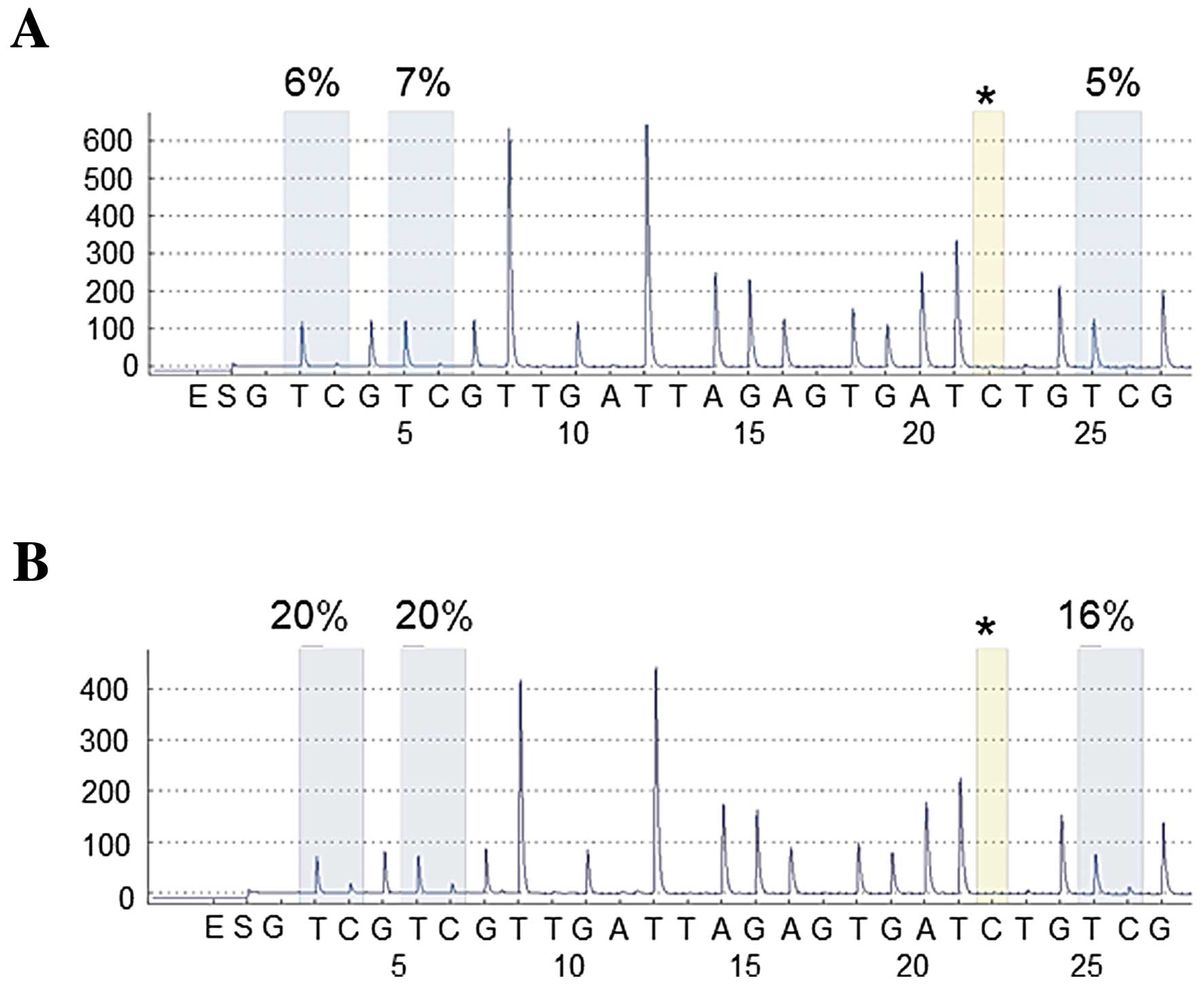
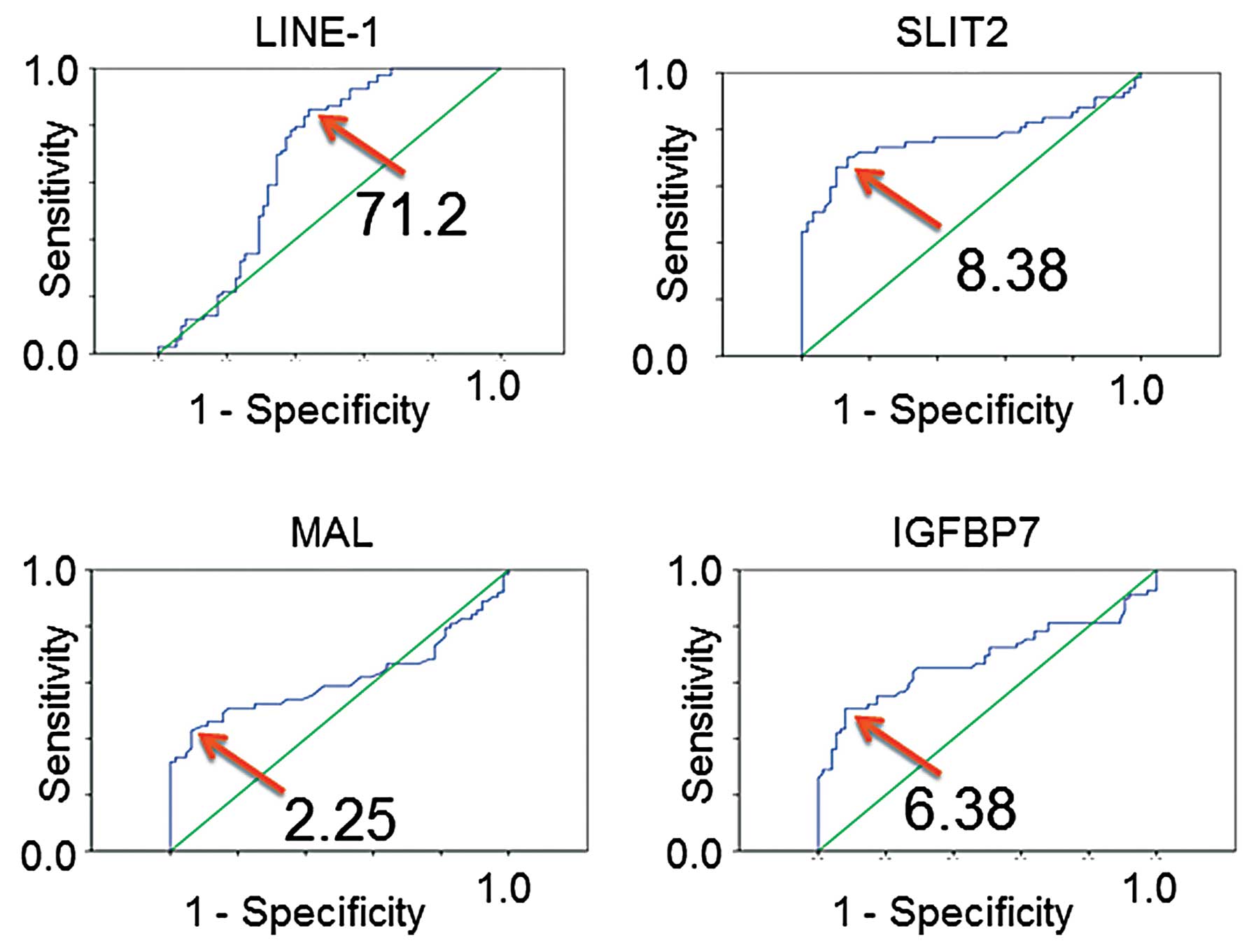
|
1
|
Takai D and Jones PA: Comprehensive
analysis of CpG islands in human chromosomes 21 and 22. Proc Natl
Acad Sci USA. 99:3740–3745. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suzuki M and Yoshino I: Aberrant
methylation in non-small cell lung cancer. Surg Today. 40:602–607.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gaudet F, Hodgson JG, Eden A, et al:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herman JG, Graff JR, Myohanen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lo YM, Wong IH, Zhang J, Tein MS, Ng MH
and Hjelm NM: Quantitative analysis of aberrant p16 methylation
using real-time quantitative methylation-specific polymerase chain
reaction. Cancer Res. 59:3899–3903. 1999.PubMed/NCBI
|
|
6
|
Xiong Z and Laird PW: COBRA: a sensitive
and quantitative DNA methylation assay. Nucleic Acids Res.
25:2532–2534. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suzuki M, Toyooka S, Shivapurkar N, et al:
Aberrant methylation profile of human malignant mesotheliomas and
its relationship to SV40 infection. Oncogene. 24:1302–1308. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takahashi T, Shivapurkar N, Riquelme E, et
al: Aberrant promoter hypermethylation of multiple genes in
gallbladder carcinoma and chronic cholecystitis. Clin Cancer Res.
10:6126–6133. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tamura H, Suzuki M, Moriya Y, et al:
Aberrant methylation of N-methyl-D-aspartate receptor type 2B
(NMDAR2B) in non-small cell carcinoma. BMC Cancer. 11:2202011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tian L, Suzuki M, Nakajima T, et al:
Clinical significance of aberrant methylation of prostaglandin E
receptor 2 (PTGER2) in nonsmall cell lung cancer: association with
prognosis, PTGER2 expression, and epidermal growth factor receptor
mutation. Cancer. 113:1396–1403. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ronaghi M: Pyrosequencing sheds light on
DNA sequencing. Genome Res. 11:3–11. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sepulveda AR, Jones D, Ogino S, et al: CpG
methylation analysis - current status of clinical assays and
potential applications in molecular diagnostics: a report of the
Association for Molecular Pathology. J Mol Diagn. 11:266–278. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iwagami S, Baba Y, Watanabe M, et al:
Pyrosequencing assay to measure LINE-1 methylation level in
esophageal squamous cell carcinoma. Ann Surg Oncol. 19:2726–2732.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamaji H, Iizasa T, Koh E, et al:
Correlation between interleukin 6 production and tumor
proliferation in non-small cell lung cancer. Cancer Immunol
Immunother. 53:786–792. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suzuki M, Sunaga N, Shames DS, Toyooka S,
Gazdar AF and Minna JD: RNA interference-mediated knockdown of DNA
methyltransferase 1 leads to promoter demethylation and gene
re-expression in human lung and breast cancer cells. Cancer Res.
64:3137–3143. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baba Y, Huttenhower C, Nosho K, et al:
Epigenomic diversity of colorectal cancer indicated by LINE-1
methylation in a database of 869 tumors. Mol Cancer. 9:1252010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshida K, Yatabe Y, Park JY, et al:
Prospective validation for prediction of gefitinib sensitivity by
epidermal growth factor receptor gene mutation in patients with
non-small cell lung cancer. J Thorac Oncol. 2:22–28. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moses LE, Shapiro D and Littenberg B:
Combining independent studies of a diagnostic test into a summary
ROC curve: data-analytic approaches and some additional
considerations. Stat Med. 12:1293–1316. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Saito K, Kawakami K, Matsumoto I, Oda M,
Watanabe G and Minamoto T: Long interspersed nuclear element 1
hypomethylation is a marker of poor prognosis in stage IA non-small
cell lung cancer. Clin Cancer Res. 16:2418–2426. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dallol A, Da Silva NF, Viacava P, et al:
SLIT2, a human homologue of the Drosophila Slit2 gene, has
tumor suppressor activity and is frequently inactivated in lung and
breast cancers. Cancer Res. 62:5874–5880. 2002.
|
|
21
|
Kwon YJ, Lee SJ, Koh JS, et al:
Genome-wide analysis of DNA methylation and the gene expression
change in lung cancer. J Thorac Oncol. 7:20–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suzuki M, Wada H, Yoshino M, et al:
Molecular characterization of chronic obstructive pulmonary
disease-related non-small cell lung cancer through aberrant
methylation and alterations of EGFR signaling. Ann Surg Oncol.
17:878–888. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Puertollano R, Martin-Belmonte F, Millan
J, et al: The MAL proteolipid is necessary for normal apical
transport and accurate sorting of the influenza virus hemagglutinin
in Madin-Darby canine kidney cells. J Cell Biol. 145:141–151. 1999.
View Article : Google Scholar
|
|
24
|
Mimori K, Shiraishi T, Mashino K, et al:
MAL gene expression in esophageal cancer suppresses motility,
invasion and tumorigenicity and enhances apoptosis through the Fas
pathway. Oncogene. 22:3463–3471. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lind GE, Ahlquist T, Kolberg M, et al:
Hypermethylated MAL gene - a silent marker of early colon
tumorigenesis. J Transl Med. 6:132008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Buffart TE, Overmeer RM, Steenbergen RD,
et al: MAL promoter hypermethylation as a novel prognostic marker
in gastric cancer. Br J Cancer. 99:1802–1807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Overmeer RM, Henken FE, Bierkens M, et al:
Repression of MAL tumour suppressor activity by promoter
methylation during cervical carcinogenesis. J Pathol. 219:327–336.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ruan W, Xu E, Xu F, et al: IGFBP7 plays a
potential tumor suppressor role in colorectal carcinogenesis.
Cancer Biol Ther. 6:354–359. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vizioli MG, Sensi M, Miranda C, et al:
IGFBP7: an oncosuppressor gene in thyroid carcinogenesis. Oncogene.
29:3835–3844. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mutaguchi K, Yasumoto H, Mita K, et al:
Restoration of insulin-like growth factor binding protein-related
protein 1 has a tumor-suppressive activity through induction of
apoptosis in human prostate cancer. Cancer Res. 63:7717–7723.
2003.
|
|
31
|
Smith P, Nicholson LJ, Syed N, et al:
Epigenetic inactivation implies independent functions for
insulin-like growth factor binding protein (IGFBP)-related protein
1 and the related IGFBPL1 in inhibiting breast cancer phenotypes.
Clin Cancer Res. 13:4061–4068. 2007. View Article : Google Scholar
|
|
32
|
Ye F, Chen Y, Knosel T, et al: Decreased
expression of insulin-like growth factor binding protein 7 in human
colorectal carcinoma is related to DNA methylation. J Cancer Res
Clin Oncol. 133:305–314. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Y, Cui T, Knosel T, Yang L, Zoller K
and Petersen I: IGFBP7 is a p53 target gene inactivated in human
lung cancer by DNA hypermethylation. Lung Cancer. 73:38–44. 2011.
View Article : Google Scholar : PubMed/NCBI
|