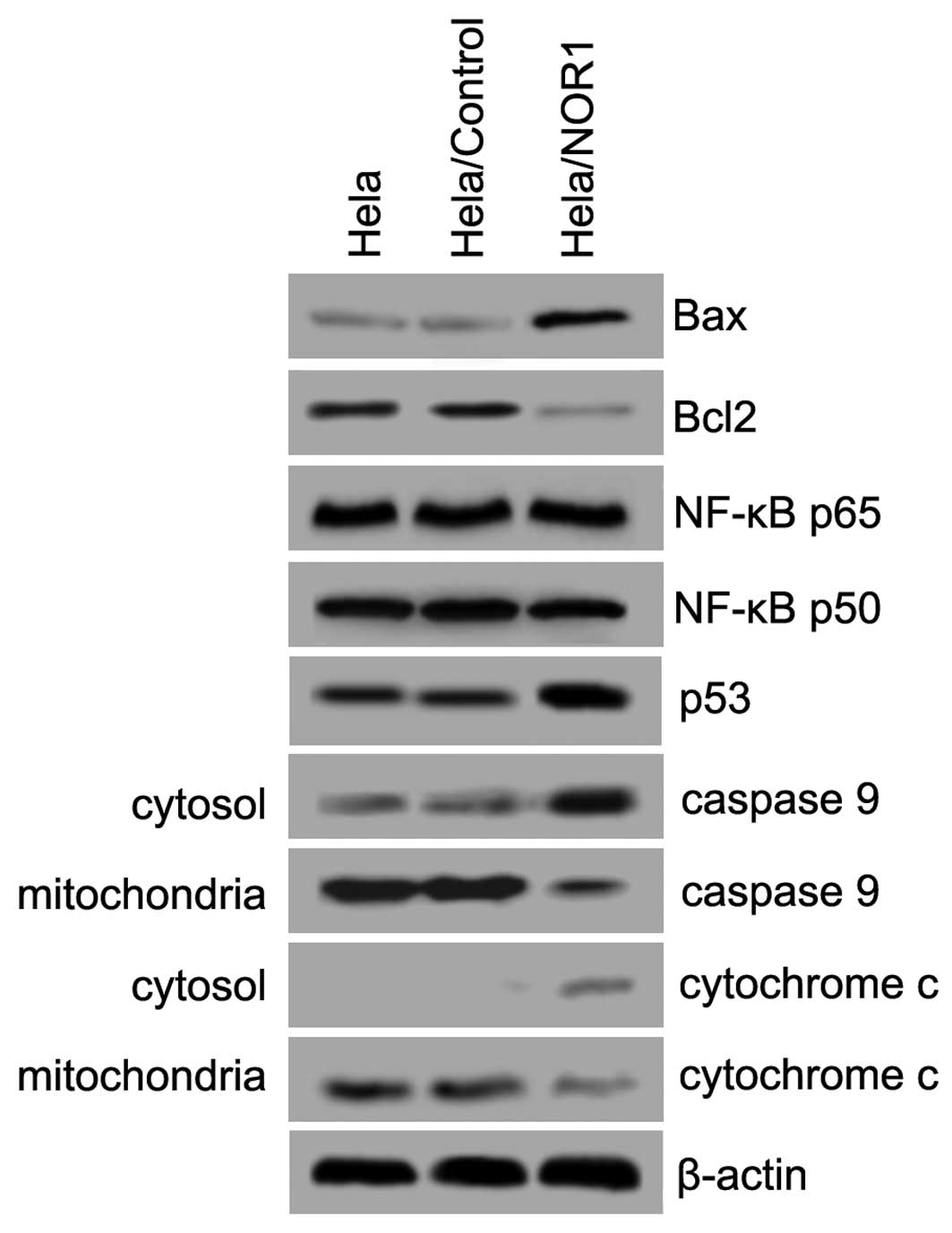
Apoptosis-related gene expression in
NOR1-induced apoptosis
We next investigated pathways that are potentially
involved in NOR1-induced apoptosis. Western blot assays were
performed using HeLa/NOR1 cell lysates to examine the expression
levels of Bax, Bcl-2, p53, and NF-κB and the release of cytochrome
c and caspase 9 from the mitochondria to cytosol. As shown in
Fig. 5, NOR1 upregulation
significantly led to an increased expression of Bax and p53 and a
decreased expression of Bcl-2. However, NOR1 overexpression did not
influence the expression of NF-κB p65 or NF-κB p50. Notably,
overexpressed NOR1 caused a decrease in expression of mitochondrial
cytochrome c and caspase 9, causing a concomitant increase in
expression of cytosolic cytochrome c and caspase 9. Collectively,
these results imply that NOR1-induced apoptosis in HeLa cells is
mitochondria dependent.
Intracellular localization of NOR1 in
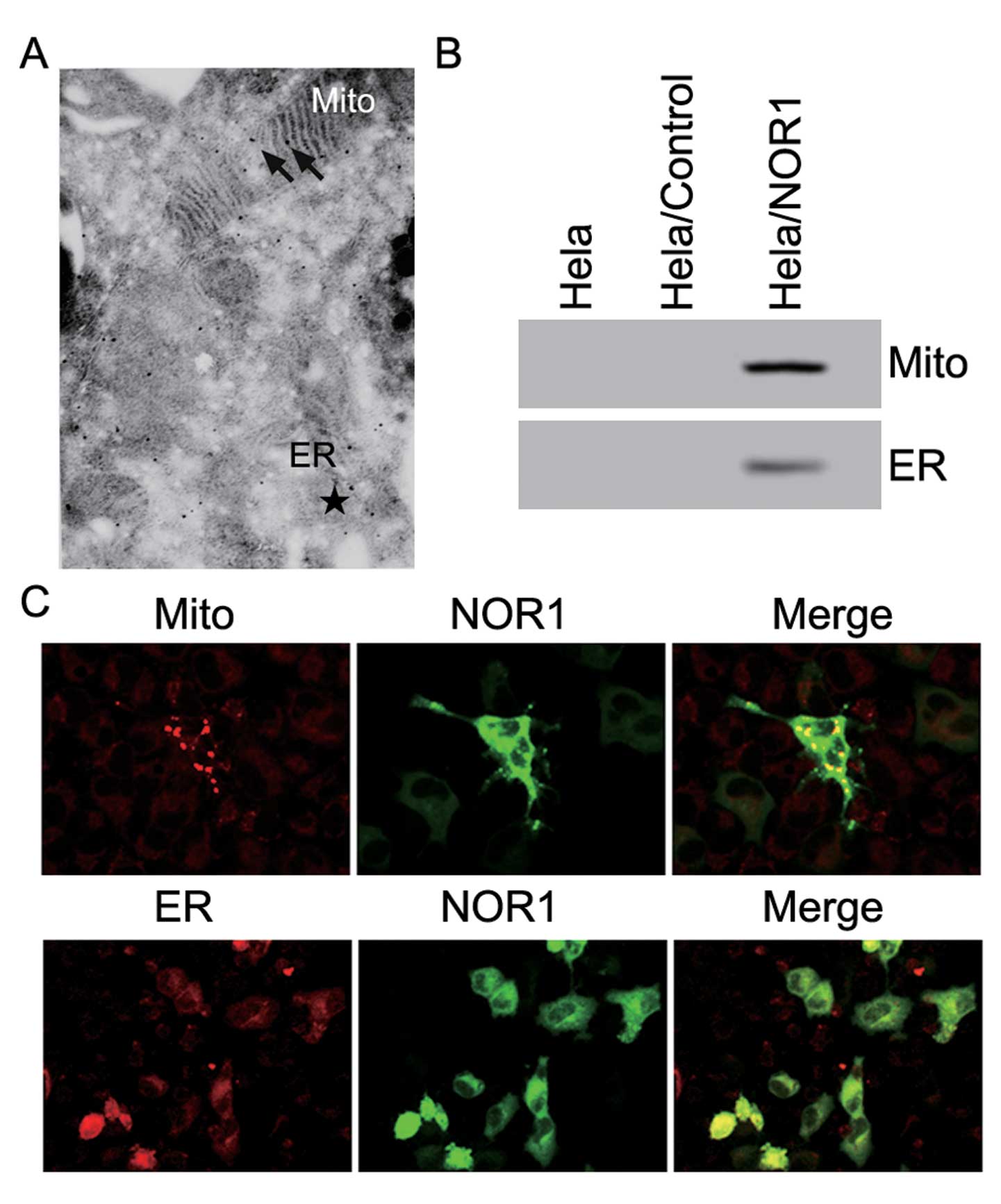
HeLa cells
To further investigate the intracellular
localization of NOR1 and its relationship with mitochondria, immune
electron microscopy analysis was used. Immune electron microscopic
analysis using an anti-His antibody confirmed that NOR1 is a
cytoplasmic protein, which partially localizes to the mitochondria
and ER (Fig. 6A). Next, the
expression of NOR1 in the mitochondria and ER was assessed by
western blotting (Fig. 6B). NOR1
protein was present in both the mitochondria and ER. pGFP-N1-NOR1
vectors were transiently transfected into HeLa cells, and after 24
hours, fluorescence microscopy was used to examine the subcellular
localization of the NOR1 protein and mitochondria or ER in HeLa
cells. Staining of HeLa/GFP-NOR1 cells with Mitotracker and
ER-Tracker Red showed that NOR1 was localized to the cytoplasm and
partly colocalized with the mitochondria and ER (Fig. 6C). Taken together, these results
suggest that NOR1 is a cytoplasmic protein, which partly localizes
to the mitochondria and ER.
Discussion
The human NOR1 gene maps to human chromosome 1p34.2,
which is a locus that is most frequently found to be lost in
primary NPC biopsies (11–14). Previous studies have also revealed
that the deletions of chromosomal arm 1p is frequent in
oligodendroglial tumors, and this has been associated with a
sensitivity to radio- and chemotherapy and favorable prognosis
(15,16). A high frequency of the loss of
heterozygosity of 1p34.2 was associated with a higher grade of
breast cancer in older women (17).
Promoter hypermethylation-mediated silencing of tumor suppressor
genes (TSGs) is a hallmark of oncogenesis. The NOR1 promoter region
was found to be frequently methylated in NPC (3) and leukemia (4), and this may also lead to lower NOR1
expression in cancer cells. To understand whether NOR1 expression
contributes to various types of cancers, NOR1 mRNA expression
levels were quantified using the Cancer Profiling Array and TMAs.
The results indicated underexpression of NOR1 mRNA in NPC, lung,
liver, gastric, colon and rectum cancers and suggest that its low
expression may contribute to the pathogenesis of various
cancers.
To better understand the antitumor roles of NOR1, we
constructed the recombinant expression vector pCDNA3.1-myc-his-NOR1
and transfected it into NPC 6-10B and CCA HeLa cells, which appear
to have low levels of NOR1 expression. Our study shows that NOR1
overexpression in NPC and CCA cells effectively inhibits cell
proliferation and colony formation, arrests cells in S phase, and
induces apoptosis in vitro, indicating a key role of NOR1 in
the regulation of cell proliferation and apoptosis, which is
consistent with its function as a tumor suppressor.
Moreover, our study is the first to prove the
apoptosis-relative molecules involved in NOR1-induced apoptosis.
Bcl-2 family members also play a critical role in the regulation of
apoptosis. The Bcl-2 family, which is composed of both
pro-apoptotic molecules (Bax, Bcl-Xs, Bak, Bid, Bad, Bim, and Bik)
and anti-apoptotic molecules (Bcl-2, Bcl-XL, Bcl-W, Mcl-1, and A1),
controls the release of mitochondrial cytochrome c by modulating
the permeability of the outer mitochondrial membrane (18,19).
Herein, we showed that NOR1 may impact apoptosis through the
activation or repression of downstream target genes. NOR1
overexpression attenuated the upregulation of Bax and decreased
Bcl-2 expression, which resulted in mitochondrial dysfunction.
Western blot analysis of cytochrome c and caspase 9 showed that
NOR1 overexpression induced cytochrome c and caspase 9 release from
the mitochondria to the cytosol. These results suggest that
NOR1-induced apoptosis is mitochondria dependent. Furthermore,
immune electron microscopy, western blotting and fluorescence
staining analysis confirmed that NOR1 is a cytoplasmic protein that
is partly localized to the mitochondria and ER. The role of the
mitochondria-dependent apoptotic pathway in NOR1-induced apoptosis
in cancer cells needs to be studied further.
Besides the Bcl-2 family members and the
mitochondria-dependent apoptotic pathway, NOR1 can also upregulate
p53 protein expression. Previous studies have suggested that p53 is
a central mediator for organizing cell responses to various stress
and anticancer molecules with apoptosis, G1-arrest, and DNA repair
(20). Nuclear factor-κB (NF-κB) is
also known for its anti-apoptotic function of transcriptional
regulation of various anti-apoptotic genes involved in survival
signaling (21,22), and the relationship between p53 and
NF-κB has been reported in recent years (23). p53 and the NF-κB-linked pathway were
reported to be involved in various antitumor molecules that induced
apoptosis (24–26). However, in this study, NOR1
overexpression did not influence the expression of NF-κB p65 and
NF-κB p50. These data might be helpful to understand the molecular
mechanism underlying NOR1 function in cancer cells, while the
relationship between p53 in NOR1-induced apoptosis still needs to
be further studied.
In conclusion, our findings demonstrate that NOR1 is
underexpressed in NPC, lung, liver, gastric, colon and rectum
cancers. Our results from the functional study of NOR1 at the
cellular level further confirms that NOR1 functions as a tumor
suppressor by inhibiting proliferation, preventing colony formation
and promoting apoptosis. NOR1 is a cytoplasmic protein that is
partially located in the mitochondria and endoplasmic reticulum,
and the mitochondria-dependent apoptotic pathway is involved in
NOR1-induced apoptosis. However, the precise mechanism behind the
antitumor effects of NOR1 needs to be investigated further.
References
|
1
|
Nie X, Zhang B, Li X, et al: Cloning,
expression, and mutation analysis of NOR1, a novel human gene
down-regulated in HNE1 nasopharyngeal carcinoma cell line. J Cancer
Res Clin Oncol. 129:410–414. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xiang B, Yi M, Wang L, et al: Preparation
of polyclonal antibody specific for NOR1 and detection of its
expression pattern in human tissues and nasopharyngeal carcinoma.
Acta Biochim Biophys Sin. 41:754–762. 2009. View Article : Google Scholar
|
|
3
|
Li W, Li X, Wang W, et al: NOR1 is an
HSF1- and NRF1-regulated putative tumor suppressor inactivated by
promoter hypermethylation in nasopharyngeal carcinoma.
Carcinogenesis. 32:1305–1314. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kroeger H, Jelinek J, Estecio MR, et al:
Aberrant CpG island methylation in acute myeloid leukemia is
accentuated at relapse. Blood. 112:1366–1373. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calmels S, Ohshima H, Rosenkranz H, McCoy
E and Bartsch H: Biochemical studies on the catalysis of
nitrosation by bacteria. Carcinogenesis. 8:1085–1088. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reznik G, Mohr U and Kruger FW:
Carcinogenic effects of Di-n-propylnitrosamine,
beta-hydroxypropyl-n-propylnitrosamine, and
methyl-n-propylnitrosamine on Sprague-Dawlay rats. J Natl Cancer
Inst. 54:937–943. 1975.PubMed/NCBI
|
|
7
|
Lijinsky W and Taylor HW: Carcinogenicity
of methylated nitrosopiperidines. Int J Cancer. 16:318–322. 1975.
View Article : Google Scholar
|
|
8
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lo KW, To KF and Huang DP: Focus on
nasopharyngeal carcinoma. Cancer Cell. 5:423–428. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin YP, Nicholas K, Ball FR, McLaughlin B
and Bishai FR: Detection of Norwalk-like virus and specific
antibody by immune-electron microscopy with colloidal gold immune
complexes. J Virol Methods. 35:237–253. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan J, Fang Y and Liang Q: Frequent
chromosomal gain of 4q and loss of 1p in primary nasopharyngeal
carcinoma. Zhonghua Zhong Liu Za Zhi. 23:208–210. 2001.(In
Chinese).
|
|
12
|
Fang Y, Guan X, Guo Y, et al: Analysis of
genetic alterations in primary nasopharyngeal carcinoma by
comparative genomic hybridization. Genes Chromosomes Cancer.
30:254–260. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shao JY, Wang HY, Huang XM, et al:
Genome-wide allelotype analysis of sporadic primary nasopharyngeal
carcinoma from southern China. Int J Oncol. 17:1267–1275.
2000.PubMed/NCBI
|
|
14
|
Lo KW, Teo PM, Hui AB, et al: High
resolution allelotype of microdissected primary nasopharyngeal
carcinoma. Cancer Res. 60:3348–3353. 2000.PubMed/NCBI
|
|
15
|
Gadji M, Fortin D, Tsanaclis AM and Drouin
R: Is the 1p/19q deletion a diagnostic marker of
oligodendrogliomas? Cancer Genet Cytogenet. 194:12–22. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tews B, Roerig P, Hartmann C, et al:
Hypermethylation and transcriptional downregulation of the CITED4
gene at 1p34.2 in oligodendroglial tumours with allelic losses on
1p and 19q. Oncogene. 26:5010–5016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chunder N, Mandal S, Basu D, Roy A,
Roychoudhury S and Panda CK: Deletion mapping of chromosome 1 in
early onset and late onset breast tumors - a comparative study in
eastern India. Pathol Res Pract. 199:313–321. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jung JY and Kim WJ: Involvement of
mitochondrial- and Fas-mediated dual mechanism in
CoCl2-induced apoptosis of rat PC12 cells. Neurosci
Lett. 371:85–90. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gross A, McDonnell JM and Korsmeyer SJ:
BCL-2 family members and the mitochondria in apoptosis. Genes Dev.
13:1899–1911. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kruse JP and Gu W: Modes of p53
regulation. Cell. 137:609–622. 2009. View Article : Google Scholar
|
|
21
|
Wang CY, Mayo MW, Korneluk RG, Goeddel DV
and Baldwin AS Jr: NF-kappaB antiapoptosis: induction of TRAF1 and
TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation.
Science. 281:1680–1683. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kucharczak J, Simmons MJ, Fan Y and
Gelinas C: To be, or not to be: NF-kappaB is the answer - role of
Rel/NF-kappaB in the regulation of apoptosis. Oncogene.
22:8961–8982. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stoffel A and Levine AJ: Actvation of
NF-kappaB by the API2/MALT1 fusions inhibits p53 dependant but not
FAS induced apoptosis: a directional link between NF-kappaB and
p53. Cell Cycle. 3:1017–1020. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yin Y, Chen W, Tang C, et al: NF-kappaB,
JNK and p53 pathways are involved in tubeimoside-1-induced
apoptosis in HepG2 cells with oxidative stress and G(2)/M cell
cycle arrest. Food Chem Toxicol. 49:3046–3054. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maldonado ME, Bousserouel S, Gosse F,
Lobstein A and Raul F: Implication of NF-kappaB and p53 in the
expression of TRAIL-death receptors and apoptosis by apple
procyanidins in human metastatic SW620 cells. Biomedica.
30:577–586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pratheeshkumar P, Sheeja K and Kuttan G:
Andrographolide induces apoptosis in B16F-10 melanoma cells by
inhibiting NF-kappaB-mediated bcl-2 activation and modulating
p53-induced caspase-3 gene expression. Immunopharmacol
Immunotoxicol. 34:143–151. 2012. View Article : Google Scholar : PubMed/NCBI
|