Introduction
Colorectal cancer (CRC) is a common malignant tumor
worldwide, and over 1.2 million new cases and 608,700 deaths were
estimated to have occurred in 2008 (1). The incidence of CRC in China has
increased rapidly since the 1980s (2,3).
Currently, CRC is the fifth leading cause of cancer-related deaths
(4).
CRC can be divided into two types based on genetic
abnormalities (5,6). The major type is the chromosomal
instability phenotype, which consists of more than 85% of CRCs and
is characterized by frequent chromosomal imbalances. The minor type
is the microsatellite instability phenotype, which exhibits
microsatellite instability owing to DNA replication errors and
comprises <15% of CRCs. The genomic instability of the two types
can lead to DNA copy number aberrations.
Cancers occur as a result of the accumulation of
genetic alterations that are associated with carcinogenesis
(5,7). Thus, the study of the cancer genome by
high-resolution and high-throughput technology, for example
array-based comparative genomic hybridization (array-CGH), not only
has the ability to clarify the relationship between genomic
abnormalities and clinicopathological factors, but may also
optimize the treatment of patients by using their cancer genome
information. Although several DNA copy number aberrations have been
reported to have linkage with clinicopathological characteristics
of patients with CRC (8–10), the available information is still
limited particularly in Chinese patients.
The present study identified frequent DNA copy
number changes in rectal cancer samples from Chinese patients and
candidate target genes using integrated analysis of the genome and
gene expression data of NCI-60 cell lines. We evaluated the genetic
changes associated with lymph node metastasis, tumor stage and
distant metastasis using the methods of frequency plot comparison
together with statistical analysis.
Materials and methods
Study design
This study was conducted to identify genomic changes
associated with clinicopathological factors and candidate targets
of most frequent gains and losses of rectal cancer. We determined
the genetic aberrations in 48 rectal carcinomas using Agilent 60K
Human Genome CGH microarray and screened those linked with
clinicopathological characteristics. We then compared the gene
expression profiling of CRC cell lines with or without gains of 8q,
13q, 17q, 20p, 20q or losses of 8p, 11q, 18q and identified genes
whose expression was linked with DNA copy number changes.
Patients and samples
Fresh tissues from 48 rectal carcinoma patients were
collected at the Department of Pathology, Cancer Hospital, Peking
Union Medical College and Chinese Academy of Medical Sciences,
Beijing, China. None of the patients received either irradiation or
chemotherapy prior to surgery. All of the samples used in this
study were residual specimens after diagnostic sampling. Every
patient signed a separate informed consent form for sampling and
research. Ethics approval was obtained from the Ethics Committee of
Cancer Institute and Hospital, Chinese Academy of Medical Sciences.
Representative tumor regions were excised by experienced
pathologists. The clinicopathologic characteristics of the patients
are summarized in Table I.
 | Table IClinicopathological characteristics of
the rectal cancer patients in the array-CGH study. |
Table I
Clinicopathological characteristics of
the rectal cancer patients in the array-CGH study.
| Patient no. | Gender | Age (years) | TNM | pStage |
|---|
| 1 | M | 67 | T3N1M0 | III B |
| 2 | M | 58 | T3N1M0 | III B |
| 3 | M | 43 | T4N2M1 | III C |
| 4 | F | 63 | T3N2M0 | III C |
| 5 | F | 69 | T4N2M0 | III C |
| 6 | F | 52 | T3N2M0 | III C |
| 7 | M | 56 | T3N2M1 | III C |
| 8 | F | 47 | T4N2M0 | III C |
| 9 | F | 61 | T4N2M0 | III C |
| 10 | M | 41 | T3N2M1 | III C |
| 11 | M | 58 | T3N2M1 | IV |
| 12 | F | 62 | T3N1M1 | IV |
| 13 | F | 36 | T3N0M1 | IV |
| 14 | M | 59 | T3N0M1 | IV |
| 15 | M | 49 | T3N1M1 | IV |
| 16 | M | 53 | T3N2M1 | IV |
| 17 | M | 75 | T3N2M1 | IV |
| 18 | F | 79 | T3N2M0 | III C |
| 19 | F | 22 | T4N2M0 | III C |
| 20 | F | 70 | T4N2M0 | III C |
| 21 | M | 77 | T4N2M0 | III C |
| 22 | M | 36 | T3N1M0 | III B |
| 23 | M | 49 | T3N0M0 | II A |
| 24 | M | 45 | T4N0M0 | II B |
| 25 | M | 61 | T3N0M0 | II A |
| 26 | M | 51 | T3N0M0 | II A |
| 27 | F | 66 | T3N0M1 | II A |
| 28 | M | 59 | T3N0M1 | II A |
| 29 | M | 45 | T3N0M0 | II A |
| 30 | M | 72 | T3N0M0 | II A |
| 31 | M | 63 | T3N0M0 | II A |
| 32 | M | 58 | T3N0M0 | II A |
| 33 | M | 68 | T3N0M0 | II A |
| 34 | M | 63 | T3N0M0 | II A |
| 35 | M | 63 | T3N0M0 | II A |
| 36 | M | 55 | T3N0M0 | II A |
| 37 | M | 42 | T3N0M0 | II A |
| 38 | F | 56 | T3N0M0 | II A |
| 39 | F | 52 | T4N0M0 | II B |
| 40 | M | 45 | T4N0M0 | II B |
| 41 | M | 47 | T4N0M0 | II B |
| 42 | M | 62 | T4N0M0 | II B |
| 43 | M | 64 | T3N1M0 | III B |
| 44 | F | 53 | T4N1M0 | III B |
| 45 | F | 59 | T3N1M0 | III B |
| 46 | F | 49 | T3N1M0 | III B |
| 47 | F | 39 | T3N1M0 | III B |
| 48 | M | 63 | T3N1M1 | III B |
Genomic DNA extraction
Genomic DNA was isolated from tumor tissues using
the Qiagen DNeasy Blood and Tissue kit as described by the
manufacturer (Qiagen, Hilden, Germany). Tumor cell content of all
the samples was >50% as determined by H&E staining.
Array-based CGH
Array-CGH experiments were performed using standard
Agilent protocols (Agilent Technologies, Santa Clara, CA, USA).
Commercial human genomic DNA (Promega, Warrington, UK) was used as
reference. For each CGH hybridization, 400 ng of reference genomic
DNA and the same amount of tumor DNA were digested with AluI
and RSAI restriction enzymes (Promega). The digested
reference DNA fragments were labeled with cyanine 3-dUTP and the
tumor DNA with cyanine 5-dUTP (Agilent Technologies). After
clean-up, reference and tumor DNA probes were mixed and hybridized
onto an Agilent 60K human genome CGH microarray (Agilent
Technologies) for 24 h. Washing, scanning and data extraction
procedures were performed following standard protocols.
Microarray data analysis
Microarray data were analyzed using Agilent Genomic
Workbench (Agilent Technologies), CGH ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html) and
MD-SeeGH (www.flintbox.ca). Agilent Genomic Workbench was
used to calculate the log2ratio for every probe and identified
genomic aberrations. Mean Log2ratio of all probes in a chromosome
region between 0.25 and 0.75 was classified as a genomic gain,
>0.75 as high-level DNA amplification, <−0.25 as a hemizygous
loss, and <−0.75 as a homozygous deletion. Pathway enrichment
analyses were performed by CGH ArrayTools.
Integration analysis of DNA copy number
and gene expression data of the NCI 60 cell lines
The DNA copy number and gene expression data of the
NCI 60 cell lines were obtained from CellMiner (http://discover.nci.nih.gov/cellminer).
We selected the data sets of aCGH Agilent 44K and Agilent mRNA for
analysis. The genetic changes of seven CRC cell lines (including
colo205, HCT_116, HCT_15, KM12, HCC_2998, HT29 and SW_620) were
analyzed and divided into the gain/loss group and no change group.
The differentially expressed genes between the two groups were
indentified with a cutoff of a 2-fold change using GeneSpring GX
(Agilent Technologies).
Oncomine data analysis
The mRNA expression of genes which had a >5-fold
change in expression between the gain/loss CRC cell lines and the
no change cell lines was analyzed using Oncomine database
(https://www.oncomine.org/resource/login.html). Details
of standardized normalization techniques and statistical methods
can be found on the the Oncomine website. The data of the
interested genes in different types of cancer were collected and
then their expression status in CRC was analyzed.
Statistical analysis
The Chi-square test was used to analyze the
significance of correlation between genomic aberrations and
clinical factors. Differences were considered significant at
P<0.05.
Results
Recurrent copy number alterations in
rectal carcinoma detected by array-CGH
Forty eight samples of rectal carcinoma were
analyzed in this study and all of them had genomic changes. The
most frequent genomic aberrations were gains of 8q24.3,
20q11.21-q13.32, 20q13.33, and losses of 8p23.3-p12, 17p13.1-p12
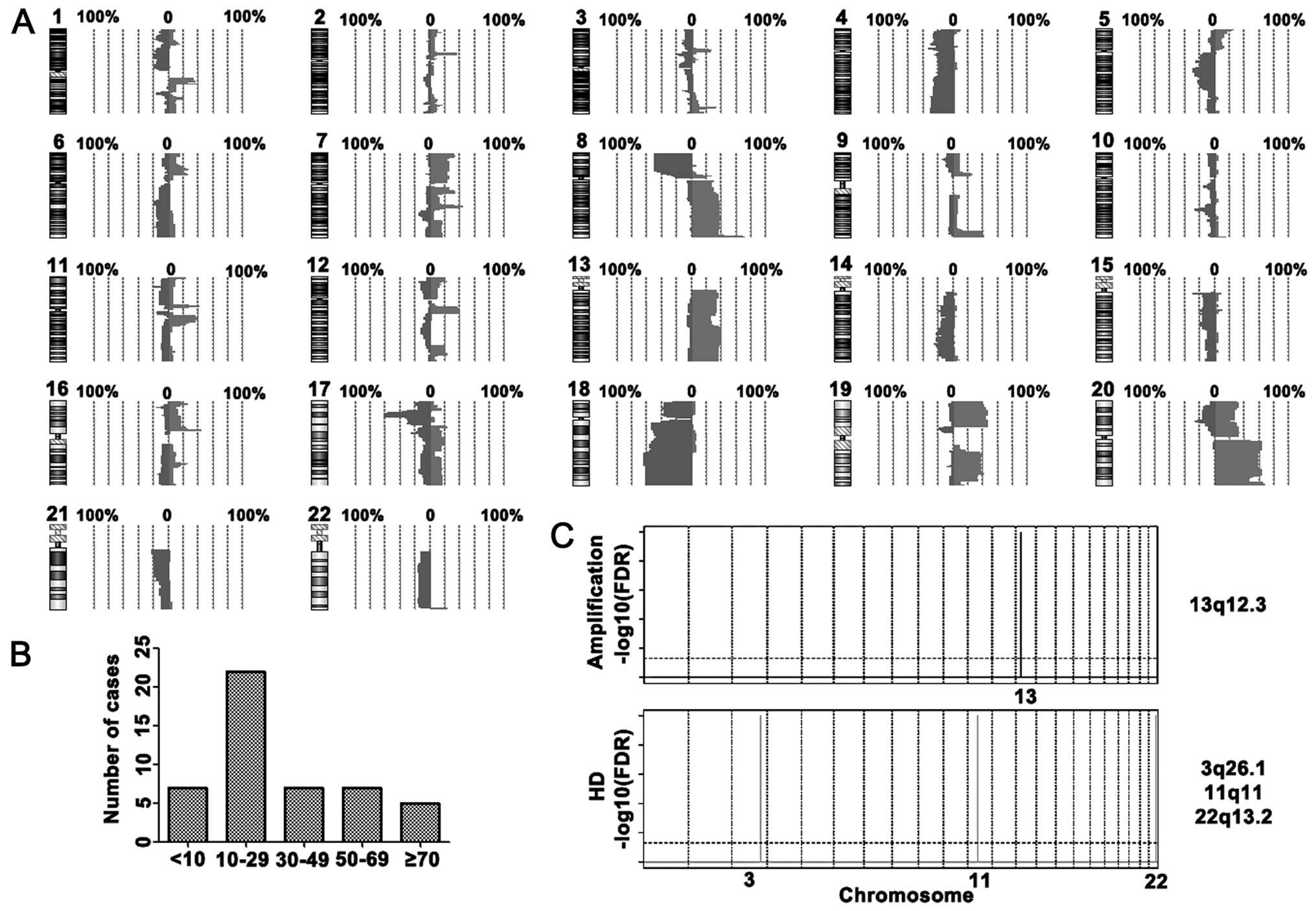
and 18q11.2-q23 (Fig. 1A).
High-level amplifications were detected at 14 chromosome regions
including 7p22.3-p21.3, 7q22.1, 8p11.21, 8p11.23, 8q22.1, 8q24.3,
13q12.2, 13q14.2-q14.3, 13q31.3, 16p11.2, 19p13.2, 20p11.23,
20q11.21-q11.23 and 20q13.33. Homozygous deletions were identified
in 5q14.3, 8p23.3-p21.2, 15q11.2, 16p13.2, 17p13.1-p12, 18q21.2 and
20p12.1 (Table II). After
analyzing the number of changes in rectal cancer, we found that
nearly half of cases in the array-CGH study had 10 to 29 genetic
alterations (Fig. 1B). GISTIC
analysis showed that a copy number increase of MTUS2 (13q12.3) and
decrease of C3orf57 (3q26.1), SPRYD5 (11q11), OR5W2 (11q11) and
MKL1 (22q13.2) were significant in the rectal cancer cases
(Fig. 1C).
 | Table IIHigh-level amplifications and
homozygous deletions in the rectal cancer cases. |
Table II
High-level amplifications and
homozygous deletions in the rectal cancer cases.
| Changes | Cytoband | Start | End | No. of probes | No. of cases | Candidates |
|---|
| Amp | 7q22.1 | 99852752 | 100767476 | 42 | 5 | MUC17 |
| 7p22.3-p21.3 | 524935 | 7428910 | 128 | 3 | FSCN1 |
| 8q24.3 | 145210837 | 145782038 | 24 | 11 | FOXH1 |
| 8q22.1 | 95061529 | 97342088 | 45 | 4 | CDH17 |
| 8p11.23 | 39378051 | 39461834 | 3 | 8 | ADAM5P, ADAM3A |
| 8p11.21 | 42816942 | 42849186 | 3 | 4 | THAP1, RNF170 |
| 13q12.2 | 27095352 | 27439560 | 11 | 4 | CDX2 |
| 13q14.2-q14.3 | 46790942 | 51899157 | 117 | 4 | ALG11 |
| 13q31.3 | 91075476 | 91176960 | 4 | 4 | GPC5 |
| 16p11.2 | 29890929 | 31412127 | 78 | 4 | MAPK3 |
| 19p13.2 | 11141557 | 11548932 | 29 | 3 | CNN1 |
|
20q11.21-q11.23 | 29501535 | 35014201 | 155 | 10 | |
| 20q13.33 | 60008660 | 62320720 | 85 | 6 | PTK6, TNFRSF6B |
| 20p11.23 | 18692429 | 20864752 | 42 | 3 | |
| HD | 5q14.3 | 87722621 | 90788169 | 48 | 3 | MEF2C |
| 8p23.3-p21.2 | 211611 | 27199611 | 467 | 3 | DLC1, PCM1 |
| 15q11.2 | 18835660 | 20010618 | 12 | 5 | |
| 16p13.2 | 6492886 | 6860972 | 9 | 3 | A2BP1 |
| 17p13.1-p12 | 11089880 | 15073870 | 69 | 3 | MAP2K4 |
| 18q21.2 | 46764796 | 47107764 | 9 | 3 | SMAD4 |
| 18q21.2 | 48023000 | 48423627 | 8 | 3 | DCC |
| 20p12.1 | 14772372 | 14939552 | 4 | 4 | MACROD2 |
Genomic changes associated with lymph
node metastasis, tumor stage and distant metastasis
We compared the frequencies of genomic aberrations
in the rectal cancer patients subdivided accoding to cases with or
without lymph node metastasis, with tumor stages II or III-IV, and
with or without distant metastasis using MD-SeeGH software. The
results showed that gains of 7p, 13q and losses of 4p, 4q and 8p
were more frequent in the rectal cancers with lymph node metastasis
(Fig. 2A). The frequencies of 7p,
13q gain and 4q loss were higher in stage III-IV cases when
compared with the frequencies in stage II cases (Fig. 2B). The largest differences were
detected in copy number changes of 4q and 17q, with more frequent
4q loss and 17q gain in the cases with distant metastasis (Fig. 2C). We analyzed these candidate
genomic regions with clinical factors by Chi-square test, and found
that losses of 4p16.1-p15.31, 8p21.1-p12 and gains of 7p12.3-p12.1
and 13q33.1-q34 were associated with positive lymph node metastasis
and advanced clinical stage (stages III and IV). Moreover, loss of
4q34.3-q35.1 was linked only with advanced stage (stages III and
IV). We also found that the patients with distant metastasis had
more frequent 17q24.2–25.3 gain (Table III).
 | Figure 2Frequency plot comparison. (A)
Frequency plot comparison between pN1 and pN0. Red, pN0 group;
green, pN1 group; yellow, shared by two groups. (B) Frequency plot
comparison between pStage II and pStage III-IV. Red, pStage II
group; green, pStage III-IV group; yellow, shared by two groups.
(C) Frequency plot comparison between pM1 and pM0. Red, pM0 group;
green, pM1 group; yellow, shared by two groups. The presentation is
per array probe; gains are represented by the colors on the right,
and losses are represented by the colors on the left. Vertical blue
line represents 100% of the samples. Red arrows highlight the
chromosomal areas with different frequency in two groups. |
 | Table IIIGenomic aberrations linked with
clinicopathological characteristics of the rectal cases. |
Table III
Genomic aberrations linked with
clinicopathological characteristics of the rectal cases.
| | pN status | Distant
metastasis | Stage |
|---|
| |
|
|
|
|---|
| Cytoband | Change | Positive | Negative | P-value | Positive | Negative | P-value | Stage II | Stages III and
IV | P-value |
|---|
| 4p16.1-p15.31 | Loss | 9 | 2 | 0.036 | 4 | 7 | 0.430 | 1 | 10 | 0.013 |
| No loss | 17 | 20 | | 9 | 28 | | 19 | 18 | |
| 4q34.3-q35.1 | Loss | 11 | 4 | 0.072 | 6 | 9 | 0.175 | 3 | 12 | 0.040 |
| No loss | 15 | 18 | | 7 | 26 | | 17 | 16 | |
| 7p12.3-p12.1 | Gain | 12 | 1 | 0.001 | 4 | 9 | 0.726 | 1 | 12 | 0.004 |
| No gain | 14 | 21 | | 9 | 26 | | 19 | 16 | |
| 8p21.1-p12 | Loss | 16 | 5 | 0.007 | 5 | 16 | 0.653 | 5 | 16 | 0.027 |
| No loss | 10 | 17 | | 8 | 19 | | 15 | 12 | |
| 13q33.1-q34 | Gain | 13 | 3 | 0.008 | 3 | 13 | 0.358 | 3 | 13 | 0.023 |
| No gain | 13 | 19 | | 10 | 22 | | 17 | 15 | |
| 17q24.2-q25.3 | Gain | 4 | 4 | 0.796 | 6 | 2 | 0.001 | 2 | 6 | 0.295 |
| No gain | 22 | 18 | | 7 | 33 | | 18 | 22 | |
Candidate target genes of gains and
losses in rectal carcinoma
We performed an integrated analysis of the array-CGH
dataset and the gene expression profiling dataset of CRC cells of
NCI-60 to identify the candidate target genes of genomic gains and
losses. The expression level of CA2, PDP1, ANGPT1, LOC346887, MAL2,
NOV, TRIB1 and ZNF572 was higher in cell lines with an 8q gain than
without. We also analyzed candidate target genes of gains in 13q,
17q, 20p and 20q and of loss in 18q (Table IV) and found that PDP1 (8q), TRIB1
(8q), C13orf27 (13q), FOXA2 (20p), PMEPA1 (20q) and PHACTR3 (20q)
were overexpressed not only in CRC but also in other types of
cancers (Table V). Three genes in
18q (FHOD, SMAD4 and BCL2) presented underexpression in several
types of cancers including CRC (Table
V).
 | Table IVCandidate target genes of gains and
losses in rectal cancer. |
Table IV
Candidate target genes of gains and
losses in rectal cancer.
| Change | Cytoband | Genes (>5-fold
change) |
|---|
| Gain | 8q | CA2, PDP1, ANGPT1,
LOC346887, MAL2, NOV, TRIB1, ZNF572 |
| 13q | OXGR1, PCDH9,
EFNB2, C13orf27, LMO7, ARL11 |
| 17q | TTYH2, RAB37,
MXRA7, SLC26A11, MGAT5B |
| 20p | FOXA2, C20orf56,
SLC24A3, CHGB, C20orf194, NRSN2 |
| 20q | PMEPA1, PCK1,
SULF2, LOC100240735, WFDC2, PHACTR3 |
| Loss | 8p | Not found |
| 11q | Not found |
| 18q | FHOD3, PSTPIP2,
SMAD4, MBD2, BCL2, ST8SIA5 |
 | Table VCandidate targets in the Oncomine
database. |
Table V
Candidate targets in the Oncomine
database.
| | Colorectal
cancer | |
|---|
| |
| |
|---|
| Cytoband | Gene | Up | Down | Change in other
cancers |
|---|
| 8q gain | PDP1 | 8 | 0 | Cervical cancer,
gastic cancer, head and neck cancer, kidney cancer, leukemia,
lymphoma, melanoma, pancreatic cancer |
| TRIB1 | 1 | 0 | Brain and CNS
cancer, breast cancer, esophageal cancer, head and neck cancer,
leukemia, lymphoma, melanoma, prostate cancer |
| ZNF572 | 2 | 0 | No |
| 13q gain | OXGR1 | 4 | 1 | No |
| C13orf27 | 5 | 0 | Cervical
cancer |
| 20p gain | FOXA2 | 6 | 1 | Esophageal
cancer |
| 20q gain | PMEPA1 | 9 | 0 | Bladder cancer,
breast cancer, esophageal cancer, gastric cancer, head and neck
cancer, lung cancer, pancreatic cancer |
| PHACTR3 | 2 | 0 | Brain and CNS
cancer, pancreatic cancer |
| 18q loss | FHOD3 | 0 | 1 | Bladder cancer,
brain and CNS cancer, breast cancer, kidney cancer, prostate
cancer |
| SMAD4 | 0 | 2 | Lymphoma |
| BCL2 | 0 | 14 | Bladder cancer,
brain and CNS cancer, breast cancer, head and neck cancer,
leukemia, lymphoma, ovarian cancer, prostate cancer, sarcoma |
Pathways enriched for copy number
alterations
Pathway enrichment analysis using KEGG database was
applied to the CGH data, and we found four pathways enriched in
genes with gain and three pathways enriched in genes with loss. The
genomic gains in rectal carcinoma changed the pathways of nitrogen
metabolism, oxidative phosphorylation, cell cycle and maturity
onset diabetes of young. However, cytokine-cytokine receptor
interaction, MAPK signaling pathway, and dentatorubropallidoluysian
atrophy (DRPLA) pathways were changed by the genomic losses
(Table VI).
 | Table VIPathways enriched in the array-CGH
data. |
Table VI
Pathways enriched in the array-CGH
data.
| Aberration | Pathway | Description | No. of genes | Gene symbol | P-value |
|---|
| Gain | hsa00910 | Nitrogen
metabolism | 22 | CA6, CTH, CA14,
GLUL, GLS, CPS1, AMT, ASNS, CA8, CA13, CA1, CA3, CA2, CA9, GLUD1,
GLS2, HAL, CA12, CA7, CA5A, CA4, CA11 | 0.003 |
| Gain | hsa00190 | Oxidative
phosphorylation | 109 | SDHB, NDUFS5,
ATP6V0B, UQCRH, ATP5F1, NDUFS2, SDHC, ATP6V1G3, ATP6V1C2,
COX7A2L | 0.017 |
| Gain | hsa04110 | Cell cycle | 108 | MAD2L2, SFN, HDAC1,
CDC20, CDKN2C, ORC1, GADD45A, CDC7, CDC14A, TGFB2 | 0.025 |
| Gain | hsa04950 | Maturity onset
diabetes of the young | 24 | PKLR, NR5A2,
NEUROD1, SLC2A2, HES1, NKX6-1, GCK, BHLHA15, PAX4, MNX1, HNF4G,
MAFA, NEUROG3, HHEX, PAX6, IAPP, HNF1A, PDX1, ONECUT1, HNF1B,
FOXA3, NKX2-2, FOXA2, HNF4A | 0.044 |
| Loss | hsa04060 | Cytokine-cytokine
receptor interaction | 240 | TNFRSF18, TNFRSF4,
TNFRSF14, TNFRSF25, TNFRSF9, TNFRSF8, TNFRSF1B, IL22RA1, IL28RA,
CSF3R | 0.001 |
| Loss | hsa04010 | MAPK signaling
pathway | 256 | CASP9, PLA2G2E,
PLA2G2A, PLA2G5, PLA2G2D, PLA2G2F, CDC42, STMN1, RPS6KA1,
MAP3K6 | 0.011 |
| Loss | hsa05050 |
Dentatorubropallidoluysian atrophy
(DRPLA) | 14 | RERE, CASP8, MAGI1,
CASP3, MAGI2, WWP1, CASP7, CASP1, GAPDH, ATN1, WWP2, BAIAP2, INSR,
ITCH | 0.038 |
Discussion
The biological properties of cancers are different
in patients presenting with different clinical parameters such as
invasive depth, lymph node metastasis, distant metastasis,
differentiation and clinical stage. Thus, the optimal treatment
should be based on an individual cancer. Biomarkers can improve the
accuracy of determining the clinical parameters that are predictors
of prognosis and indicators of a response to treatment.
By applying array-CGH to rectal carcinoma samples of
Chinese parients, we screened the genomic aberrations associated
with clinical parameters using frequency comparison. The results
showed that losses of 4p16.1-p15.31, 8p21.1-p12 and gains of
7p12.3-p12.1 and 13q33.1-q34 were associated with positive lymph
node metastasis and advanced clinical stage (stages III and IV).
Loss of 4q34.3-q35.1 was a marker of advanced stage (stages III and
IV). We also found that the patients with distant metastasis had a
more frequent gain in 17q24.2–25.3. Chromosome 4 was found to be
the most frequent loss region in cancers including cervical,
esophageal, lung, head and neck, gastric and CRC (11–16).
The incidence of 4q loss was much higher in pulmonary metastatic
tissues when compared with primary cancer tissues and LOH of the
D4S1534 locus (4q) in primary tissues was significantly linked with
liver metastasis (17,18). Our results also suggested that loss
of 4q in primary rectal tissues was a candidate predictor of lymph
node metastasis. This indicates that target genes of the loss of 4q
play important roles in lymphatic invasion and tumor progression,
to which further investigation should be addressed.
The correlation of 17q with clinicopathological
factors of CRC was not explicit. Diep et al(7) found that gain of 17q was correlated
with the transition from a primary tumor to liver metastasis, while
Knosel et al(16) reported
that more deletion at 17q were observed in lung metastasis than
primary tumors (19). Our results
showed that the gain of 17q was more frequent in rectal cancer
patients with distant metastasis when compared with patients
without metastasis. Additional independent validation assays should
be performed to reveal the correlation of 17q and metastasis.
Our results revealed that the alteration in
expression of PDP1 (8q), TRIB1 (8q), C13orf27 (13q), FOXA2 (20p),
PMEPA1 (20q), PHACTR3 (20q), FHOD (18q), SMAD4 (18q) and BCL2 (18q)
occurred in CRC and other types of cancer, with a consistent copy
number increase or decrease. To date, there is no report concerning
the function of PDP1, C13orf27 and FHOD in cancer. Our results
indicate the need to study these genes in rectal carcinogenesis.
TRIB1 is a mammalian homolog of tribbles, an evolutionarily
conserved Drosophila protein family that regulates protein
degradation. In myeloid leukemogenesis, TRIB1 was found to be
overexpressed and was a key mediator between the RTK-MAPK pathway
and the C/EBP transcription factor (20,21).
FOXA2 was found to function as a suppressor of tumor metastasis by
inhibition of epithelial-to-mesenchymal transition (EMT). Loss of
FOXA2 expression due to epigenetic silencing was frequent in lung
cancer (22,23). PMEPA1 is a TGF-β inducible gene and
encodes an NEDD4 E3 ubiquitin ligase binding protein. PMEPA1 was
found to be overexpressed in prostate, breast, renal cell, stomach
and rectal carcinomas (24–26). PHACTR3 was identified as a
PP1-binding protein and was selectively expressed in the brain.
PHACTR3 was found to be overexpressed in 20% of non-small cell lung
cancer (NSCLC), and was associated with reduced survival time of
patients. In advanced neoplasia the methylation level of PHACTR3
was 70-fold higher than that in normal colon mucosa, and the
sensitivity and specificity in stool assay were 55 and 95%,
respectively (27). Loss of SMAD4
expression was reported as a predictor of liver metastasis in CRC,
and patients with reduced SMAD4 expression presented with a poor
prognosis (28–30). Transgenic expression of SMAD4 was
found to significantly reduce the oncogenic potential of SW620 and
MC38 cell lines (31). Overall,
these genes may be the target genes of genomic gains and losses in
rectal cancer. Further research should be addressed to elucidate
the roles of the candidate genes in rectal carcinogenesis.
In summary, the genomic aberrations identified in
the present study can be suggested as candidate biomarkers with
which to predict the clinical outcome of patients with rectal
carcinoma and may be expected to serve to individualize the
treatment of rectal cancer. Our study identified several candidate
target genes of the most common gains and losses in rectal cancer,
and our findings provide information to explore the role of these
genes in the development and progression of rectal cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (30950013) and the Special Public
Health Fund of China (200902002-5).
References
|
1
|
Ministry of Health, PCR. Chinese Health
Statistical Digest. 2010, Available from: http://www.moh.gov.cn/publicfiles//business/htmlfiles/zwgkzt/ptjty/digest2010/index.html.
|
|
2
|
Alazzouzi H, Alhopuro P, Salovaara R,
Sammalkorpi H, Jarvinen H, Mecklin JP, Hemminki A, Schwartz S Jr,
Aaltonen LA and Arango D: SMAD4 as a prognostic marker in
colorectal cancer. Clin Cancer Res. 11:2606–2611. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Basseres DS, D’Alo F, Yeap BY, Lowenberg
EC, Gonzalez DA, Yasuda H, Dayaram T, Kocher ON, Godleski JJ,
Richards WG, Meyerson M, Kobayashi S, Tenen DG, Halmos B and Costa
DB: Frequent downregulation of the transcription factor Foxa2 in
lung cancer through epigenetic silencing. Lung Cancer. 77:31–37.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bockmuhl U, Schmidt S, Petersen S and
Petersen I: Deletion of chromosome 10q - a marker for metastasis of
head-neck carcinomas? Laryngorhinootologie. 79:81–85. 2000.(In
German).
|
|
5
|
Bosch LJ, Oort FA, Neerincx M, Khalid-de
Bakker CA, Terhaar sive Droste JS, Melotte V, Jonkers DM, Masclee
AA, Mongera S, Grooteclaes M, Louwagie J, van Criekinge W, Coupe
VM, Mulder CJ, van Engeland M, Carvalho B and Meijer GA: DNA
methylation of phosphatase and actin regulator 3 detects colorectal
cancer in stool and complements FIT. Cancer Prev Res. 5:464–472.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dellas A, Torhorst J, Jiang F, Proffitt J,
Schultheiss E, Holzgreve W, Sauter G, Mihatsch MJ and Moch H:
Prognostic value of genomic alterations in invasive cervical
squamous cell carcinoma of clinical stage IB detected by
comparative genomic hybridization. Cancer Res. 59:3475–3479.
1999.PubMed/NCBI
|
|
7
|
Diep CB, Kleivi K, Ribeiro FR, Teixeira
MR, Lindgjaerde OC and Lothe RA: The order of genetic events
associated with colorectal cancer progression inferred from
meta-analysis of copy number changes. Genes Chromosomes Cancer.
45:31–41. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Giannini G, Ambrosini MI, Di Marcotullio
L, Cerignoli F, Zani M, MacKay AR, Screpanti I, Frati L and Gulino
A: EGF- and cell-cycle-regulated STAG1/PMEPA1/ERG1.2 belongs to a
conserved gene family and is overexpressed and amplified in breast
and ovarian cancer. Mol Carcinog. 38:188–200. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu N, Roth MJ, Emmert-Buck MR, Tang ZZ,
Polymeropolous M, Wang QH, Goldstein AM, Han XY, Dawsey SM, Ding T,
Giffen C and Taylor PR: Allelic loss in esophageal squamous cell
carcinoma patients with and without family history of upper
gastrointestinal tract cancer. Clin Cancer Res. 5:3476–3482.
1999.PubMed/NCBI
|
|
11
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
12
|
Jiang JK, Chen YJ, Lin CH, Yu IT and Lin
JK: Genetic changes and clonality relationship between primary
colorectal cancers and their pulmonary metastases - an analysis by
comparative genomic hybridization. Genes Chromosomes Cancer.
43:25–36. 2005. View Article : Google Scholar
|
|
13
|
Jiang LX, Xu J, Wang ZW, Li DP, Peng ZH,
Gao JJ, He L and Zheng HT: Tumor suppress genes screening analysis
on 4q in sporadic colorectal carcinoma. World J Gastroenterol.
14:5606–5611. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kawakami M, Yamaguchi T, Takahashi K,
Matsumoto H, Yasutome M, Horiguchi S, Hayashi Y, Funata N and Mori
T: Assessment of SMAD4, p53, and Ki-67 alterations as a predictor
of liver metastasis in human colorectal cancer. Surg Today.
40:245–250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kimura Y, Noguchi T, Kawahara K, Kashima
K, Daa T and Yokoyama S: Genetic alterations in 102 primary gastric
cancers by comparative genomic hybridization: gain of 20q and loss
of 18q are associated with tumor progression. Mod Pathol.
17:1328–1337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Knosel T, Schluns K, Dietel M and Petersen
I: Chromosomal alterations in lung metastases of colorectal
carcinomas: associations with tissue specific tumor dissemination.
Clin Exp Metastasis. 22:533–538. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lagerstedt KK, Staaf J, Jonsson G, Hansson
E, Lonnroth C, Kressner U, Lindstrom L, Nordgren S, Borg A and
Lundholm K: Tumor genome wide DNA alterations assessed by array CGH
in patients with poor and excellent survival following operation
for colorectal cancer. Cancer Inform. 3:341–355. 2007.PubMed/NCBI
|
|
18
|
Lei T, Chen WQ, Zhang SW, Lei TH, Ying Q,
He ZY and Wang XH: Prevalence trend of colorectal cancer in 10
cities and counties in China from 1988 to 2002. Zhonghua Zhong Liu
Za Zhi. 31:428–433. 2009.(In Chinese).
|
|
19
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instability in colorectal cancers. Nature. 386:623–627.
1997. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instabilities in human cancers. Nature. 396:643–649. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li HL, Gao YT, Zheng Y, Zhang W, Gao LF,
Xu B and Xiang YB: Incidence trends of colorectal cancer in urban
Shanghai, 1973–2005. Zhonghua Yu Fang Yi Xue Za Zhi. 43:875–879.
2009.(In Chinese).
|
|
22
|
Losi L, Bouzourene H and Benhattar J: Loss
of Smad4 expression predicts liver metastasis in human colorectal
cancer. Oncol Rep. 17:1095–1099. 2007.PubMed/NCBI
|
|
23
|
Nakao M, Kawauchi S, Furuya T, Uchiyama T,
Adachi J, Okada T, Ikemoto K, Oga A and Sasaki K: Identification of
DNA copy number aberrations associated with metastases of
colorectal cancer using array CGH profiles. Cancer Genet Cytogenet.
188:70–76. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nakao M, Kawauchi S, Uchiyama T, Adachi J,
Ito H, Chochi Y, Furuya T, Oga A and Sasaki K: DNA copy number
aberrations associated with the clinicopathological features of
colorectal cancers: Identification of genomic biomarkers by
array-based comparative genomic hybridization. Oncol Rep.
25:1603–1611. 2011.
|
|
25
|
Petersen S, Aninat-Meyer M, Schluns K,
Gellert K, Dietel M and Petersen I: Chromosomal alterations in the
clonal evolution to the metastatic stage of squamous cell
carcinomas of the lung. Br J Cancer. 82:65–73. 2000.PubMed/NCBI
|
|
26
|
Rae FK, Hooper JD, Nicol DL and Clements
JA: Characterization of a novel gene, STAG1/PMEPA1, upregulated in
renal cell carcinoma and other solid tumors. Mol Carcinog.
32:44–53. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rothlisberger B, Heizmann M, Bargetzi MJ
and Huber AR: TRIB1 overexpression in acute myeloid leukemia.
Cancer Genet Cytogenet. 176:58–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Y, Shu G, Yuan X, Jing N and Song J:
FOXA2 functions as a suppressor of tumor metastasis by inhibition
of epithelial-to-mesenchymal transition in human lung cancers. Cell
Res. 21:316–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu LL, Shanmugam N, Segawa T, Sesterhenn
IA, McLeod DG, Moul JW and Srivastava S: A novel androgen-regulated
gene, PMEPA1, located on chromosome 20q13 exhibits high level
expression in prostate. Genomics. 66:257–263. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yokoyama T, Kanno Y, Yamazaki Y, Takahara
T, Miyata S and Nakamura T: Trib1 links the MEK1/ERK pathway in
myeloid leukemogenesis. Blood. 116:2768–2775. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang B, Halder SK, Kashikar ND, Cho YJ,
Datta A, Gorden DL and Datta PK: Antimetastatic role of Smad4
signaling in colorectal cancer. Gastroenterology. 138:969–980.
2010. View Article : Google Scholar : PubMed/NCBI
|