Introduction
Nasopharyngeal carcinoma (NPC) is a distinct
malignancy that differs in epidemiology from other types of
cancers, most frequently occurring in South-Eastern Asia (1). Despite advances in the therapy of NPC,
no significant improvements have been observed in terms of overall
survival and relapse-free survival due to local or regional failure
and recurrence (2). Thus, the
mechanism underlying the oncogenesis and development of NPC remains
to be elucidated.
Heparanase (HPSE), an endo-β-D-glucuronidase, is
able to cleave heparan sulfate proteoglycans (HPSG) within the
extracellular matrix (ECM), basement membrane (BM) or on the
cellular surface, influencing cell growth and invasion of many
malignancies by regulating the shedding of HPSG (3). Elevated expression of HPSE was
observed in head and neck tumors, pancreatic tumors, hepatocellular
carcinoma, and esophageal carcinomas (4). Inhibition of HPSE in lymphoma, glioma,
breast carcinoma and melanoma cells displayed significantly
attenuated adhesion and invasion capacities (5). In addition, gene silencing of HPSE
efficiently suppresses the proliferation, invasion, metastasis and
angiogenesis of gastric cancer cells (6). Silencing of HSPE also resulted in the
remarked inhibition of the growth of prostate tumors (7).
A previous study demonstrated that HPSE
overexpression in nasopharyngeal carcinoma tissues is inversely
correlated with patient survival (8); however, it remains largely unknown
whether inhibition of HPSE expression affects the biological
function of nasopharyngeal carcinoma cells. This study analyzed the
effects of an shRNA targeting HPSE on cultured NPC cells. Our
results indicated that downregulation of HPSE expression abolished
the growth and invasion of NPC cells.
Materials and methods
Cell lines and culture conditions
Nasopharyngeal carcinoma cells C666-1, 6-10B and
CNE-2 were kindly provided by the Oncology Laboratory of Nanfang
Hospital (Guangzhou, China). All cells were maintained in DMEM with
10% FBS (both from Invitrogen/Gibco).
RNA isolation and qRT-PCR
Total cellular RNA was isolated from 3 NPC cell
lines using TRIzol reagent (Invitrogen) according to the
manufacturer’s instructions and was quantified using a UV
spectrophotometer. RNA (2 μg) was reverse transcribed using the
Access Reverse Transcription system (Promega, Madison, WI, USA)
according to the standard protocol. In brief, reaction mixtures
(total volume, 20 μl) containing 500 ng cDNA were amplified to a
final concentration of 250 nM using 10 μl of the 2X Brilliant
SYBR-Green QPCR Master Mix kit (Stratagene). The primers were: HPSE
forward, 5′-GAATGGACGGACTGCTAC-3′; HPSE reverse,
5′-CCAAAGAATACTTGCCTCA-3′ (GenBank: NM_006665.5) (6); β-actin forward, 5′-TAAGAAGCTGCTG
TGCTACG-3′; β-actin reverse, 5′-GACTCGTCATACTCC TGCTT-3′ (GenBank:
NM_001101). Morever, thermal cycling conditions were as follows:
94°C for 5 min and 45 cycles at 94°C for 30 sec, followed by 60°C
for 30 sec and 72°C for 45 sec. Experiments were performed in
triplicate in the same reaction. Target genes and the β-actin gene
were amplified in the same reaction. The results of the relative
quantitation were analyzed by comparison of 2-ΔΔCt.
Vector construction and transfection
Three pairs of shRNAs were designed (GenePharma)
according to the HPSE sequence in the GenBank (NM_006665.5) to
verify the specific effect of HPSE on the biological function of
NPC, as shown in Table I. Scrambled
shRNA (pGPU6/GFP/Neo-shNC) was set as the negative control. Each
vector contained the neomycin-resistance gene to provide neomycin
resistance in mammalian cells. Efficiency of interference was
evaluated by qRT-PCR and by western blot analysis. The chosen
constructed pGPU6/GFP/Neo-HPSE-shRNAs were introduced into CNE-2
cells using Lipofectamine reagent (Invitrogen). Stable cell lines
selected with media containing 400 μg/ml G418, were named
CNE-2/shHPSE and CNE-2/NC. HPSE, ERK, p-RAF, p-ERK and p-MEK
expression levels for each group were detected using western blot
analysis.
 | Table IOligonucleotide sequences of the
HPSE-specific shRNAs. |
Table I
Oligonucleotide sequences of the
HPSE-specific shRNAs.
| Name | Sense/antisense
strands of the shRNAs | Target nucleotide
sites |
|---|
| HPSE-shRNA1 |
5′-CACCGCTCTGTAGATGTGCTATACATTCAAGAGATGTATAGCACATCTACAGAGCTTTTTTG-3′/
5′-GATCCAAAAAAGCTCTGTAGATGTGCTATACATCTCTTGAATGTATAGCACATCTACAGAGC-3′ | 602–663 |
| HPSE-shRNA2 |
5′-CACCGCATCACTACTATTTGAATGGTTCAAGAGACCATTCAAATAGTAGTGATGCTTTTTTG-3′/
5′-GATCCAAAAAAGCATCACTACTATTTGAATGGTCTCTTGAACCATTCAAATAGTAGTGATGC-3′ | 984–1045 |
| HPSE-shRNA3 |
5′-CACCGCAAGTGGATAAATACCTTCTTTCAAGAGAAGAAGGTATTTATCCACTTGCTTTTTTG-3′/
5′-GATCCAAAAAAGCAAGTGGATAAATACCTTCTTCTCTTGAAAGAAGGTATTTATCCACTTGC-3′ | 1518–1579 |
Western blot analysis
Cell samples were lysed in a lysis buffer (Beyotime,
Jiangsu, China) after collection from a 100-mm dish and disruption,
respectively. Proteins (20 μg) were resolved on 10% SDS-PAGE and
transferred to PVDF membranes. Western blot analysis was performed
using antibodies against HPSE, phospho-RAF, phospho-ERK and
phospho-MEK with anti-β-actin as the control. The blocking steps
and dilutions for the assessment of all proteins were performed in
5% bovine serum albumin. After incubation with horseradish
peroxidase-conjugated antibodies (Amersham Pharmacia), the labeled
proteins were detected with an ECL-Plus detection system (Amersham
Pharmacia). The anti-HPSE antibody was from Abcam, anti
phospho-MEK, anti phospho-RAF, phospho-ERK and total ERK antibodies
were from Cell Signaling. The anti-β-actin was from Santa Cruz
Biotechnology (Santa Cruz, CA, USA).
Cell proliferation assay
CNE-2, CNE-2/shHPSE and CNE-2/NC cells were prepared
at a concentration of 1×104 cells/ml and incubated for
1, 2, 3, 4, 5, 6 and 7 days, respectively.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay was performed by adding 20 μl of MTT (Promega) for 4 h. The
supernatants were removed and 150 μl of DMSO (Sigma) was added to
each well. Fifteen minutes later, the absorbance value (OD) of each
well was measured with a microplate reader set at 490 nm. All
experiments were performed in triplicate.
Plate clone formation assay
Approximately 1×102 cells were added to
each well (3-cm in diameter) of a 6-well culture plate, and each
cell group consisted of three wells. After incubation at 37°C for
12 days, the cells were washed twice with PBS and stained with
Giemsa solution. The number of colonies containing ≥50 cells was
counted under a microscope. Plate clone formation efficiency =
(number of colonies/number of cells inoculated) × 100%. Three
individual experiments were carried out.
Cell motility assay
CNE-1, CNE-2/shHPSE and CNE-2/NC (1×105)
cells in 0.5 ml serum-free medium were placed in the upper chamber
of a transwell, whereas the lower chamber was loaded with 0.8 ml
medium containing 10% FBS. The cells that migrated to the lower
chamber were stained with 0.5% crystal violet, and the total cell
number was counted after 24 h of incubation at 37°C with 5%
CO2.
Cell invasion assay
Upper chambers of a 24-well Transwell plate (Corning
Incorporated) were coated with 50% Matrigel (BD Biosciences) in
phosphate-buffered saline. CNE-2, CNE-2/shHPSE and CNE-2/NC cells
were incubated in the upper chamber. After a 24-h incubation,
invaded cells were stained with 0.5% crystal violet, examined by
bright field microscopy, and photographed. The invasion rate was
quantified by counting the number of invaded cells in five random
fields per chamber under a fluorescence microscope. Data values
were summarized from three independent experiments.
Tumorigenicity assay
CNE-2/shHPSE and CNE-2/NC (5×106) cells
were injected subcutaneously in the flank of 6- to 7-week-old nude
mice. Tumor growth was evaluated for 3 weeks after injection and
every 7 days thereafter. The tumor volume was determined by
measuring the largest (a) and the smallest (b) axis using a
caliper, and calculated as V = 0.5ab2. Mice handling and
experimental procedures followed institutional guidelines. All
experimental procedures and protocols were approved by our
Institutional Animal Care and Use Committee.
Statistical analysis
Unless otherwise stated, the data are presented as
means ± standard error of the mean (SEM). Statistical significance
(P<0.05) was determined by the t-test or analysis of variance
(ANOVA) followed by assessment of differences using SPSS 16.0
software (SPSS Inc., Chicago, IL, USA).
Results
Expression of HPSE in three
nasopharyngeal carcinoma cell lines
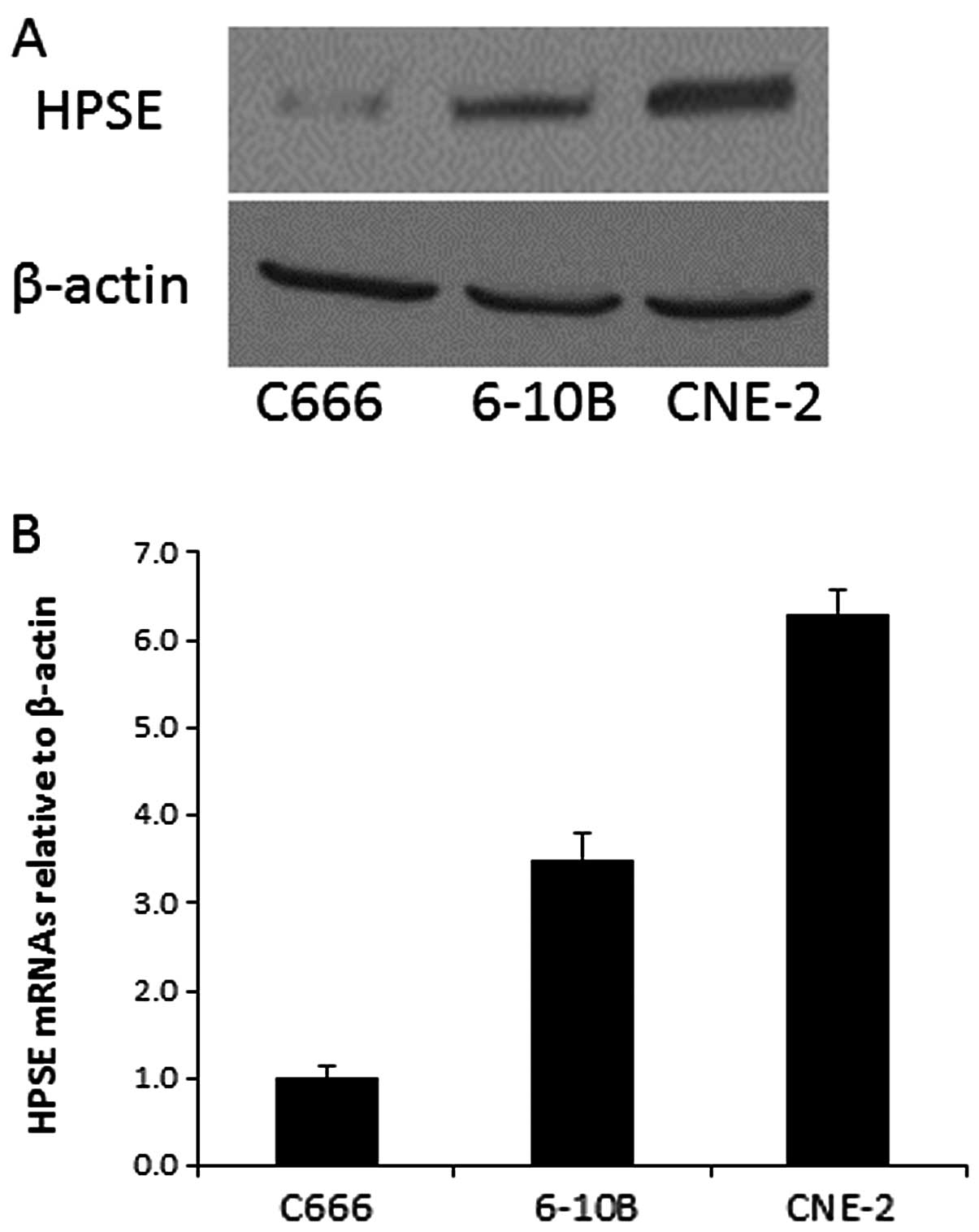
To investigate the expression of HPSE in
nasopharyngeal carcinoma cells, we performed qRT-PCR and western
blotting of C666, 6-10B and CNE-2 cells. The mRNA and protein
levels of HPSE in the three cell lines are shown in Fig. 1. The data showed that CNE-2 cells
expressed the highest level of HPSE mRNA and protein, and thus, was
chosen to further study the biological function of HPSE by RNA
interference.
The vector stably expressing HPSE shRNA
causes effective and specific downregulation of HPSE
expression
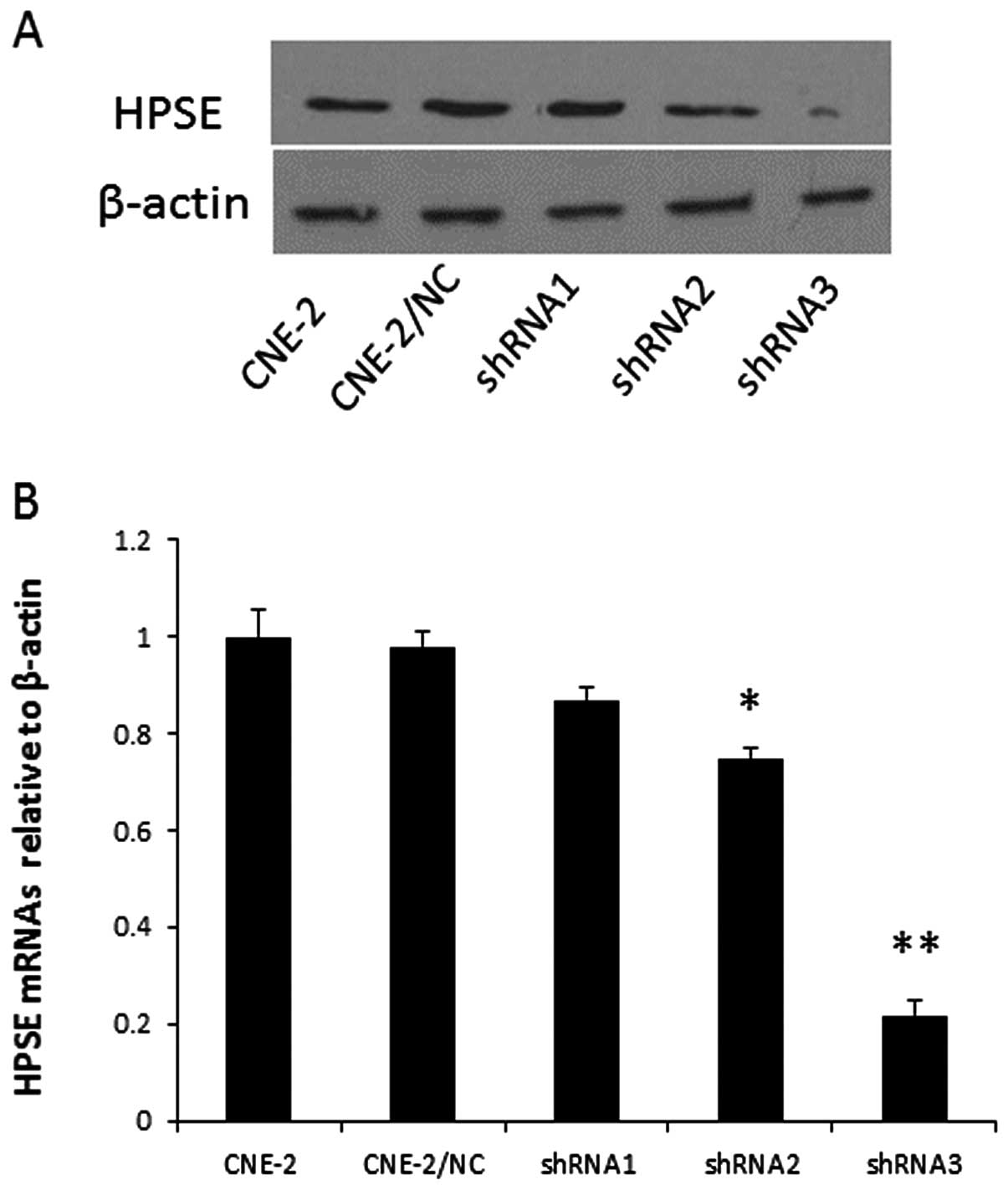
The knock down efficiencies of the different
HPSE-specific shRNAs in CNE-2 cells were first evaluated using
qRT-PCR. Relative HPSE mRNA levels in the individual stable
transfectants were normalized against the mRNA level of an internal
control gene, β-actin, performed in the same run. As shown in
Fig. 2A, cells transfected with
pGPU6-HPSE-shRNA3 showed a significantly reduced level of HPSE
protein when compared with that of the vector control and the
negative transfectants, respectively. A reduction in protein was
not detectable in cells transfected with pGPU6-HPSE-shRNA1 and
pGPU6-HPSE-shRNA2. In addition, qRT-PCR analysis (Fig. 2B) showed a marked reduction in the
HPSE mRNA levels in the CNE-2 cell line transfecting with
HPSE-shRNA3. The above results demonstrated that expression of HPSE
was downregulated specifically and effectively by the specific HPSE
shRNA, and that different shRNAs showed striking differences in
silencing efficiency. Thus, CNE-2 cells transfected with
pGPU6-HPSE-shRNA3 were named CNE-2/shHPSE, and CNE-2 cells
transfected with pGPU6-shNC were named CNE-2/NC.
Effect of HPSE on cell proliferation of
CNE-2 cells
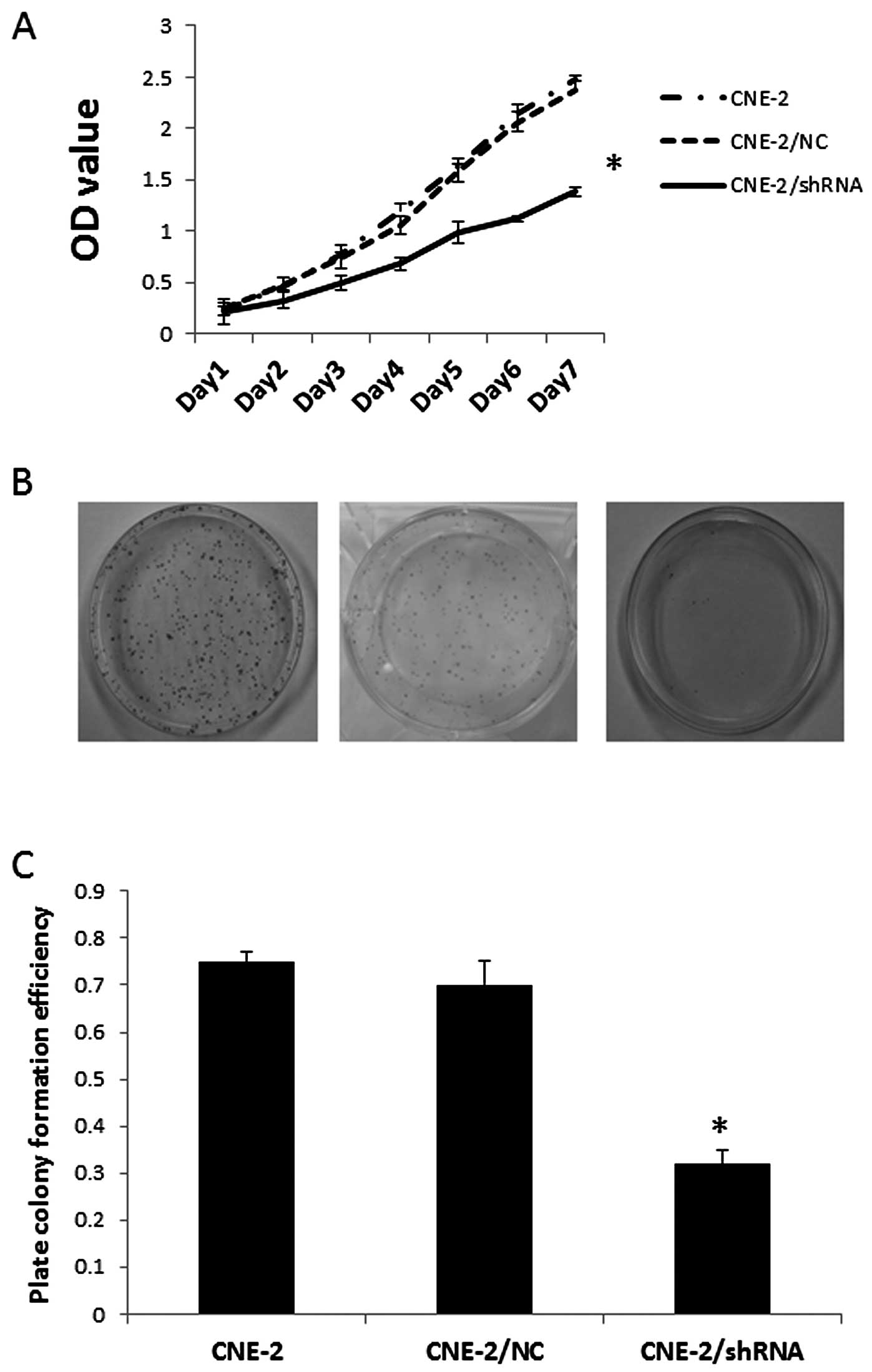
The proliferative activity of tumor cells is
important in the invasion/metastasis of tumors. After identifying
the most effective HPSE-specific shRNA transfectant (CNE-2/shHPSE),
we immediately examined cell proliferative activity of the
transfected cells using MTT assay. As known in Fig. 3A, the growth of CNE-2 cells in
vitro was markedly inhibited after the transfection of
pGPU6-HPSE-shRNA3 (P<0.05). This indicates a positive relation
between the expression of HPSE and the rate of NPC cell growth.
Effect of the silencing of HPSE using
shRNA on the colony formation potential of CNE-2 cells
In order to document the ability of CNE-2/shHPSE
cells to form colonies, single-cell suspensions were plated at a
density of 100 cells in 30-mm culture dishes. As shown in Fig. 3B, after 12 days, CNE-2/shHPSE cells,
compared with CNE2 and MHCC97-H/negative-shRNA cells, exhibited a
significant reduction in their ability to form colonies, and their
ability to form colonies was correlated with HPSE expression
(P<0.05) (Fig. 3C).
Effect of HPSE on the migration and
invasion of CNE-2 cells
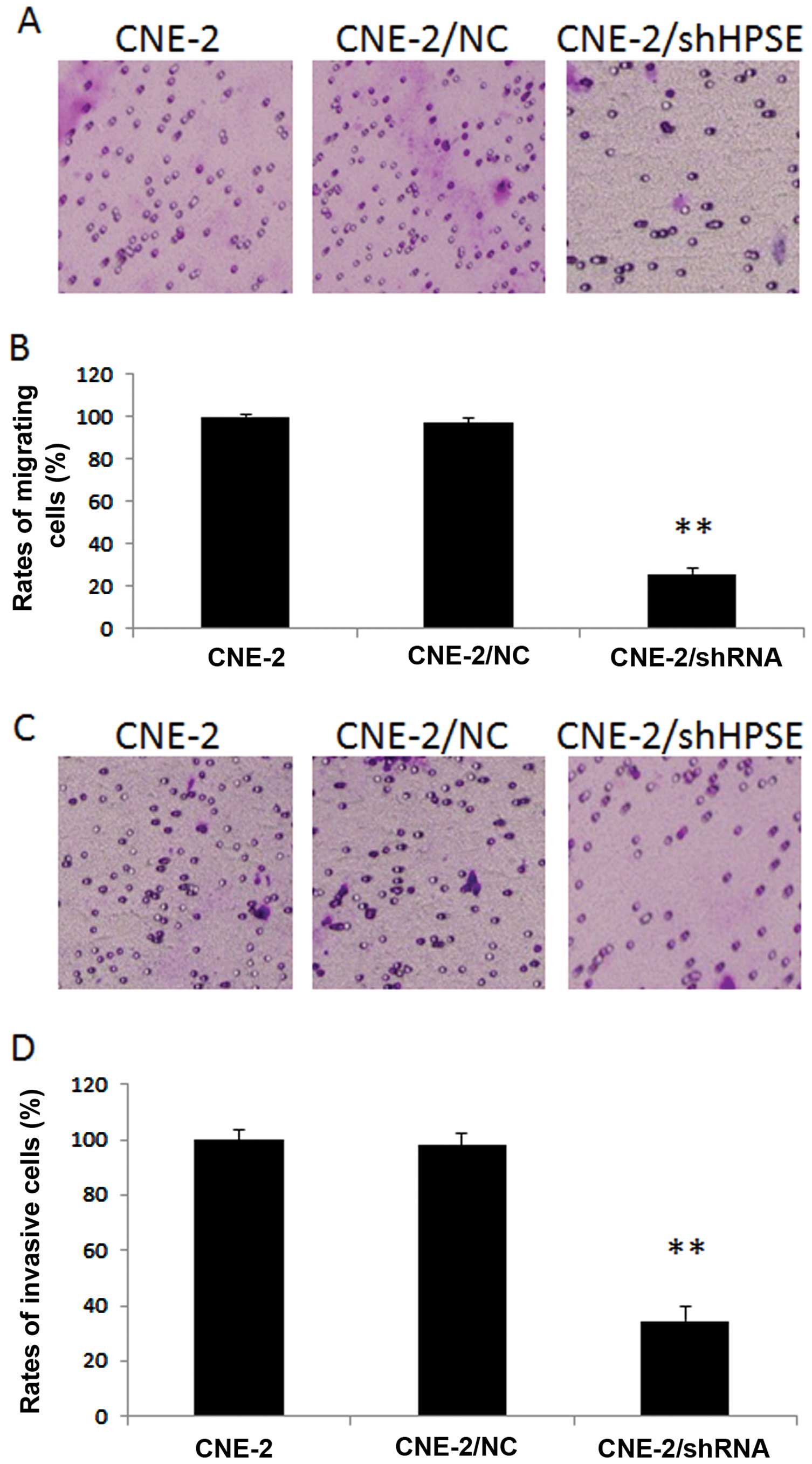
CNE-2/shHPSE cells displayed a marked decrease in
the migratory ability as comparing with either CNE-2/NC or CNE/2
cells (P<0.01, respectively). As shown in Fig. 4A and B, the migration rate of the
CNE-2/shHPSE cells was 25.9%, while the rates of the CNE-2 and
CNE-2-NC groups were 100 and 97%, respectively. Inhibition of HPSE
caused significantly attenuated migration of CNE-2 cells.
Invasion assays were carried out using
Matrigel-coated Transwell culture chambers. CNE-2/shHPSE cells
exhibited much lower invasive abilities than either the CNE-2 or
CNE-2/NC cells (P<0.01, respectively) (Fig. 4C and D). The invasion rates of the
CNE-2, CNE-2/NC and CNE-2/shHPSE cells were 100, 98 and 34.6%,
respectively. Suppression of HSPE led to significantly reduced
invasive ability of the CNE-2 cells.
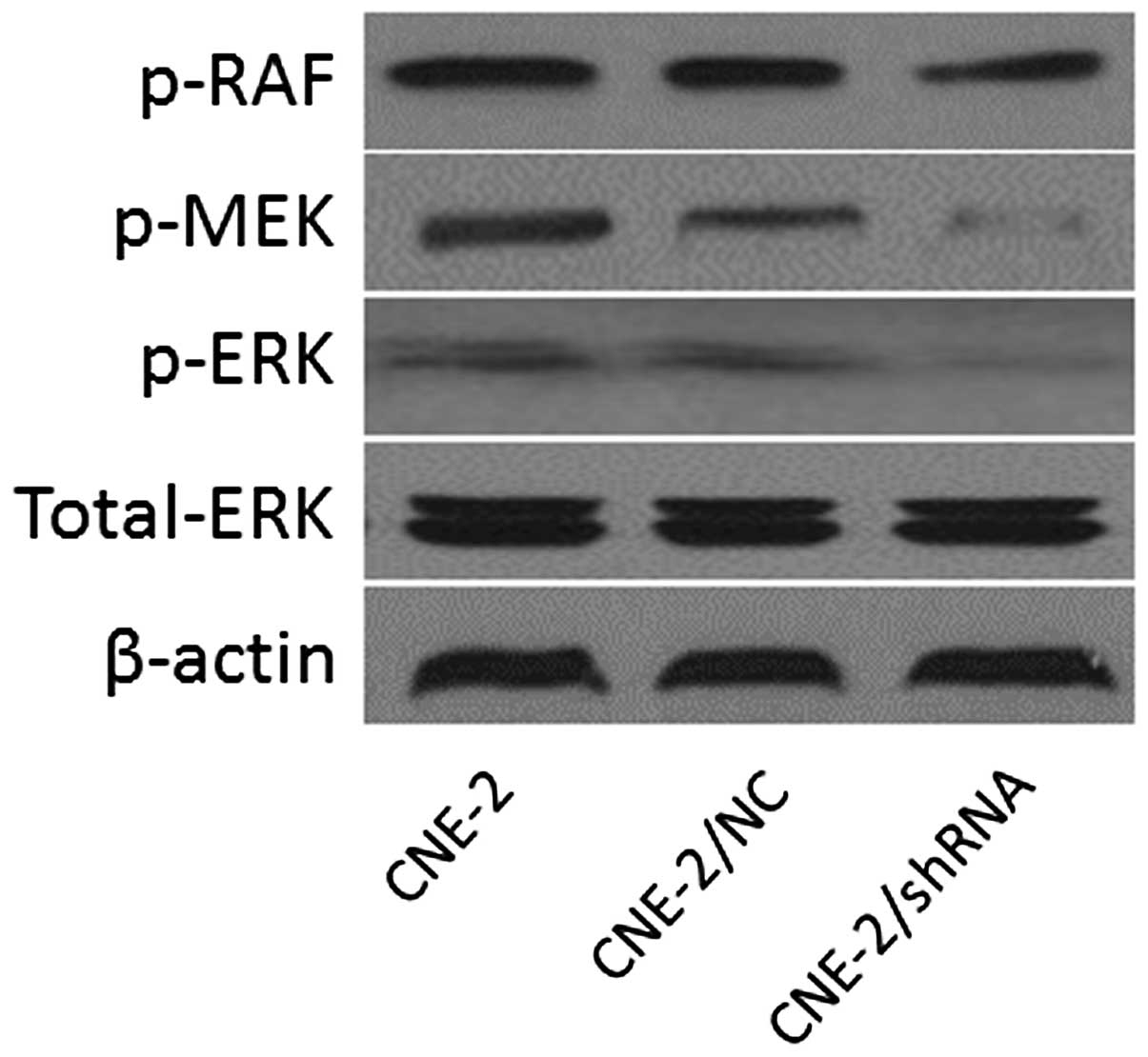
Effects of HSPE inhibition on the
expression of MARK transduction
Western blot analysis was applied to detect the
expression of components of MARK transduction in CNE/2, CNE-2/NC
and CNE-2/shHPSE cells. Inhibition of HPSE resulted in a
significant decrease in p-RAF, p-RAF and p-MEK expression. Protein
levels of p-RAF, p-ERK and p-MEK in the CNE-2/shHPSE cells were
lower than these levels in the control groups (CNE-2 and CNE-2/NC)
(Fig. 5).
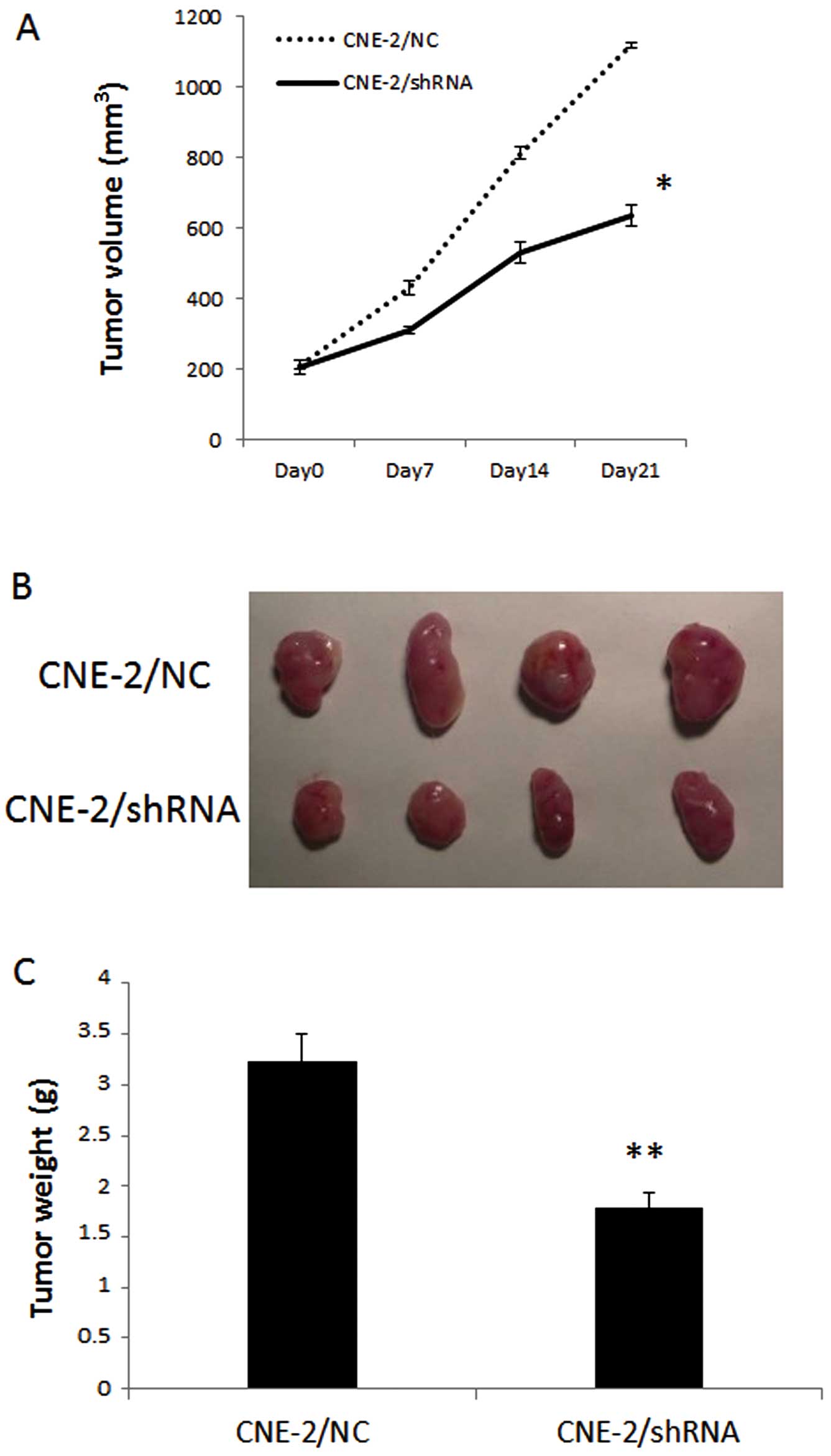
HPSE gene silencing suppresses cell
proliferation in vivo
The effect of HPSE on in vivo tumor growth
was assessed by the subcutaneous injection of CNE-2/shHPSE and
CNE-2/NC cells in nude mice for 21 days. As shown in Fig. 6, a marked reduction in tumor size in
the CNE-2/shHPSE groups was observed as compared with that of the
control group (P<0.05). By day 21 after cell injection, the
average tumor weight (n=4) of the CNE-2/shHPSE and CNE-2/NC groups
was 1.78±0.16 and 3.23±0.27 g, respectively (P<0.01), indicating
that knockdown of HPSE in NPC cells reduced their tumorigenic
potential.
Discussion
In the present study, we showed that overexpression
of HPSE has an important role in the progression of NPC. These
findings suggest that HPSE is a major contributor to the
proliferation, invasion and migration of NPC cells.
HPSE enzymatic activity, capable of cleaving
glucuronidic linkages and releasing polysaccharide chains resistant
to further degradation by the enzyme, was first identified by Ogren
and Lindahl (9). The activity of
HPSE was shown to be related with the metastatic potential of tumor
cells such as B16 melanoma and T-lymphoma (10,11).
These early observations gained substantial support from many
subsequent studies that clearly indicate that HPSE not only
enhances cell dissemination, but also promotes the establishment of
a vascular network which accelerates primary tumor growth and
provides a gateway for invading metastatic cells (12,13).
In addition, the clinical significance of the enzyme in tumor
development emerged from a systematic evaluation of HPSE expression
in primary human tumors, including those of the bladder (14), colon (15), stomach (16), breast (17), oral cavity (18), esophagus (19), pancreas (20) and brain (21), and in multiple myeloma and acute
myeloid leukemia (22,23). It has also been reported that HPSE
is upregulated in essentially all human tumors examined. Notably,
increased HPSE levels are often associated with reduced patient
survival, increased carcinoma metastasis and high microvessel
density (12,13).
Since HPSE plays an indispensable role in the
progression of cancer, numerous studies have focused on the
development of HPSE inhibitors, including neutralizing antibodies,
peptides, small molecules, modified non-anticoagulant species of
heparin, as well as several other polyanionic molecules, such as
laminaran sulfate, suramin and PI-88. These inhibitors that reduce
HPSE expression in cancer cells could significantly decrease their
metastatic properties, signifying the importance of HPSE in cancer
cell dissemination (24–27). However, because of the diverse
biologic activities of these compounds, the mechanism of their
antitumor activity and its relation to HPSE inhibition are not
straightforward. Moreover, these compounds may produce several
non-specific and undesirable effects when they interact with the
molecules which are distributed on the cell surface (15–17).
Thus, novel approaches are needed to inhibit the effect of HPSE in
cancer progression.
RNAi is a specific, potent, and highly successful
approach for loss-of-function studies in virtually all eukaryotic
organisms (28). It has been
identified in several reports that HPSE shRNA significantly
downregulates both HPSE protein and mRNA expression, and inhibits
proliferation and invasion of tranfected cells in different tumor
models, such as breast cancer (29), hepatocellular carcinoma (30) and gastric tumors (31). In the present study, CNE-2 was
chosen as the RNA interfering subject, as it demonstrated increased
levels of HPSE as compared with other NPC cell lines. The
cytological results indicated a relationship between attenuated
HPSE expression and cell proliferation, and together with further
in vivo experiments, support our hypothesis that HPSE may be
a direct factor contributing to the carcinogenesis and development
of NPC by regulating cellular proliferation. In addition, low
expression of HPSE is also associated with a marked decrease in
invasive capability of CNE-2 cells in vitro. The above
results suggest a possible reason for the poor outcomes observed in
NPC patients overexpressing HPSE.
The MAPK signaling pathway is a major determinant in
the control of diverse cellular processes such as proliferation,
survival, differentiation and motility and is associated with tumor
development (32). Our results
indicate that the anti-proliferative effects of HPSE on NPC cells
are partially mediated by MAPK signaling. Suppression of HPSE in
NPC cells overexpressing HPSE inhibited cell proliferation
associated with a decrease in phosphorylation of RAF, MEK and ERK.
However, we noted that even after knockdown of HPSE, the NPC cells
still possessed the capabilities of invasion and metastasis. We
believe that other factors, such as uPA and MMPs, also influence
these characteristics of NPC, which warrants further
investigation.
In summary, we demonstrated that suppression of HPSE
results in MAPK signaling pathway inactivation in NPC cells,
consequently inhibiting cell proliferation and invasion. Together
with previous reports that interference of HPSE expression
regulates at least two major signaling pathways (Wnt/β-catenin and
TGF-β) in cancer, we suggest that interference of HPSE expression
and function exert broad-spectrum biologic effects that may be
beneficial for the treatment of NPC. Further studies of the
mechanisms by which HPSE regulates multiple signaling pathways may
reveal other points of intervention for this important target.
Acknowledgements
We gratefully thank Chengwei Lv from the Cancer
Biotherapy Center for the collection and maintenance of the CNE-2,
6-10B and C666 cell lines used in this research. The project was
supported by the National Natural Science Foundation of China
(grant no. 81001212), the Science and Technology Foundation of
Zhejiang Province (grant no. 2012C33011), Foundation of State Key
Laboratory of Oncology in South China (grant no. HN2011-03),
Zhejiang Provincial Chinese Medicine Research Foundation (grant no.
2011ZQ012), and the Natural Science Foundation of Guangdong (grant
no. s2012010008209).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sanderson RD, Yang Y, Suva LJ and Kelly T:
Heparan sulfate proteoglycans and heparanase - partners in
osteolytic tumor growth and metastasis. Matrix Biol. 23:341–352.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Simizu S, Ishida K and Osada H: Heparanase
as a molecular target of cancer chemotherapy. Cancer Sci.
95:553–558. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Edovitsky E, Elkin M, Zcharia E, Peretz T
and Vlodavsky I: Heparanase gene silencing, tumor invasiveness,
angiogenesis, and metastasis. J Natl Cancer Inst. 96:1219–1230.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng L, Jiang G, Mei H, et al: Small RNA
interference-mediated gene silencing of heparanase abolishes the
invasion, metastasis and angiogenesis of gastric cancer cells. BMC
Cancer. 10:332010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lerner I, Baraz L, Pikarsky E, et al:
Function of heparanase in prostate tumorigenesis: potential for
therapy. Clin Cancer Res. 14:668–676. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bar-Sela G, Kaplan-Cohen V, Ilan N,
Vlodavsky I and Ben-Izhak O: Heparanase expression in
nasopharyngeal carcinoma inversely correlates with patient
survival. Histopathology. 49:188–193. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ogren S and Lindahl U: Cleavage of
macromolecular heparin by an enzyme from mouse mastocytoma. J Biol
Chem. 250:2690–2697. 1975.PubMed/NCBI
|
|
10
|
Nakajima M, Irimura T, Di Ferrante D, Di
Ferrante N and Nicolson GL: Heparan sulfate degradation: relation
to tumor invasive and metastatic properties of mouse B16 melanoma
sublines. Science. 220:611–613. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vlodavsky I, Fuks Z, Bar-Ner M, Ariav Y
and Schirrmacher V: Lymphoma cell-mediated degradation of sulfated
proteoglycans in the subendothelial extracellular matrix:
relationship to tumor cell metastasis. Cancer Res. 43:2704–2711.
1983.PubMed/NCBI
|
|
12
|
Ilan N, Elkin M and Vlodavsky I:
Regulation, function and clinical significance of heparanase in
cancer metastasis and angiogenesis. Int J Biochem Cell Biol.
38:2018–2039. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vreys V and David G: Mammalian heparanase:
what is the message? J Cell Mol Med. 11:427–452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao W, Wang XS, Niu HT, Wang LL, Han BM
and Xia SJ: Clinical relevance of heparanase mRNA expression in
bladder cancer and its usefulness as a detection marker in voided
urine. Mol Med Rep. 2:327–331. 2009.PubMed/NCBI
|
|
15
|
Nobuhisa T, Naomoto Y, Ohkawa T, et al:
Heparanase expression correlates with malignant potential in human
colon cancer. J Cancer Res Clin Oncol. 131:229–237. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen JQ, Zhan WH, He YL, et al:
Relationship between heparanase mRNA expression in human gastric
cancer and its clinicopathological features. Zhonghua Zhong Liu Za
Zhi. 26:609–611. 2004.(In Chinese).
|
|
17
|
Theodoro TR, de Matos LL, Sant AA, et al:
Heparanase expression in circulating lymphocytes of breast cancer
patients depends on the presence of the primary tumor and/or
systemic metastasis. Neoplasia. 9:504–510. 2007. View Article : Google Scholar
|
|
18
|
Leiser Y, Abu-El-Naaj I, Sabo E, et al:
Prognostic value of heparanase expression and cellular localization
in oral cancer. Head Neck. 33:871–877. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brun R, Naroditsky I, Waterman M, et al:
Heparanase expression by Barrett’s epithelium and during esophageal
carcinoma progression. Mod Pathol. 22:1548–1554. 2009.
|
|
20
|
Quiros RM, Rao G, Plate J, et al: Elevated
serum heparanase-1 levels in patients with pancreatic carcinoma are
associated with poor survival. Cancer. 106:532–540. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hong X, Nelson KK, DeCarvalho AC and
Kalkanis SN: Heparanase expression of glioma in human and animal
models. J Neurosurg. 113:261–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kelly T, Miao HQ, Yang Y, et al: High
heparanase activity in multiple myeloma is associated with elevated
microvessel density. Cancer Res. 63:8749–8756. 2003.PubMed/NCBI
|
|
23
|
Bitan M, Polliack A, Zecchina G, et al:
Heparanase expression in human leukemias is restricted to acute
myeloid leukemias. Exp Hematol. 30:34–41. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nakajima M, DeChavigny A, Johnson CE,
Hamada J, Stein CA and Nicolson GL: Suramin. A potent inhibitor of
melanoma heparanase and invasion. J Biol Chem. 266:9661–9666.
1991.PubMed/NCBI
|
|
25
|
Miao HQ, Liu H, Navarro E, Kussie P and
Zhu Z: Development of heparanase inhibitors for anti-cancer
therapy. Curr Med Chem. 13:2101–2111. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miao HQ, Elkin M, Aingorn E,
Ishai-Michaeli R, Stein CA and Vlodavsky I: Inhibition of
heparanase activity and tumor metastasis by laminarin sulfate and
synthetic phosphorothioate oligodeoxynucleotides. Int J Cancer.
83:424–431. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karoli T, Liu L, Fairweather JK, et al:
Synthesis, biological activity, and preliminary pharmacokinetic
evaluation of analogues of a phosphosulfomannan angiogenesis
inhibitor (PI-88). J Med Chem. 48:8229–8236. 2005. View Article : Google Scholar
|
|
28
|
Fjose A, Ellingsen S, Wargelius A and Seo
HC: RNA interference: mechanisms and applications. Biotechnol Annu
Rev. 7:31–57. 2001. View Article : Google Scholar
|
|
29
|
Zhang ZH, Chen Y, Zhao HJ, Xie CY, Ding J
and Hou YT: Silencing of heparanase by siRNA inhibits tumor
metastasis and angiogenesis of human breast cancer in vitro and in
vivo. Cancer Biol Ther. 6:587–595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Li L, Wang Y, et al:
Downregulating the expression of heparanase inhibits the invasion,
angiogenesis and metastasis of human hepatocellular carcinoma.
Biochem Biophys Res Commun. 358:124–129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang G, Zheng L, Pu J, et al: Small RNAs
targeting transcription start site induce heparanase silencing
through interference with transcription initiation in human cancer
cells. PLoS One. 7:e313792012. View Article : Google Scholar
|
|
32
|
De Luca A, Maiello MR, D’Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16(Suppl 2):
S17–S27. 2012.PubMed/NCBI
|