Introduction
The tumor necrosis factor-related apoptosis-inducing
ligand (TRAIL), one member of the tumor necrosis factor (TNF)
superfamily, binds to its death receptors or decoy receptors on the
cell surface in the form of homotrimers and triggers TRAIL-induced
apoptosis or inhibits the apoptotic pathway (1). Once TRAIL binds to its death receptors
(DR4 and DR5), FAS-associated protein with death domain (FADD) and
caspase-8 or caspase-10 are recruited to death domain motifs of the
receptors, and the death-inducing signal complex (DISC) is formed,
which eventually allows spontaneous cell apoptosis (2). In recent years, TRAIL and its
receptors have been considered as promising targets for cancer
therapy, and several molecular drugs targeting the TRAIL-induced
apoptosis pathway are already in clinical use (3).
The MAPK-activating death domain protein (MADD) is
encoded by an mRNA formed through the post-transcriptional splicing
of exons 16, 21 and 26 of the insulinoma-glucagonoma clone 20
(IG20) gene (4). It is a 183-kDa
protein containing 1647 amino acids and can be phosphorylated by
protein kinase B (Akt), a serine/threonine protein kinase, at three
highly conserved amino acid sites: S70, T173 and T1041 (5). MADD is expressed at a low level in
normal cells, a relatively higher level in the fetal brain and
kidney as well as adult testis, ovarian, brain and heart tissues,
and a markedly elevated level in malignant tumors of the pancreas,
ovary, kidney, lung and breast (6,7). After
TRAIL treatment, endogenous MADD binds to the TRAIL death receptor
DR4/DR5 cytoplasmic tail by its C-terminal death domain and
prevents cell apoptosis by inhibiting caspase-8 activation
(8). Overexpression of MADD
enhances TNF-α-induced activation of nuclear factor-κB (NF-κB) and
mitogen-activated protein kinase, which eventually contributes to
cancer cell survival (9,10). Obviously, MADD is a key downstream
mediator in the TRAIL-induced apoptosis pathway and plays important
roles in enhancing cell proliferation and reducing cell
apoptosis.
Lung cancer, the primary malignant tumor of the
lungs, is one of the leading causes of cancer-related death. It
originates from the bronchial mucosa and is histologically divided
into non-small cell lung cancer (NSCLC, approximately 75–80% of
lung cancer cases) and small-cell lung cancer (less than 20% of
lung cancer cases). NSCLC is further divided into squamous cell
carcinoma (SCC), adenocarcinoma (ADC) and large-cell carcinoma.
This study investigated MADD expression in normal, SCC and ADC
tissues of the lungs and its effects on proliferation and apoptosis
of lung ADC cells. It was found that MADD was expressed at a high
level and played an important anti-apoptotic role in lung ADC,
which means that MADD may be a novel and effective target for
therapy of lung ADC aimed at the TRAIL-induced apoptosis
pathway.
Materials and methods
Immunohistochemistry (IHC)
All formalin-fixed and paraffin-embedded tissue
samples were retrieved from the Department of Pathology of the Qilu
Hospital of Shandong University. The tissue samples were procured
between July 2009 and June 2011. All archival hematoxylin and eosin
(H&E)-stained slides for each patient were reviewed by two
pathologists. For the use of the clinical materials for research
purposes, prior patient consent and approval from the Institutional
Research Ethics Committee were obtained. All of the diagnoses were
made following the Pathology and Genetics of Tumors of the Lung of
the World Health Organization Classification of Tumors.
Clinicopathological classification and staging were determined
according to the American Joint Committee on Cancer criteria.
Paraffin sections were baked at 60°C for 30 min,
de-paraffinized in xylene, and hydrated in descending
concentrations of alcohol. Endogenous peroxidase activity was
blocked with 0.3% (v/v) hydrogen peroxide in methanol for 30 min.
Antigen retrieval was carried out by heating in a microwave oven
with 0.01 M citrate buffer, pH 6.0, for 10 min. The sections were
then incubated with 5% bovine serum albumin for 30 min at room
temperature and subsequently with rabbit anti-MADD primary antibody
(provided by Dr B.S. Prabhakar and Dr L.C. Li of the Department of
Neurology at Illinois University School of Medicine) at 4°C
overnight in a humidified container. Equal volumes of
phosphate-buffered saline (PBS, pH 7.4) and rabbit serum were added
instead of the primary antibody as blank and negative controls,
respectively. The sections were then incubated with horseradish
peroxidase (HRP)-conjugated secondary antibody (Beijing Zhongshan
Golden Bridge Biological Technology Co., Ltd., Beijing, China) at
37°C for 30 min and stained using a DAB detection kit (Beijing
Zhongshan Golden Bridge Biological Technology Co., Ltd.) followed
by hematoxylin counterstain and differentiation in HCl-alcohol.
After dehydration, the gum mounted sections were photographed under
an inverted microscope (BX51 TRF, Olympus, Japan). The integrated
optic density (IOD) of the positive area in each microphotograph
was assessed using immunohistochemical analysis software (Image-Pro
Plus 6.0).
Cell culture
The human lung adenocarcinoma A549 cell line and the
human embryonic kidney (HEK) 293T cell line were purchased from the
China Center for Type Culture Collection (CCTCC, Shanghai, China).
The A549 and HEK293T cells were separately cultured in Ham’s F12
medium (Thermo Fisher Scientific Inc., Rockford, IL, USA) and DMEM
(high glucose) (Hyclone, Logan, UT, USA), both of which contained
10% fetal bovine serum (FBS, Hyclone), 100 U/ml penicillin and 100
μg/ml streptomycin, at 37°C in a humidified atmosphere with 5%
CO2. At 80–90% confluence, the cells were subcultured at
a ratio of 1:3 with 0.25% trypsin.
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from A549 cells using TRIzol
reagent (DBI, Biosciences, USA). Reverse transcription was carried
out with 1 μg of total RNA using a ReverTra Ace qPCR RT kit
(Toyobo, Osaka, Japan). Briefly, RNA and random primers in a 12-μl
reaction mixture were put on ice immediately after incubation at
65°C for 5 min. Then, cDNAs were synthesized in a 20-μl reaction
mixture containing ReverTra Ace, dNTP mix, and RNase inhibitor at
30°C for 10 min followed by 42°C for 60 min, 90°C for 5 min, and
4°C for 5 min. For PCR amplification, 2 μl of cDNA was added to a
50-μl reaction mixture containing 25 μl 2X Taq PCR Master mix (DBI,
Bioscience, USA) and 0.01 μM of each primer. The reaction mixture
was first incubated at 50°C for 30 min and 94°C for 2 min.
Subsequently, 30 cycles of PCR (denaturation at 94°C for 30 sec,
annealing at 55°C for 30 sec, and extension at 72°C for 1 min) were
carried out with a final incubation at 72°C for 7 min. Two primer
sets, F1/R1 (5′-CGGGACTCTGACTCCGAACCTAC-3′ and
5′-GCGGTTCAGCTTGCTCAGGAC-3′) and F2/R2
(5′-CTGCAGGTGACCCTGGAAGGGATC-3′ and
5′-TGTACCCGGGTCAGCTAGAGACAGGCC-3′) were separately used to amplify
the desired fragments of the IG20 gene as previously reported
(4). The primers (F/R) for
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were provided in
the ReverTra Ace qPCR RT kit. PCR products were separated on a 5%
polyacrylamide gel. The gel was stained with ethidium bromide and
photographed under a UV transilluminator (Alpha Imager™ 2200, San
Leandro, CA, USA).
Plasmid transfection
A549 cells were seeded into 6-well plates at a
density of 2×105 cells/well and cultured in 2 ml Ham’s
F-12 medium containing 10% FBS without antibiotics for 24 h in a
CO2 incubator. Lipofectamine 2000 (10 μl) (Invitrogen,
Carlsbad, CA, USA) and 4 μg pEYFP or pEYFP-MADD plasmid DNA
(provided by Prabhakar and Li as previously noted) were separately
diluted in 250 μl portions of Opti-MEM reduced serum medium
(Invitrogen). After 5 min of incubation at room temperature, the
diluted Lipofectamine 2000 and plasmid DNA were mixed together. The
DNA-liposome complex was incubated for 20 min at room temperature
and then directly added to the cultured cells in 2 ml of Opti-MEM
medium. The culture medium was replaced with Ham’s F-12 medium
containing 10% FBS without antibiotics after transfection for 6 h
in the CO2 incubator. Expression of MADD in the
transfected cells was detected using western blot assay after 48 h
of incubation.
Lentiviral vector transfection
HEK293T cells were seeded in 100-mm plates at a
density of 1.5×106 cells/plate and cultured for 24 h.
Lentiviruses were produced by co-transfecting 16 μg pNL-SIN-GFP-SCR
or pNL-SIN-GFP-MID lentiviral vectors (provided by Prabhakar and Li
as previously noted) and 32 μl Lenti-Pac HIV Mix (GeneCopoeia,
Rockville, MD, USA) into HEK293T cells. The medium was replaced
with fresh medium after transfection for 16 h. The
lentivirus-containing supernatant was harvested after incubation
for 40 h and filtered using a 0.45-μm filter. The optimal viral
titer was determined by transfecting HEK293T cells using a
previously described method (11).
A549 cells were seeded in 6-well plates at a density of
5×105 cells/well and cultured in 2-ml complete medium.
After incubation for 24 h, the medium was replaced with serum-free
medium containing 8 μg/ml Polybrene (Sigma, St. Louis, MO, USA),
and the viral supernatant was added to the cells in multiples of
infection of 10. After 6 h of incubation for transfection, the
medium was replaced with complete medium, the cells were incubated
for a further 48 h, and expression of MADD in the transfected cells
was detected using western blot assay.
Protein determination and western blot
analysis
The transfected cells were lysed with RIPA lysis
buffer (Thermo Fisher Scientific Inc.). After centrifugation at
13,000 × g for 20 min at 4°C, protein concentrations in the cell
lysates were determined using a BCA protein assay kit (Beyotime,
Jiangsu, China). Samples corresponding to 20 μg of protein were
fractionated by 8% sodium dodecyl sulfate polyacrylamide gel
electrophoresis and then transferred to a polyvinylidene difluoride
membrane. The membrane was blocked with TBST buffer (20 mM
Tris-buffered saline with 0.1% Tween-20) containing 5% non-fat dry
milk for 1 h at room temperature and incubated with primary
antibodies targeting β-actin (Beijing Zhongshan Golden Bridge
Biological Technology Co., Ltd.) and MADD at 4°C overnight. The
next day, the membrane was washed 3 times for 5 min each time with
TBST buffer and incubated with HRP-conjugated secondary antibody at
room temperature for 1 h. After the membrane was washed once with
TBS (20 mM Tris-buffered saline) for 15 min and twice with TBST for
5 min each at room temperature, MADD and β-actin on the membrane
were detected using Pierce ECL Western blotting substrate (Thermo
Fisher Scientific Inc.) by a chemiluminescence apparatus
(FluorChem®Q, Cell Biosciences, Santa Clara, CA,
USA).
MTT assay
A549 cells were seeded into 96-well plates at a
density of 5×103 cells/well (for plasmid transfection)
or 1×104 cells/well (for lentiviral vector transfection)
and incubated for 24 h. The A549 cells were respectively
transfected with pEYFP and pEYFP-MADD plasmids as well as
pNL-SIN-GFP-SCR and pNL-SIN-GFP-MID lentiviral vectors using the
methods as described above. After transfection for 48 h, 10 μl of 5
mg/ml 3-(4,5-dimethylthiazol-2-y1)-2,5-diphenytetrazolium bromide
(MTT, Sigma) was added to each well, and the cultures were
incubated for an additional 4 h. Then formazan crystals were
dissolved by the addition of dimethyl sulfoxide (DMSO, Sigma)
solution. The absorbance of each well was measured at 490 nm (Model
680, Bio-Rad Laboratories, Hercules, CA, USA).
Flow cytometric (FCM) analysis
Cell apoptosis was detected by flow cytometry using
an Annexin V-FITC/PI apoptosis detection kit (KeyGen, China). A549
cells were seeded into a 6-well plate at 2×105 or 4×
cells/well and transfected with pEYFP-MADD plasmids or
pNL-SIN-GFP-MID lentiviral vectors as described in the Plasmid
transfection and Lentiviral vector transfection sections. After
incubation for 48 h, the transfected cells were collected, washed
twice with ice-cold PBS, and resuspended in 500-μl binding buffer.
Annexin V/FITC (5 μl) and 5 μl PI were sequentially added to the
cell suspension. The cell suspension was incubated at room
temperature in the dark for 15 min, and the cells were immediately
analyzed on a FACS flow cytometer (Becton-Dickinson, Franklin
Lakes, NJ, USA). Annexin V/FITC-positive and PI-negative cells were
scored as apoptotic.
Statistical analysis
All of the experiments were conducted at least three
times. Data are shown as the means ± SD. Statistical significance
was determined by statistical analysis software SPSS 17.0.
P<0.05 was considered to indicate a statistically significant
result.
Results
Expression of MADD in normal lung tissues
and NSCLC
In this study, the clinicopathological features of
68 samples of tissues of the lungs were analyzed. These included 20
samples of normal tissues, 31 samples of SCC tissues and 17 samples
of ADC tissues. There was no significant difference in gender and
age among the three groups. Clinicopathological characteristics of
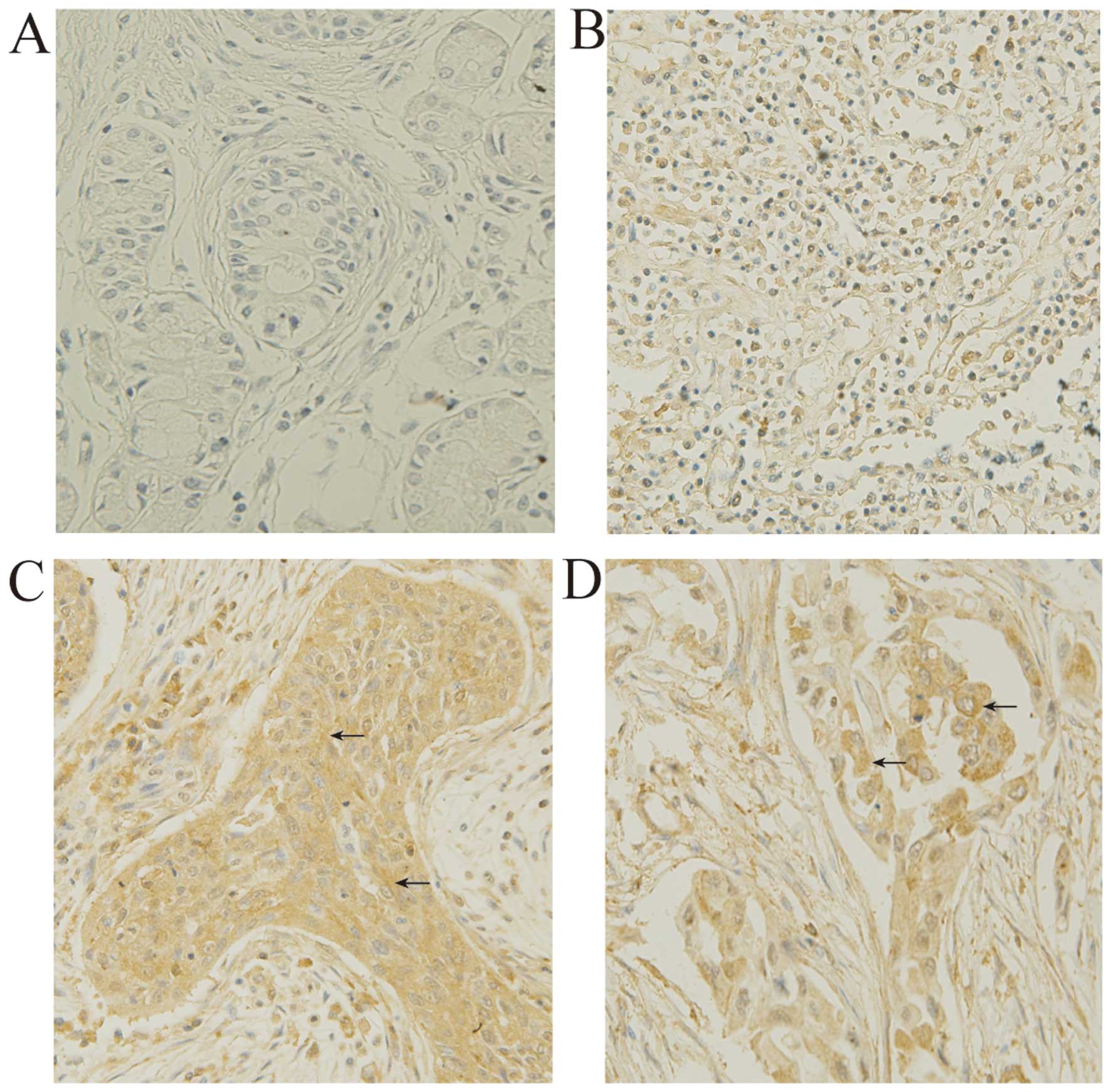
the tumor tissues are summarized in Table I. IHC results are shown in Fig. 1 and Table II. MADD was expressed in both
normal lung and NSCLC tissues. IOD analysis revealed that the
expression level of MADD in ADC or SCC was higher than that in the
normal lung tissues (P<0.01). The mean IOD values of the ADC and
SCC tissues were 1.95 and 1.61 times that of normal lung tissues,
respectively. It was also found that ADC expressed more MADD than
SCC (P<0.01). The mean IOD of ADC tissues was 1.22 times that of
SCC tissues. In addition, IOD analysis showed that levels of MADD
expression in both the ADC and SCC tissues were not related to
differentiation and stage of ADC and SCC.
 | Table IClinicopathological characteristics of
the tumor tissues. |
Table I
Clinicopathological characteristics of
the tumor tissues.
| Tumor
characteristics | No. of cases |
|---|
| Histology |
| SCC | 31 |
| ADC | 17 |
| Degree of
differentiation |
| High | 7 |
| Moderate | 20 |
| Low | 21 |
| Stage |
| I | 14 |
| II | 25 |
| III | 9 |
 | Table IIIOD of the IHC images. |
Table II
IOD of the IHC images.
| Tissues | No. of cases | IOD (means ± SD) |
|---|
| Normal | 20 | 16.419±0.606 |
| SCC | 31 |
26.358±0.759a |
| ADC | 17 |
30.065±0.962a,b |
Expression of the IG20 gene in A549
cells
As MADD was expressed at a relatively higher level
in ADC, all subsequent studies were carried out using the human
lung adenocarcinoma cell line A549. First, RT-PCR assay was applied
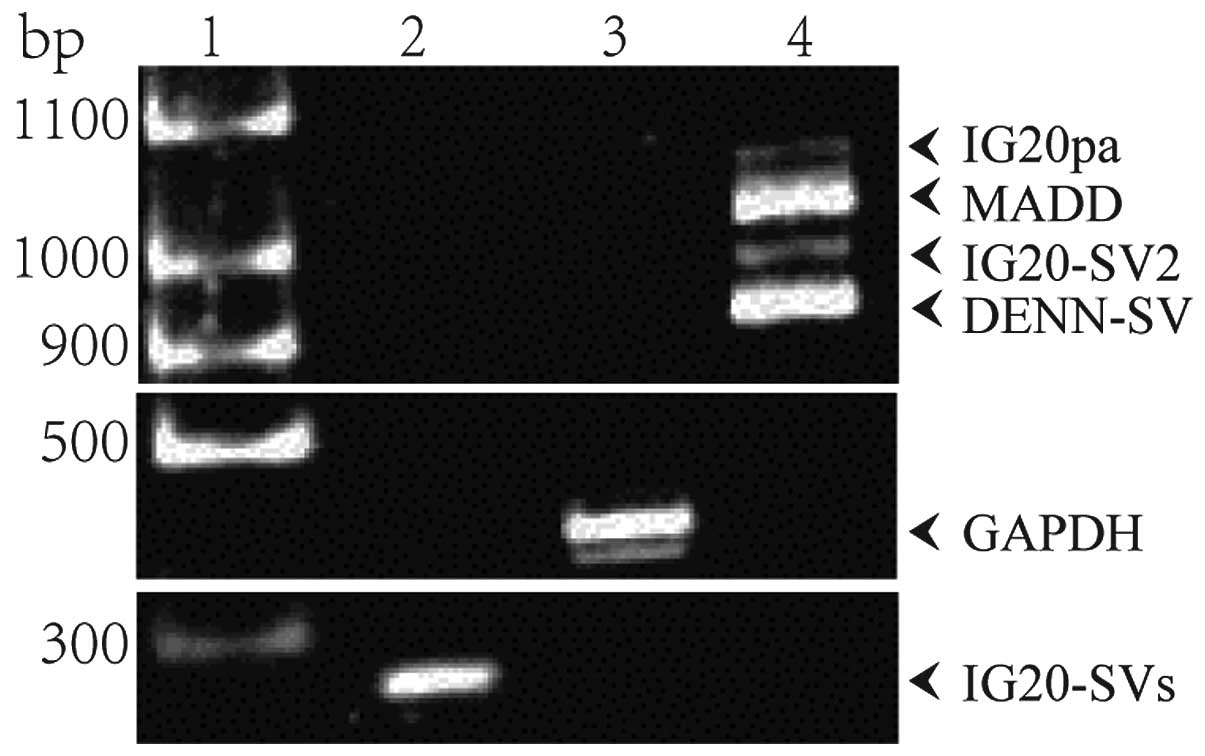
to examine mRNA expression of the IG20 gene in A549 cells since
MADD is a splice variant of IG20. RT-PCR results are shown in
Fig. 2. Four spliced transcripts,
~1 kb in length, were amplified with the primer pair F1 and R1 that
targets exons 13L and 16 of the IG20 gene (Fig. 2, lane 4). These are the transcripts
encoding IG20pa, MADD, IG20-SV2 and DENN-SV. With the primer pair
F2 and R2 targeting exon 34 of the IG20 gene, only one fragment of
~300 bp was amplified (Fig. 2, lane
2), which means that the transcripts encoding KIAA0358 and IG20-SV4
were undetectable in A549 cells. These results indicated that the
four splice variants of IG20, which included MADD, were expressed
in A549 cells.
MADD expression in transfected A549
cells
In order to assess the function of MADD in A549
cells, we introduced the exogenous MADD gene into the cells through
transfection and silencing of MADD expression using RNA
interference (RNAi). The expression of MADD in the transfected A549
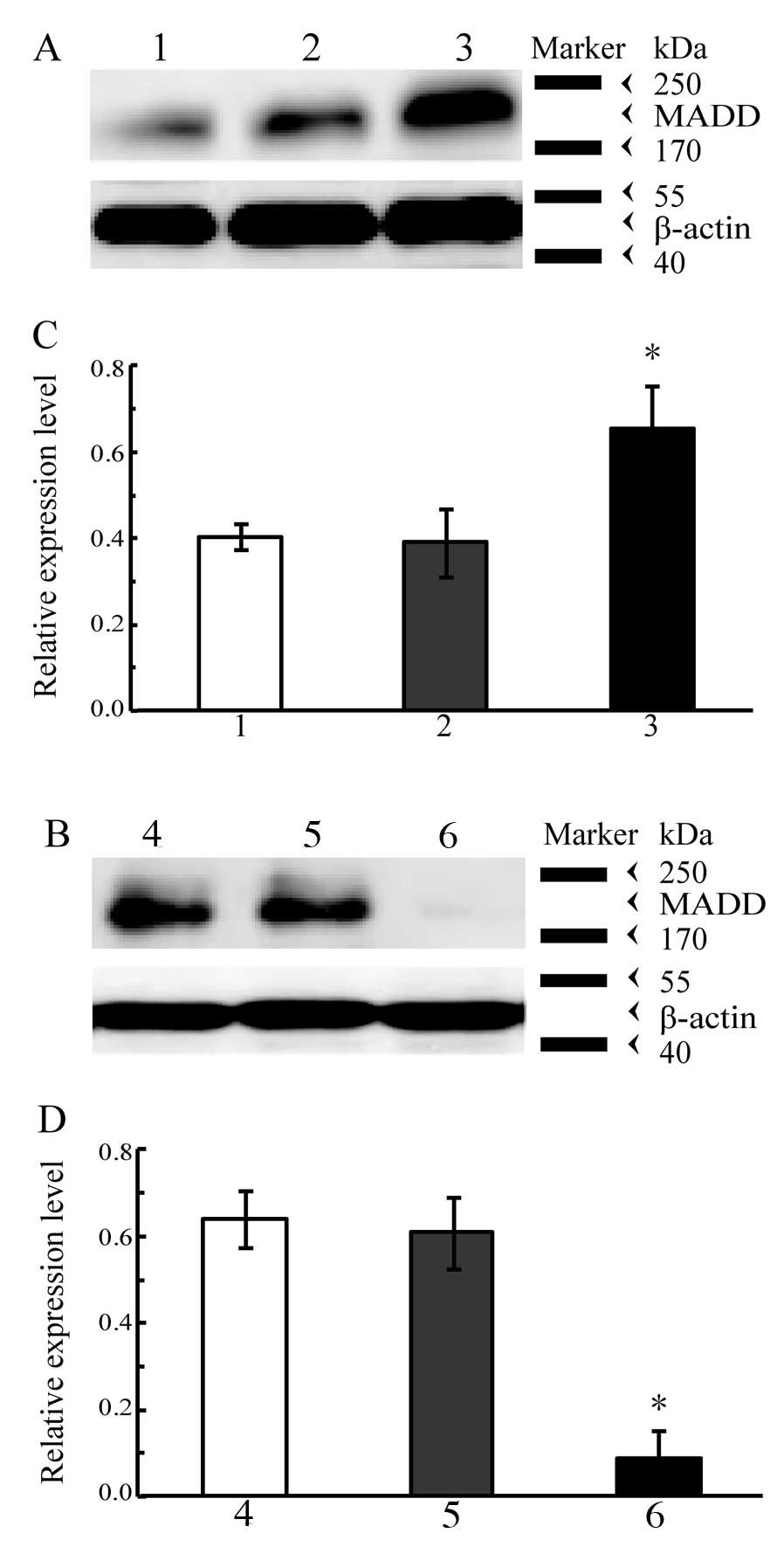
cells was determined by western blot analysis. The results of
western blot analysis are shown in Fig.
3. The level of MADD obviously increased in the A549 cells
containing the exogenous MADD gene (P<0.01 vs. control, Fig. 3A and C) whereas it was markedly
decreased in the A549 cells subjected to RNAi targeting of MADD
(P<0.01 vs. control, Fig. 3B and
D); both introduction of the exogenous MADD gene and RNAi
targeting of MADD effectively altered the MADD expression levels in
A549 cells.
Effects of MADD on A549 cell
proliferation
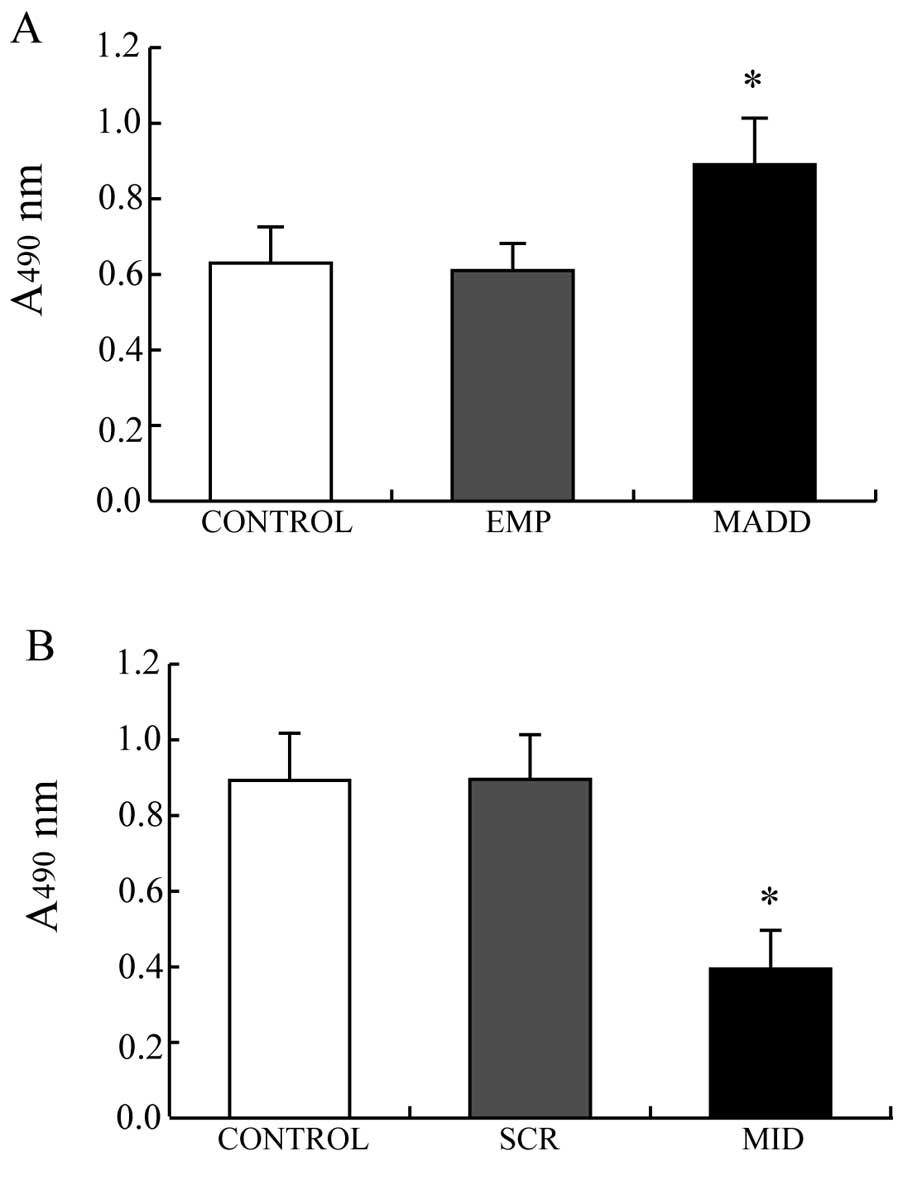
MTT assay was performed to observe the effects of
MADD on the proliferation of A549 cells. As showed in Fig. 4, the A549 cells transfected with the
pEYFP-MADD plasmids, in which MADD was overexpressed, demonstrated
greater viability than the control cells (P<0.01, Fig. 4A) while the viability of the A549
cells transfected with the pNL-SIN-GFP-MID lentiviral vectors, with
silenced expression of MADD, was lower than that of the control
cells (P<0.01, Fig. 4B).
Obviously, MADD promotes proliferation of A549 cells.
Effects of MADD on the apoptosis of A549
cells
To further investigate the mechanisms by which MADD
promotes proliferation of A549 cells, FCM was utilized to evaluate
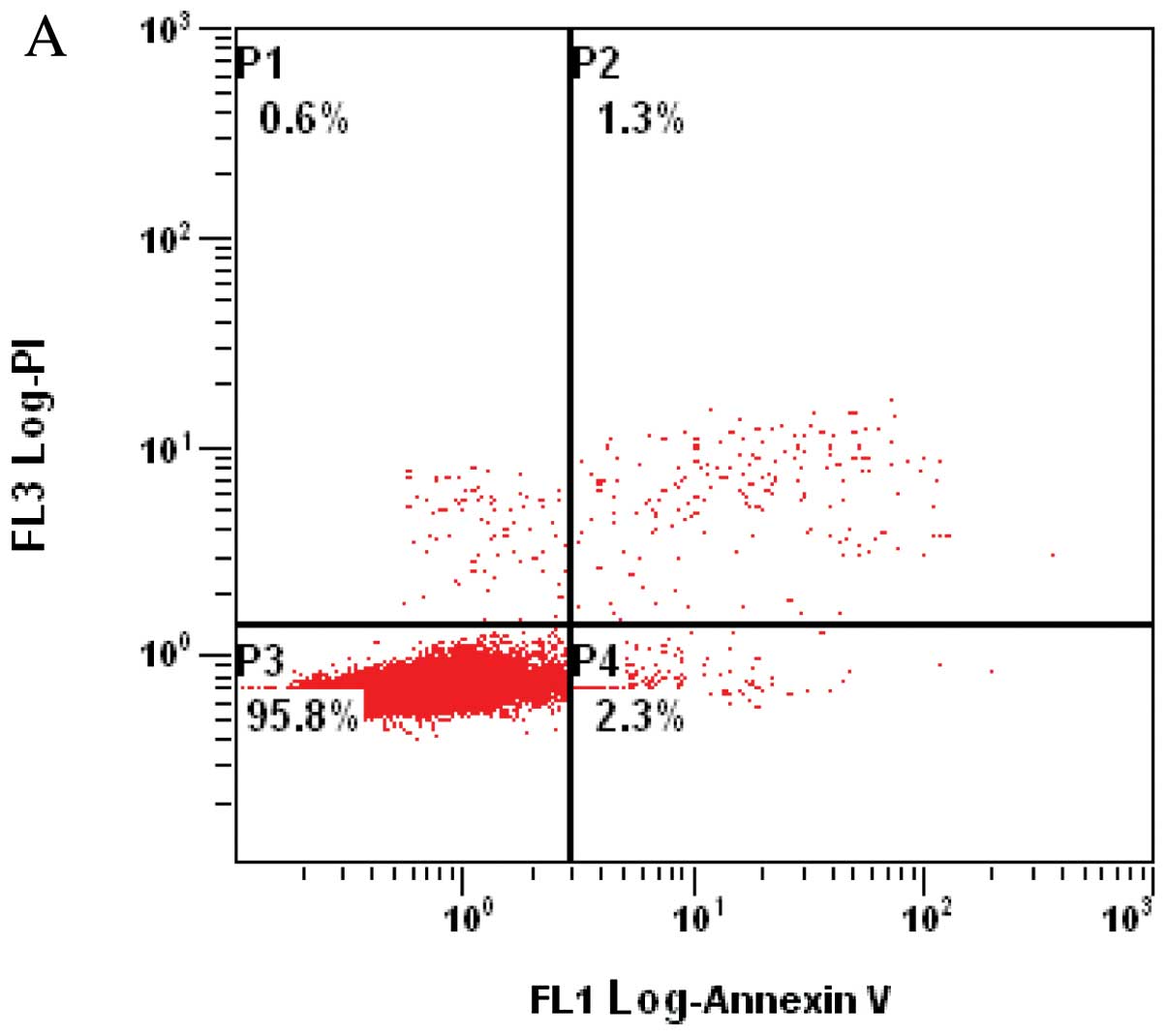
the apoptosis of A549 cells. FCM results are shown in Table III and Fig. 5. The apoptosis rate of control cells
was 2.4±0.5%. The apoptosis rate of the A549 cells transfected with
the pEYFP-MADD plasmids decreased to 1.4±0.6%, whereas that of the
A549 cells transfected with the pNL-SIN-GFP-MID lentiviral vectors
increased to 6.7±1.5%. These results demonstrated that
overexpression of MADD effectively inhibits A549 cell apoptosis,
and down-modulation of MADD obviously enhances apoptosis, which
suggests that in A549 cells, MADD distinctly affects apoptosis and
functions as an inhibitor of apoptosis.
 | Table IIIApoptosis rate of A549 cells,
n=3. |
Table III
Apoptosis rate of A549 cells,
n=3.
| Group | Apoptosis rate (%)
(means ± SD) |
|---|
| Control | 2.4±0.5 |
| Overexpressing
MADD | 1.4±0.6a |
| Silencing MADD | 6.7±1.5a |
Discussion
The worldwide incidence of lung cancer has rapidly
increased in recent years. The exact pathogenesis of lung cancer is
still unclear. Studies have revealed that the development of lung
cancer is closely associated with abnormal alterations in
oncogenes, tumor-suppressor genes, cytokines and their receptors
and cell adhesion molecules (12–14).
For early-stage lung cancer without overt metastasis, surgery is
the optimal choice while it has great limitations for
advanced-stage or metastatic lung cancer. However, the fact is that
most patients with lung cancer are diagnosed with advanced-stage
disease or metastasis. Although radiotherapy and chemotherapy are
also feasible therapies for lung cancer, severe side effects and
drug tolerance hamper their application. Thus, it is of vital
practical significance to explore new therapeutic targets and
approaches for lung cancer.
TRAIL is a promising candidate for cancer therapy as
it can selectively induce apoptosis in tumor cells and effectively
kill them but not normal cells (15). Interaction between TRAIL and its
cell surface receptors DR4/DR5 can result in intracellular
recruitment of FADD and caspase-8/10 and formation of DISC, which
can eventually trigger apoptosis of tumor cells (2). DR1 and DR2, two receptors of TRAIL
that are only expressed on normal cell membranes, are considered as
decoy receptors since they have no or only part of a death domain.
As a result, after binding to TRAIL, DR1 and DR2 fail to transmit
apoptotic signals and hardly induce apoptosis in normal cells
(16,17). At present, some molecular antitumor
drugs targeting the TRAIL-induced apoptosis pathway have been
developed, which can kill tumor cells without obvious toxicity and
side effects to normal cells (18).
However, tumor cells can easily acquire resistance to TRAIL therapy
(19). Overexpression of decoy
receptors and invalid mutations of TRAIL receptors together with
loss of caspase-8 expression due to CASP gene methylation can cause
TRAIL resistance in tumor cells (20,21).
Additionally, NF-κB is always activated downstream in the TRAIL
pathway and may more easily cause tumor cell metastasis (22). Hence, TRAIL strategy for cancer
therapy needs further improvement.
The IG20 gene was first discovered in β cells of
human pancreas islets by Cunningham in 1996 (23). It is located on human chromosome
11p11, having 5995 bp and containing 36 exons (6,24). By
alternative splicing of exons 13L, 16, 21, 26 and 34, it can encode
at least six protein isoforms: KIAA0358, IG20-SV4, IG20pa, MADD,
DENN and IG20-SV2 (4). KIAA0358 and
IG20-SV4 are mainly expressed in the central nervous system such as
the cerebral cortex, hippocampus and spinal cord and are involved
in neurotransmission (25–28) and neuro-degeneration (29). IG20pa, IG20-SV2, MADD and DENN-SV
are expressed in almost all normal human tissues at low levels and
participate in the TRAIL-mediated signaling pathway (4). IG20pa and IG20-SV2 have no tissue and
cell specificity. MADD and DENN-SV proteins are expressed at higher
levels compared to other protein isoforms (30) and are obviously increased in tumor
cells and tissues (30–33), which suggests that they play
important roles not only in survival of normal cells but in
pathogenesis and development of tumors.
According to previous studies, continuous expression
of MADD can render cells more resistant to TRAIL-induced apoptosis
(30). Phosphorylated MADD can
specifically interact with TRAIL receptors to suppress cell
apoptosis by means of inhibiting the cleavage of procaspase-8
(8). Antisense abrogation of MADD
expression makes tumor cells more susceptible to extracellular
apoptotic stimuli such as TNF-α, TRAIL, etoposide and vinblastine
(24) as well as γ-irradiation
(34). MADD expression of tumor
cells markedly differs from that of normal cells (4). Therefore, studies of the functions of
MADD and regulatory mechanisms of its expression will help find
novel and effective targets for TRAIL therapy against tumors.
To date, the roles of MADD in lung cancer have not
been elucidated. This study mainly investigated expression of MADD
in lung ADC tissues and its function in lung ADC cells. IHC
analysis was used to observe MADD expression in normal and NSCLC
tissues of the lungs. The results of IHC revealed that SCC and ADC
tissues expressed MADD at a higher level when compared with normal
lung tissues, which implies that MADD may play an important role in
the pathogenesis of NSCLC. It was also found that ADC tissues had a
higher MADD level than SCC tissues, indicating that ADC and SCC
have different pathogenic mechanisms, and MADD may play a more
vital role in ADC development. Hence, we used human lung
adenocarcinoma A549 cells to further investigate the function of
MADD in lung cancer. RT-PCR results showed that A549 cells
expressed MADD, thus A549 cells were transfected with pEYFP-MADD
plasmids or pNL-SIN-GFP-MID lentiviral vectors to increase or
decrease MADD expression. MADD expression in the transfected A549
cells and cell proliferation were detected by means of western blot
analysis and MTT assay, respectively. Results demonstrated that
A549 cells overexpressing MADD exhibited an increased survival rate
whereas silencing of MADD expression inhibited A549 cell viability.
Since MADD has no obvious effect on the cell cycle (35), we applied FCM to access the
influence of MADD on A549 cell apoptosis and found that MADD
obviously affected the apoptosis of A549 cells. The apoptosis rate
of MADD-overexpressing A549 cells was only 58.3% of that of the
control cells, whereas the apoptosis rate of the MADD-silenced A549
cells was 2.8 times that of control cells, indicating that MADD
strongly inhibits the apoptosis of A549 cells. These results
elucidated the function of MADD in lung ADC and provides an
important foundation and a promising future for improving the
effects of TRAIL therapy on lung ADC by regulation and control of
MADD expression and activity.
Acknowledgements
We would like to express our sincere appreciation
and gratitude to Dr B.S. Prabhakar and Dr L.C. Li of the Department
of Neurology at Illinois University School of Medicine for their
generosity in providing us with the primary antibody against MADD
and the pEYFP and pEYFP-MADD plasmids as well as the
pNL-SIN-GFP-SCR and pNL-SIN-GFP-MID lentiviral vectors.
References
|
1
|
Niemoeller OM and Belka C: Radiotherapy
and TRAIL for cancer therapy. Cancer Lett. July 12–2011.(Epub ahead
of print).
|
|
2
|
Johstone RW, Frew AJ and Smyth MJ: The
TRAIL apoptotic pathway in cancer onset, progression and therapy.
Nat Rev Cancer. 10:782–798. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kruyt FA: TRAIL and cancer therapy. Cancer
Lett. 263:14–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Al-Zoubi AM, Efimova EV, Kaithamana S,
Martinez O, El-Idrissi Mel-ADogan RE and Prabhakar BS: Contrasting
effects of IG20 and its splice isoforms, MADD and DENN-SV, on tumor
necrosis factor alpha-induced apoptosis and activation of caspase-8
and -3. J Biol Chem. 276:47202–47211. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li P, Jayarama S, Ganesh L, et al:
Akt-phosphorylated mitogen-activated kinase-activating death domain
protein (MADD) inhibits TRAIL-induced apoptosis by blocking
Fas-associated death domain (FADD) association with death receptor
4. J Biol Chem. 285:22713–22722. 2010. View Article : Google Scholar
|
|
6
|
Chow VT, Lim KM and Lim D: The human DENN
gene: genomic organization, alternative splicing, and localization
to chromosome 11p11.21–p11.22. Genome. 41:543–552. 1998.PubMed/NCBI
|
|
7
|
Efimova EV, Al-Zoubi AM, Martinez O, et
al: IG20, in contrast to DENN-SV, (MADD splice variants) suppresses
tumor cell survival, and enhances their susceptibility to apoptosis
and cancer drugs. Oncogene. 23:1076–1087. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mulherkar N, Prasad KV and Prabhakar BS:
MADD/DENN splice variant of the IG20 gene is a negative regulator
of caspase-8 activation. Knockdown enhances TRAIL-induced apoptosis
of cancer cells. J Biol Chem. 282:11715–11721. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mulherkar N, Ramaswamy M, Mordi DC and
Prabhakar BS: MADD/DENN splice variant of the IG20 gene is
necessary and sufficient for cancer cell survival. Oncogene.
25:6252–6261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schievella AR, Chen JH, Graham JR and Lin
LL: MADD, a novel death domain protein that interacts with the type
1 tumor necrosis factor receptor and activates mitogen-activated
protein kinase. J Biol Chem. 272:12069–12075. 1997. View Article : Google Scholar
|
|
11
|
Lee MT, Coburn GA, McClure MO and Cullen
BR: Inhibition of human immunodeficiency virus type 1 replication
in primary macrophages by using Tat- or CCR5-specific small
interfering RNAs expressed from a lentivirus vector. J Virol.
77:11964–11972. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Demirhan O, Tastemir D, Hastürk S, Kuleci
S and Hanta I: Alterations in p16 and p53 genes and chromosomal
findings in patients with lung cancer: fluorescence in situ
hybridization and cytogenetic studies. Cancer Epidemiol.
34:472–477. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee YL, Kuo WH, Lin CW, et al: Association
of genetic polymorphisms of CXCL12/SDF1 gene and its receptor,
CXCR4, to the susceptibility and prognosis of non-small cell lung
cancer. Lung Cancer. 73:147–152. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rosell R, Moran T, Queralt C, et al:
Screening for epidermal growth factor receptor mutations in lung
cancer. N Engl J Med. 361:958–967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pukac L, Kanakaraj P, Humphreys R, et al:
HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody,
induces cell death in multiple tumor types in vitro and in vivo. Br
J Cancer. 92:1430–1441. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
LeBlanc HN and Ashkenazi A: Apo2L/TRAIL
and its death and decoy receptors. Cell Death Differ. 10:66–75.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Riccioni R, Pasquini L, Mariani G, et al:
TRAIL decoy receptors mediate resistance of acute myeloid leukemia
cells to TRAIL. Haematologica. 90:612–624. 2005.PubMed/NCBI
|
|
18
|
Morizot A, Mérino D, Lalaoui N, et al:
Chemotherapy overcomes TRAIL-R4-mediated TRAIL resistance at the
DISC level. Cell Death Differ. 18:700–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L and Fang B: Mechanisms of
resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther.
12:228–237. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hopkins-Donaldson S, Ziegler A, Kurtz S,
et al: Silencing of death receptor and caspase-8 expression in
small cell lung carcinoma cell lines and tumors by DNA methylation.
Cell Death Differ. 10:356–364. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ray S, Bucur O and Almasan A:
Sensitization of prostate carcinoma cells to Apo2L/TRAIL by a Bcl-2
family protein inhibitor. Apoptosis. 10:1411–1418. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trauzold A, Siegmund D, Schniewind B, et
al: TRAIL promotes metastasis of human pancreatic ductal
adenocarcinoma. Oncogene. 25:7434–7439. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cunningham SJ: Cloning and
characterization of a novel cDNA isolated from human β cells.
Doctoral dissertation. The University of Texas, Medical Branch;
1996
|
|
24
|
Chow VT and Lee SS: DENN, a novel human
gene differentially expressed in normal and neoplastic cells. DNA
Seq. 6:263–273. 1996.PubMed/NCBI
|
|
25
|
Li LC, Sheng JR, Mulherkar N, Prabhakar BS
and Meriggioli MN: Regulation of apoptosis and caspase-8 expression
in neuroblastoma cells by isoforms of the IG20 gene. Cancer Res.
68:7352–7361. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyoshi J and Takai Y: Dual role of
DENN/MADD (Rab3GEP) in neurotransmission and neuroprotection.
Trends Mol Med. 10:476–480. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Niwa S, Tanaka Y and Hirokawa N:
KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic
regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD.
Nat Cell Biol. 10:1269–1279. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yamaguchi K, Tanaka M, Mizoguchi A, et al:
A GDP/GTP exchange protein for the Rab3 small G protein family
up-regulates a postdocking step of synaptic exocytosis in central
synapses. Proc Natl Acad Sci USA. 99:14536–14541. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Del Villar K and Miller CA:
Down-regulation of DENN/MADD, a TNF receptor binding protein,
correlates with neuronal cell death in Alzheimer’s disease brain
and hippocampal neurons. Proc Natl Acad Sci USA. 101:4210–4215.
2004.PubMed/NCBI
|
|
30
|
Kurada BR, Li LC, Mulherkar N, Subramanian
M, Prasad KV and Prabhakar BS: MADD, a splice variant of IG20, is
indispensable for MAPK activation and protection against apoptosis
upon tumor necrosis factor-alpha treatment. J Biol Chem.
284:13533–13541. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Allaire PD, Marat AL, Dall’Armi C, Di
Paolo G, McPherson PS and Ritter B: The connecdenn DENN domain: a
GEF for Rab35 mediating cargo-specific exit from early endosomes.
Mol Cell. 37:370–382. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Prabhakar BS, Mulherkar N and Prasad KV:
Role of IG20 splice variants in TRAIL resistance. Clin Cancer Res.
14:347–351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Coppola T, Perret-Menoud V, Gattesco S, et
al: The death domain of Rab3 guanine nucleotide exchange protein in
GDP/GTP exchange activity in living cells. Biochem J. 362:273–279.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lim KM, Yeo WS and Chow VT: Antisense
abrogation of DENN expression induces apoptosis of leukemia cells
in vitro, causes tumor regression in vivo and alters the
transcription of genes involved in apoptosis and the cell cycle.
Int J Cancer. 109:24–37. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Efimova E, Martinez O, Lokshin A, Arima T
and Prabhakar BS: IG20, a MADD splice variant, increases cell
susceptibility to gamma-irradiation and induces soluble mediators
that suppress tumor cell growth. Cancer Res. 63:8768–8776.
2003.PubMed/NCBI
|