1. Introduction
Ovarian cancer is the most aggressive cancer of the
female reproductive system, and a leading cause of cancer-related
mortality worldwide every year. Early-stage malignancy is
frequently asymptomatic and difficult to detect, and thus diagnosis
usually occurs after the disease has disseminated beyond the
ovaries (1). Therefore,
approximately 70% of ovarian cancer cases are diagnosed at advanced
stage and only 40% of women with this type of cancer survive 5
years (2). Although
cisplatin-centered chemotherapy, which is the current preferred
treatment modality in human ovarian cancer, significantly reduces
the mortality rates and prolongs the survival time of patients, the
main obstacle to a successful treatment for ovarian cancer is the
development of drug resistance to combined chemotherapy (3).
Drug resistance, both intrinsic and acquired,
results from a variety of factors including individual variations
in patients and somatic cell genetic differences in tumors
(4). Several molecular mechanisms
have been implicated in the increase of resistance in cellular
models of ovarian cancer. Johnson et al(5) considered that three general categories
consisting of decreased cell-associated drugs, altered drug
inactivation, and increased DNA damage tolerance/repair would be
the platinum-based resistance mechanisms in ovarian cancer.
Sorrentino et al(3) reported
that increased antiapoptotic regulator activity, growth factor
receptor deregulation, defective DNA damage response, and increased
DNA repair activity would respond to drug resistance in ovarian
cancer. It is now widely accepted that the apoptotic capacity of
cancer cells is crucial in determining the response to
chemotherapeutic agents (6). At the
same time, apoptosis is the cellular underpinning of
cisplatin-induced cell death, which associates with the expression
of specific ‘death’ genes and downregulation of ‘survival’
counterparts (7). However,
regardless of mechanisms, abnormal expression of drug
resistance-related genes often plays an important role in drug
resistance. Among all these drug resistance-related genes, tumor
suppressor genes (TSGs) are clearly the key players.
TSGs are wild-type alleles of genes which play
regulatory roles in diverse cellular activities, including cell
proliferation, differentiation, migration, cell cycle checkpoint
responses, protein ubiquitination and degradation, detection and
repair of DNA damage, mitogenic signaling, and tumor angiogenesis
(8,9), and whose loss of function contributes
to the development of cancer (9).
It has been proved that TSGs contribute to drug resistance in
several types of solid tumors. For example, TSGs including E1A,
p53, Fhit, IL-24, Fus1 and BiKDD are associated with
drug resistance in ovarian, lung and pancreatic cancer, and these
genes are potential genes for gene therapy (10). In this study, on the basis of
published reports, 15 TSGs which contributed to drug resistance in
ovarian cancer were reviewed to provide an overview of the
relationship of TSGs with ovarian cancer drug resistance;
meanwhile, protein interactions and bio-association analysis were
performed to demonstrate the associations of these TSGs and to mine
the potential drug resistance-related genes which are closely
related to the 15 TSGs.
2. Overall information on the 15 TSGs that
contribute to drug resistance in ovarian cancer
To comprehensively collect the drug
resistance-related TSGs in ovarian cancer, we first summarized a
total of 39 ovarian cancer-related TSGs from the PubMed online
database (http://www.ncbi.nlm.nih.gov/pubmed/), the online
Dragon database for exploration of Ovarian Cancer Genes (DDOC,
October, 2012) (http://apps.sanbi.ac.za/ddoc/) (11), six candidate gene lists produced by
large-scale genomic platforms on ovarian cancer from the Cancer
Genome Atlas (TCGA) (12), one
comprehensive expert review on ovarian cancer-related genes from
Nature Reviews Cancer (13) and one
bioinformatics study on ovarian cancer-related genes from PLOS One
(14), followed by an advanced
search with ‘ovarian cancer’ or ‘ovarian carcinoma’, ‘drug
resistance’ or ‘multi-drug resistance’ or ‘chemoresistance’ or
‘resistance’ and ‘name of the TSGs’ performed in the PubMed
database to acquire the drug resistance-related TSGs in ovarian
cancer.
A total of 15 TSGs including BRCA1, BRCA2,
CHEK2/Chk2, FBXO32, MLH1, SULF1, IL24/MDA-7, p16/CDKN2A,
p21/CDKN1A, p53/TP53, TP73, PDCD4, PTEN, RASSF1 and WWOX
which contributed to drug resistance in ovarian cancer were
summarized. As shown in Table I,
the modifications of the TSGs, the responding drugs, and the ways
for TSGs to regulate the drug resistance in ovarian cancer were
integrated. We observed that both genetic and epigenetic changes of
the TSGs were contributed to drug resistance in ovarian cancer, but
the latter apparently played dominant roles. As it is challenging
to treat ovarian cancer through a genetic strategy, due, in part,
to its heterogeneity, the reversibility of epigenetic mechanisms
involved in ovarian cancer opens potential new avenues for
treatment (15). The epigenomics of
ovarian cancer has become a rapidly expanding field leading to
intense investigation, and the reversion of epigenetic changes of
TSGs has already proved to be effective in reversing the drug
resistance clinically in ovarian cancer. It has been reported that
low-dose decitabine alters DNA methylation restoring sensitivity to
carboplatin in patients with heavily pre-treated ovarian cancer,
resulting in a high response rate and prolonged progression free
survival, and demethylation of the MLH1 and RASSF1a
in tumors from day 1 to 8 is positively correlated with progression
free survival (P<0.05) (16).
These results were promising and encouraged further study on
epigenetic changes of TSGs associated with drug resistance in
ovarian cancer. In addition, with the exception of FBXO32
and WWOX, the other 13 TSGs participated in the regulation
of drug resistance in ovarian cancer through certain pathways, in
particular, through apoptosis and DNA damage-related pathways.
However, regardless of pathways, the 15 TSGs responded to drug
resistance in ovarian cancer through ‘growth’ (including cell
proliferation, cell growth and cell survival) and ‘death’
(including cell death and cell apoptosis) (Table I).
 | Table IGeneral overview of the 15 TSGs that
contribute to drug resistance in ovarian cancer. |
Table I
General overview of the 15 TSGs that
contribute to drug resistance in ovarian cancer.
| TSG
abbreviation | Full name of the
TSGs | Modifications of
the TSGs | Drugs | Regulation manner
of drug resistance | Pathways associated
with drug resistance |
|---|
| BRCA1 | Breast cancer
susceptibility gene 1 | Mutation, DNA
methylation (17) | Taxol | DNA repairing
(18) | DNA damage response
(19) |
| BRCA2 | Breast cancer
susceptibility gene 2 | Mutation, DNA
methylation | Platinum | Cell survival
(17) | DNA damage response
(19) |
|
CHEK2/Chk2 | Checkpoint kinase 2
gene | Mutation (20) | Cisplatin | Inducible
degradation of CHEK2 protein (21) |
Ubiquitin-proteasome pathway (21) |
| FBXO32 | F-box only protein
32 gene | DNA
methylation | Platinum | Cell proliferation,
cell apoptosis (22) | - |
| MLH1 | DNA mismatch repair
gene 1 | DNA
methylation | Cisplatin | Mismatch repairing
(23,24) | DNA mismatch repair
(24) |
| SULF1 | Sulfatase 1
gene | DNA methylation,
Histone acethylation (25) | Platinum | Cell proliferation,
cell apoptosis | HSPG-related signal
transduction pathways (26) |
|
IL24/MDA-7 | Interleukin 24
gene | Ubiquitinated
(27) | Taxol | Cell growth, cell
apoptosis (28) |
Ubiquitin-proteasome system (27) |
|
p16/CDKN2A | Cyclin-dependent
kinase inhibitor 2A gene | Gene mutation | Platinum,
Taxol | Cell growth, cell
death | Cell cycle
(29) |
|
p21/CDKN1A | Cyclin-dependent
kinase inhibitor 1A gene | Histone
modification (30) | Cisplatin | Cell apoptosis, DNA
mismatch repair (31,32) | DNA mismatch repair
and apoptosis pathways (32) |
| PDCD4 | Programmed cell
death 4 gene | miRNA (33) | Platinum | Tumor growth,
apoptosis (34) | Death receptor
pathway (34) |
| PTEN | Phosphatase and
tensin homolog gene | miRNA (35) | Cisplatin | Cell proliferation,
cell apoptosis (36–38) | PI3K/AKT signaling
pathway, p53 signaling pathway (36–38) |
| RASSF1A | Ras association
domain family 1A gene | DNA
methylation | Taxol | Cell apoptosis,
cell survival, stabilization of microtubules (39) | Apoptosis (39) |
| TP53 | Tumor protein 53
gene | Mutation, DNA
methylation (40) | Platinum | Cell cycle, cell
apoptosis (41) | Cell cycle,
apoptosis, p53 signaling pathway (41) |
| TP73 | Tumor protein 73
gene | DNA methylation
(42) | Cisplatin | Apoptosis (43) | Apoptosis (43) |
| WWOX | WW
domain-containing oxidoreductase gene | DNA methylation
(44) | Cisplatin | Cell proliferation,
cell apoptosis (45) | - |
3. Gene/protein interaction network of the
15 TSGs
Gene/protein function predictions based on
bioinformatics analysis is a potential, feasible and valuable way
for gene/protein function mining, and numerous large-scale networks
of molecular interactions within the cell have made it possible to
go beyond one dimensional approaches to study gene/protein function
in the context of a network (46).
GeneMANIA is a web-based database and tool for prediction of
gene/protein function on the basis of multiple networks derived
from different genomic or proteomic data/sources, and it is fast
enough to predict gene/protein function with significant accuracy
(47). Protein-protein interactions
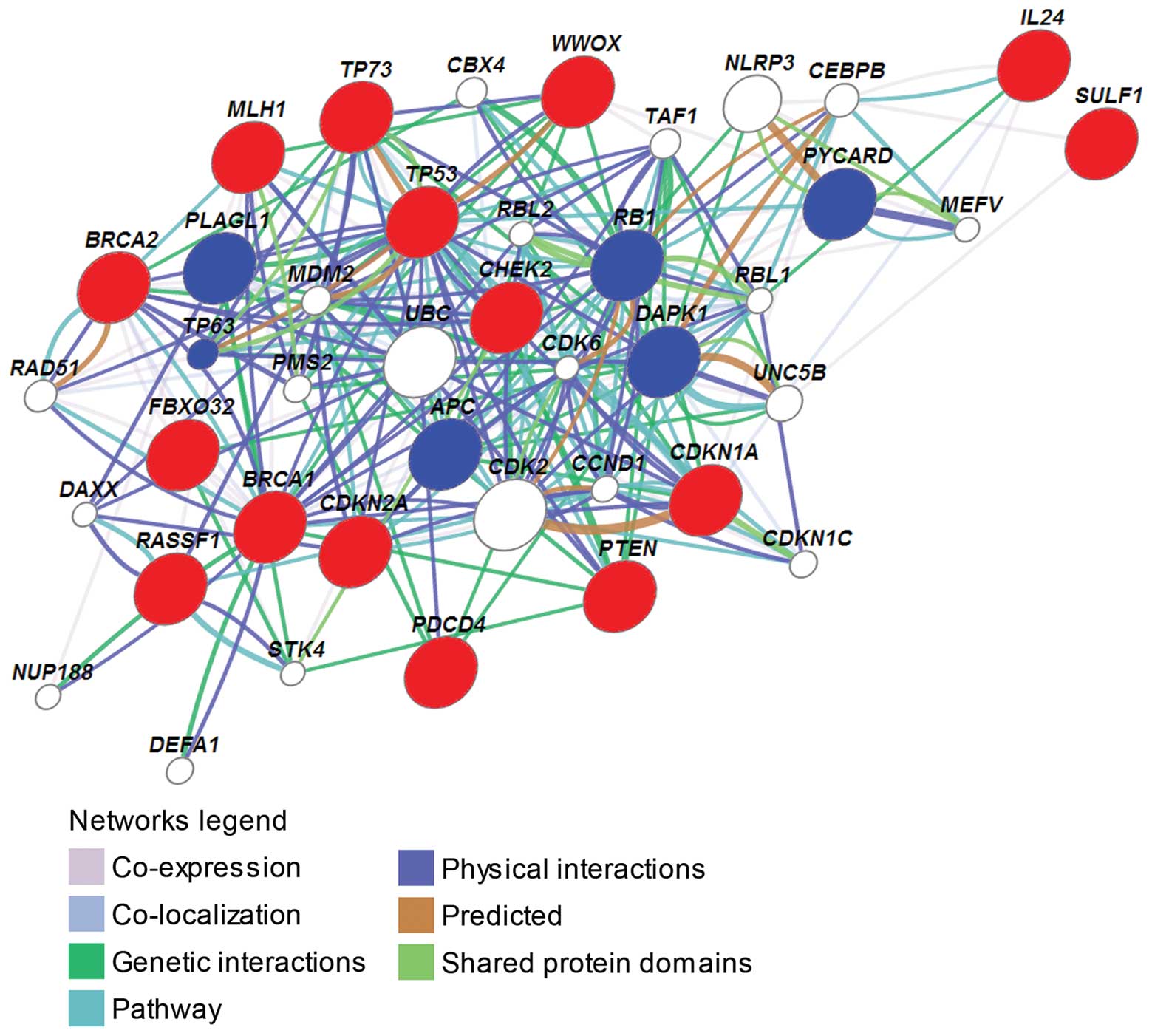
of the 15 TSGs were analyzed using the GeneMANIA online tool.
Except for IL24 and SULF1, all 15 TSGs had direct
interactions (co-expression, co-localization, genetic interactions,
shared pathway, physical interactions and shared protein domains)
or indirect interactions (through direct interactions with an
intermediate gene) with each other (Fig. 1). For example, TP53 had
direct interactions with MLH1, BRCA1, BRCA2, CHEK2, CDKN1A,
CDKN2A, PTEN, TP73 and WWOX, and it had indirect
interactions with FBXO32 and RASSF1; BRCA1 had direct
interactions with BRCA2, MLH1, CHEK2, PTEN, WWOX and p53;
RASSF1 had indirect interactions with BRCA1, CDKN1A, CDKN2A,
CHEK2, PTEN, TP53, TP73 and WWOX; PDCD4 had indirect
interactions with BRCA1, BRCA2, CDKN1A, CDKN2A, FBXO32, IL24,
MLH1, PTEN, TP53, TP73 and WWOX. These results
suggested that the 15 TSGs may contribute to drug resistance as a
whole.
There were an additional 6 TSGs, adenomatous
polyposis coli gene (APC), death associated protein kinase
gene (DAPK)1, pleiomorphic adenoma gene-like 1
(PLAGL1), retinoblastoma susceptibility gene (RB1), a
gene encoding an apoptosis-associated speck-like protein
(PYCARD), and tumor protein 63 (TP63), which had
close interactions with the 15 TSGs in the network (Table II), suggesting that these 6 TSGs
may be involved in drug resistance and would be potential drug
resistance-related TSGs in ovarian cancer. With the exception of
PLAGL1, the other 5 TSGs have been confirmed to associate
with drug resistance in several types of solid cancer. The
expression status of the APC determines the relative
sensitivity of colon cancer cells to histone deacetylase
inhibitor-induced apoptosis which may relate to drug resistance
(48); DAPK is
hypermethylated in drug-resistant non-small cell lung cancer cell
lines and head and neck squamous cell carcinoma cell lines.
Restoration of DAPK into the resistant non-small cell lung
cancer cells by stable transfection can re-sensitize the cells to
both erlotinib and cetuximab. Conversely, siRNA-mediated knockdown
of DAPK induces resistance in the parental sensitive cells.
These results demonstrate that DAPK plays important roles in
both cetuximab and erlotinib resistance (49); point mutations of the RB1
encode nuclear proteins with impaired ability to induce apoptosis
compared to wild-type pRb in vitro. Notably, three out of
four tumors harboring RB1 mutations display primary
resistance to treatment with either 5-FU/mitomycin or doxorubicin
(50); upregulated expression of
PYCARD/ASC leads to enhanced sensitivity to cisplatin,
gemcitabine and docetaxel in bladder cancer cells and pancreatic
cancer cells (51,52); TP63, with high homology with
TP53, is critical for the development of stratified
epithelial tissues such as epidermis, breast, and prostate
(53). Expression analysis in
long-term survivors reveals a significant upregulation of TP63,
PTEN, GADD45a and MAPK1 in metastatic gastric cancer,
suggesting that these genes may be involved in drug metabolism and
resistance (54).
 | Table IIThe interactions of the additional 6
TSGs with the 15 TSGs in the gene/protein interaction network. |
Table II
The interactions of the additional 6
TSGs with the 15 TSGs in the gene/protein interaction network.
| The additional 6
TSGs in the network | Member of the 15
TSGs (Direct interactions) | Member of the 15
TSGs (Indirect interactions through an intermediate gene) |
|---|
| APC | CDKN1A, PTEN,
TP53, TP73 and WWOX | CDKN2A, CHEK2,
PDCD4 and RASSF1 |
| DAPK1 | FBXO32, PDCD4,
PTEN, TP53 and TP73 | BRCA1, BRCA2,
CDKN1A, IL24, MLH1, PDCD4, PTEN, SULF1, TP53 and
TP73 |
| PLAGL1 | BRCA1 and
TP53 | CDKN1A, CDKN2A,
CHEK2, FBXO32, PDCD4, PTEN and RASSF1 |
| RB1 | BRCA1, CDKN1A,
CDKN2A, CHEK2, TP53, TP73 and WWOX | RASSF1 and
PDCD4 |
| PYCARD | TP53 | IL24 and
WWOX |
| TP63 | CHEK2, TP53
and TP73 | BRCA1, BRCA2,
CDKN1A, MLH1, PDCD4, PTEN, TP53 and TP73 |
In addition, ubiquitin C (UBC) had direct
physical interactions with 7 of the 15 TSGs, BRCA1, BRCA2,
CDKN1A, MLH1, PTEN, TP53 and TP73, and it had direct physical
interactions with other genes including MDM2, DAPK1, TP63,
PMS2 and DAXX; meanwhile, MDM2 (murine double
minute 2 gene) had direct interactions with 7 of the 15 TSGs, i.e.,
CDKN2A, CHEK2, PDCD4, RASSF1, TP53, TP73 and WWOX,
and it had direct physical interactions with other genes including
APC, CDK2, DAXX, RB1, TAF1, TP63 and UBC. Among these
other genes which had interactions with UBC and MDM2,
APC, RB1 and TP63 were TSGs associated with drug
resistance in cancer, and death domain-associated protein (DAXX)
can lead to apoptosis in cancer (55). These results indicated that
UBC and MDM2 may play roles in drug resistance in
ovarian cancer. It has previously been reported that MDM2
negatively regulates tumor suppressor p53 via binding to the
transactivation and the DNA-binding domains of p53 (56), and contributes to drug resistance in
ovarian cancer through regulation of p53 (57). UBC is low in normal kidneys
but is increased in malignant tumors in vivo and in several
established renal carcinoma cell lines, indicating that high
expressions of the UBC are important in the cancerous state
of these cells (58). Similarly,
UBC is highly expressed in human nasopharyngeal carcinoma
drug-resistant cell lines (59).
However, the studies on UBC associated with drug resistance
are limited.
Among the annotated functions in accordance with the
GeneMANIA network, eight were closely related to drug resistance
(Table III). The cell
cycle-related function which covered 8 of the 15 TSGs and 3 of the
additional 6 TSGs was annotated with the highest false discovery
rate (FDR), followed by the DNA binding-related function which also
covered 8 of the 15 TSGs and 4 of the additional 6 TSGs, suggesting
that these 3 pathways (functions) may be the major ways with which
TSGs participate in the regulation of drug resistance in ovarian
cancer. Furthermore, DNA damage and apoptosis, which both covered 6
of the 15 TSGs and TP63, may also be crucial ways for the
contribution of the TSGs to the regulation of drug resistance in
ovarian cancer.
 | Table IIIAnnotated functions of the 15 TSGs
according to the protein interaction network. |
Table III
Annotated functions of the 15 TSGs
according to the protein interaction network.
| Annotated
function | False discovery
rate | No. of the 15
TSGs | Other TSGs | Other genes |
|---|
| Cell cycle
related |
1.19e-13~9.82e-8 | 8 (BRCA1, BRCA2,
CDKN1A, CDKN2A, PTEN, RASSF1, TP53 and TP73) | APC, RB1 and
TP63 | MDM2, UBC,
CDK2, CDK6, TAF1, CCND1 and CDKNIC |
| DNA binding
related |
8.91e-6~1.13e-2 | 8 (BRCA1, BRCA2,
CDKN2A, MLH1, PDCD4, PTEN, TP53 and TP73) | APC, TP63,
RB1 and PYCARD | RAD51, PMS2,
UBC, TAF1 and NLRP3 |
| DNA damage
related |
6.05e-11~1.84e-3 | 6 (BRCA1,
CDKN1A, CDKN2A, TP53, TP73 and CHEK2) | TP63 | MDM2, UBC,
CDK2 and CCND1 |
| Apoptosis |
2.45e-6~2.45e-2 | 6 (BRCA1,
CDKN1A, CDKN2A, CHEK2, TP53 and TP73) | TP63 | |
| Signal transduction
by TP53 |
8.71e-10~3.67e-3 | 5 (BRCA1,
CDKN1A, CDKN2A, TP53 and TP73) | TP63 | MDM2, CDK2
and UBC |
| Aging | 2.5e-7~2.99e-2 | 5 (CDKN1A,
CDKN2A, CHEK2, PDCD4 and TP53) | | CDK6 |
| DNA repair |
1.43e-6~4.35e-3 | 5 (BRCA1, BRCA2,
CHEK2, MLH1 and TP73) | | RAD51, PMS2
and UBC |
| Cell growth | 6.34e-3~9.1e-2 | 3 (CDKN1A,
CDKN2A and TP53) | RB1 | |
4. Process associations of the 15 TSGs
The process associations of the 15 TSGs were
analyzed by PubGene Bio-associations (www.pubgene.org).
Pubgene is a gene/protein database and web-based tool for
literature mining. It carries out automated extraction of
experimental and theoretical biomedical knowledge from publicly
available gene and text databases to create a gene-to-gene
co-citation network for 13,712 named human genes by automated
analysis of titles and abstracts in over 10 million MEDLINE records
(60). Therefore, gene and protein
names are cross-referenced to each other and to terms that are
relevant to understanding their biological function and importance
in disease. Using PubGene process associations, we annotated the 15
TSGs to biological processes by probabilistic scoring, and the 10
most annotated processes of the 15 TSGs are shown in Table IV. It appears that most of the 15
TSGs played roles in processes of methylation/DNA methylation, DNA
damage and repair, cell cycle and apoptosis (P<0.001),
suggesting that these biological processes may be the most common
manner with which the 15 TSGs contribute to drug resistance in
ovarian cancer.
 | Table IVAnalysis of process associations of
the 15 TSGs using PubGene bio-associations. |
Table IV
Analysis of process associations of
the 15 TSGs using PubGene bio-associations.
| Annotated
process | No. of the 15
TSGs | Article
(P-value) |
|---|
| Methylation | 12 (BRCA1,
BRCA2, CDKN1A, CDKN2A, CHEK2, MLH1, PDCD4, PTEN, RASSF1,
TP53, TP73 and WWOX) | 0 |
| Mismatch
repair | 11 (BRCA1,
BRCA2, CDKN1A, CDKN2A, CHEK2, MLH1, PTEN, RASSF1, TP53,
TP73 and WWOX) | 0 |
| Cell cycle
checkpoint | 11 (BRCA1,
BRCA2, CDKN1A, CDKN2A, CHEK2, MLH1, PTEN, RASSF1, TP53,
TP73 and WWOX) | 0 |
| DNA replication
checkpoint | 3 (CDKN1A,
CHEK2 and TP53) | 0 |
| Response to DNA
damage stimulus | 10 (BRCA1,
BRCA2, CDKN1A, CDKN2A, CHEK2, MLH1, PTEN, RASSF1, TP53
and TP73) | 0 |
| DNA damage
checkpoint | 9 (BRCA1, BRCA2,
CDKN1A, CDKN2A, CHEK2, MLH1, TP53, TP73 and
WWOX) | 0 |
| Cell cycle
arrest | 14 (BRCA1,
BRCA2, CDKN1A, CDKN2A, CHEK2, FBXO32, IL24, MLH1, PDCD4,
PTEN, RASSF1, TP53, TP73 and WWOX) | 0 |
| DNA
methylation | 13 (BRCA1,
BRCA2, CDKN1A, CDKN2A, CHEK2, MLH1, PDCD4, PTEN, RASSF1,
SULF1, TP53, TP73 and WWOX) | 4.64e-191 |
| G2/M transition
checkpoint | 7 (BRCA1, BRCA2,
CDKN1A, CDKN2A, CHEK2, MLH1 and TP53) | 3.22e-164 |
| Apoptosis | 12 (BRCA1,
BRCA2, CDKN1A, CDKN2A, CHEK2, IL24, MLH1, PDCD4, PTEN,
RASSF1, TP53 and TP73) | 2.1e-119 |
5. Conclusion
TSGs play crucial roles in several aspects of cancer
development including cell cycle control, signal transduction,
angiogenesis, development and drug resistance, indicating that they
contribute to a broad array of normal and tumor-related functions.
It is proposed that TSGs provide a vast untapped resource for
anticancer therapy (8). It has been
reported that a total of 33 TSGs contribute to ovarian cancer
development through DNA damage, DNA repair, regulating
macromolecule metabolism, cell cycle, and apoptosis (14). However, an overview of the TSGs
associated with drug resistance in ovarian cancer had yet to be
reported. In this study, the 15 TSGs which are involved in drug
resistance were comprehensively reviewed. Furthermore, gene/protein
interaction and bio-association analysis were performed. The 15
TSGs may contribute to drug resistance as a whole in ovarian
cancer, since they have close interactions with each other in
accordance with the gene/protein interaction network. An additional
6 TSGs including APC, DAPK1, PLAGL1, RB1, PYCARD and
TP63, which had close interactions with the 15 TSGs
(Table II) and which were
associated with drug resistance in other types of solid cancer, may
be potential drug resistance-related genes in ovarian cancer.
UBC, which had direct physical interactions with a number of
the 15 TSGs, may be associated with drug resistance in ovarian
cancer, although its drug resistance-related functions have yet to
be reported.
DNA damage and apoptosis play important roles in
drug resistance in several solid tumors. DNA damage results in
genetic alterations that underlie drug resistance, disabled repair
and resistance to apoptosis (61).
Apoptosis plays an important role in the maintenance of
physiological homeostasis in response to stimuli, and failure of
apoptosis would result in unopposed tissue growth and, eventually,
fatal disease such as cancer (62).
It has been proved that apoptosis is involved in drug resistance in
several solid tumors including ovarian cancer. In this study, on
the basis of gene/protein-interaction and process-association
analysis, DNA damage and apoptosis were the main annotated
functions/processes for the 15 TSGs, suggesting that these two
biological processes may be the main manner for the participation
of TSGs in drug resistance in ovarian cancer. These results are
partly supported by previous studies that reported that BRCA1,
BRCA2, MLH1 and p21 contributed to ovarian cancer drug
resistance via DNA damage and repair, and p21, RASSF1, TP53
and TP73 contributed to ovarian cancer drug resistance via
apoptosis (Table I). Furthermore,
cell cycle was also a leading annotated function/process for the 15
TSGs on the basis of gene/protein-interaction and
process-association analysis. It has been proved that cell cycle
can create drug resistance and therefore reduce combination
chemotherapeutic efficacy (63). In
addition, DNA binding-related functions, which covered a total of
12 TSGs (Table III), may be
another crucial process for TSGs responding to drug resistance in
ovarian cancer. Methylation/DNA methylation was also important for
the 15 TSGs based on process association (Table IV); this finding was supported by
previous studies revealing that 9 of the 15 TSGs were regulated by
DNA methylation when they participated in the regulation of drug
resistance in ovarian cancer (Table
I). Collectively, five pathways/processes comprising DNA
damage, apoptosis, cell cycle, DNA binding, and methylation, may be
the major means by which TSGs respond to drug resistance in ovarian
cancer.
References
|
1
|
Balch C, Huang TH, Brown R and Nephew KP:
The epigenetics of ovarian cancer drug resistance and
resensitization. Am J Obstet Gynecol. 191:1552–1572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar
|
|
3
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson SW, Ozols RF and Hamilton TC:
Mechanisms of drug resistance in ovarian cancer. Cancer. 71(Suppl
2): S644–S649. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fraser M, Leung BM, Yan X, Dan HC, Cheng
JQ and Tsang BK: p53 is a determinant of X-linked inhibitor of
apoptosis protein/Akt-mediated chemoresistance in human ovarian
cancer cells. Cancer Res. 63:7081–7088. 2003.PubMed/NCBI
|
|
7
|
Cheng JQ, Jiang X, Fraser M, Li M, Dan HC,
Sun M and Tsang BK: Role of X-linked inhibitor of apoptosis protein
in chemoresistance in ovarian cancer: possible involvement of the
phosphoinositide-3 kinase/Akt pathway. Drug Resist Updat.
5:131–146. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sager R: Tumor suppressor genes: the
puzzle and the promise. Science. 246:1406–1412. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sherr CJ: Principles of tumor suppression.
Cell. 116:235–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shanker M, Jin J, Branch CD, Miyamoto S,
Grimm EA, Roth JA and Ramesh R: Tumor suppressor gene-based
nanotherapy: from test tube to the clinic. J Drug Deliv.
2011:4658452011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaur M, Radovanovic A, Essack M, et al:
Database for exploration of functional context of genes implicated
in ovarian cancer. Nucleic Acids Res. 37:D820–D823. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cancer Genome Atlas Research Network.
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar
|
|
13
|
Bast RC Jr, Hennessy B and Mills GB: The
biology of ovarian cancer: new opportunities for translation. Nat
Rev Cancer. 9:415–428. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao M, Sun J and Zhao Z: Distinct and
competitive regulatory patterns of tumor suppressor genes and
oncogenes in ovarian cancer. PLoS One. 7:e441752012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen H, Hardy TM and Tollefsbol TO:
Epigenomics of ovarian cancer and its chemoprevention. Front Genet.
2:672011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matei D, Shen C, Fang F, et al: A phase II
study of decitabine and carboplatin in recurrent platinum
(Pt)-resistant ovarian cancer (OC). J Clin Oncol. 29(Suppl): abst
5011. 2011.
|
|
17
|
Yang D, Khan S, Sun Y, Hess K, Shmulevich
I, Sood AK and Zhang W: Association of BRCA1 and BRCA2 mutations
with survival, chemotherapy sensitivity, and gene mutator phenotype
in patients with ovarian cancer. JAMA. 306:1557–1565. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou C, Smith JL and Liu J: Role of BRCA1
in cellular resistance to paclitaxel and ionizing radiation in an
ovarian cancer cell line carrying a defective BRCA1. Oncogene.
22:2396–2404. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Borst P, Rottenberg S and Jonkers J: How
do real tumors become resistant to cisplatin? Cell Cycle.
7:1353–1359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Wiltshire T, Senft J, Reed E and
Wang W: Irofulven induces replication-dependent CHK2 activation
related to p53 status. Biochem Pharmacol. 73:469–480. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang P, Gao W, Li H, Reed E and Chen F:
Inducible degradation of checkpoint kinase 2 links to
cisplatin-induced resistance in ovarian cancer cells. Biochem
Biophys Res Commun. 328:567–572. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chou JL, Su HY, Chen LY, et al: Promoter
hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and
tumor suppressor, is associated with poor prognosis in human
ovarian cancer. Lab Invest. 90:414–425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Plumb JA, Strathdee G, Sludden J, Kaye SB
and Brown R: Reversal of drug resistance in human tumor xenografts
by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene
promoter. Cancer Res. 60:6039–6044. 2000.
|
|
24
|
Strathdee G, MacKean MJ, Illand M and
Brown R: A role for methylation of the hMLH1 promoter in loss of
hMLH1 expression and drug resistance in ovarian cancer. Oncogene.
18:2335–2341. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Staub J, Chien J, Pan Y, et al: Epigenetic
silencing of HSulf-1 in ovarian cancer: implications in
chemoresistance. Oncogene. 26:4969–4978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu P, Gou M, Yi T, et al: Efficient
inhibition of an intraperitoneal xenograft model of human ovarian
cancer by HSulf-1 gene delivered by biodegradable cationic
heparin-polyethyleneimine nanogels. Oncol Rep. 27:363–370.
2012.
|
|
27
|
Gopalan B, Shanker M, Scott A, Branch CD,
Chada S and Ramesh R: MDA-7/IL-24, a novel tumor
suppressor/cytokine is ubiquitinated and regulated by the
ubiquitin-proteasome system, and inhibition of MDA-7/IL-24
degradation enhances the antitumor activity. Cancer Gene Ther.
15:1–8. 2008. View Article : Google Scholar
|
|
28
|
Xiong J, Peng ZL and Tan X: Effects of
adenoviral-mediated mda-7/IL-24 gene infection on the growth and
drug-resistance of drug-resistant. Sichuan Da Xue Xue Bao Yi Xue
Ban. 38:433–436. 2007.(In Chinese).
|
|
29
|
Kawakami Y, Hama S, Hiura M, et al:
Adenovirus-mediated p16 gene transfer changes the sensitivity to
taxanes and Vinca alkaloids of human ovarian cancer cells.
Anticancer Res. 21:2537–2545. 2001.PubMed/NCBI
|
|
30
|
Takai N and Narahara H: Histone
deacetylase inhibitor therapy in epithelial ovarian cancer. J
Oncol. 2010:4584312010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xia X, Ma Q, Li X, et al: Cytoplasmic p21
is a potential predictor for cisplatin sensitivity in ovarian
cancer. BMC Cancer. 11:3992011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Materna V, Surowiak P, Markwitz E,
Spaczynski M, Drag-Zalesinska M, Zabel M and Lage H: Expression of
factors involved in regulation of DNA mismatch repair- and
apoptosis pathways in ovarian cancer patients. Oncol Rep.
17:505–516. 2007.PubMed/NCBI
|
|
33
|
Cao Z, Yoon JH, Nam SW, Lee JY and Park
WS: PDCD4 expression inversely correlated with miR-21 levels in
gastric cancers. J Cancer Res Clin Oncol. 138:611–619. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Wang X, Song X, et al: Programmed
cell death 4 enhances chemosensitivity of ovarian cancer cells by
activating death receptor pathway in vitro and in vivo. Cancer Sci.
101:2163–2170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee S, Choi EJ, Jin C and Kim DH:
Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA
amplification contributes to cisplatin resistance in an ovarian
cancer cell line. Gynecol Oncol. 97:26–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu H, Cao Y, Weng D, et al: Effect of
tumor suppressor gene PTEN on the resistance to cisplatin in human
ovarian cancer cell lines and related mechanisms. Cancer Lett.
271:260–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yan X, Fraser M, Qiu Q and Tsang BK:
Over-expression of PTEN sensitizes human ovarian cancer cells to
cisplatin-induced apoptosis in a p53-dependent manner. Gynecol
Oncol. 102:348–355. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kassler S, Donninger H, Birrer MJ and
Clark GJ: RASSF1A and the Taxol response in ovarian cancer. Mol
Biol Int. 2012:2632672012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chmelarova M, Krepinska E, Spacek J, Laco
J, Beranek M and Palicka V: Methylation in the p53 promoter in
epithelial ovarian cancer. Clin Transl Oncol. 15:160–162. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Reles A, Wen WH, Schmider A, et al:
Correlation of p53 mutations with resistance to platinum-based
chemotherapy and shortened survival in ovarian cancer. Clin Cancer
Res. 7:2984–2997. 2001.PubMed/NCBI
|
|
42
|
Zhang YL, Guo XR, Shen DH, Cheng YX, Liang
XD, Chen YX and Wang Y: Expression and promotor methylation of p73
gene in ovarian epithelial tumors. Zhonghua Bing Li Xue Za Zhi.
41:33–38. 2012.(In Chinese).
|
|
43
|
Al-Bahlani S, Fraser M, Wong AY, Sayan BS,
Bergeron R, Melino G and Tsang BK: P73 regulates cisplatin-induced
apoptosis in ovarian cancer cells via a calcium/calpain-dependent
mechanism. Oncogene. 30:4219–4230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang W, Cui S, Ma J, Lu Q, Kong C, Liu T
and Sun Z: Cigarette smoking extract causes hypermethylation and
inactivation of WWOX gene in T-24 human bladder cancer cells.
Neoplasma. 59:216–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu YY, Li L, Li DR, Zhang W and Wang Q:
Suppression of WWOX gene by RNA interference reverses platinum
resistance acquired in SKOV3/SB cells. Zhonghua Fu Chan Ke Za Zhi.
43:854–858. 2008.(In Chinese).
|
|
46
|
Sharan R, Ulitsky I and Shamir R:
Network-based prediction of protein function. Mol Syst Biol.
3:882007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: a real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9(Suppl 1): S42008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang X and Guo B: Adenomatous polyposis
coli determines sensitivity to histone deacetylase
inhibitor-induced apoptosis in colon cancer cells. Cancer Res.
66:9245–9251. 2006. View Article : Google Scholar
|
|
49
|
Ogawa T, Liggett TE, Melnikov AA, et al:
Methylation of death-associated protein kinase is associated with
cetuximab and erlotinib resistance. Cell Cycle. 11:1656–1663. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Berge EO, Knappskog S, Geisler S, et al:
Identification and characterization of retinoblastoma gene
mutations disturbing apoptosis in human breast cancers. Mol Cancer.
9:1732010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ramachandran K, Gordian E and Singal R:
5-azacytidine reverses drug resistance in bladder cancer cells.
Anticancer Res. 31:3757–3766. 2011.PubMed/NCBI
|
|
52
|
Ramachandran K, Miller H, Gordian E,
Rocha-Lima C and Singal R: Methylation-mediated silencing of TMS1
in pancreatic cancer and its potential contribution to
chemosensitivity. Anticancer Res. 30:3919–3925. 2010.PubMed/NCBI
|
|
53
|
Tomkova K, Tomka M and Zajac V:
Contribution of p53, p63, and p73 to the developmental diseases and
cancer. Neoplasma. 55:177–181. 2008.PubMed/NCBI
|
|
54
|
Lo Nigro C, Monteverde M, Riba M, et al:
Expression profiling and long lasting responses to chemotherapy in
metastatic gastric cancer. Int J Oncol. 37:1219–1228.
2010.PubMed/NCBI
|
|
55
|
Zhang X, Gu L, Li J, et al: Degradation of
MDM2 by the interaction between berberine and DAXX leads to potent
apoptosis in MDM2-overexpressing cancer cells. Cancer Res.
70:9895–9904. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Poyurovsky MV, Katz C, Laptenko O, et al:
The C terminus of p53 binds the N-terminal domain of MDM2. Nat
Struct Mol Biol. 17:982–989. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mir R, Tortosa A, Martinez-Soler F, et al:
Mdm2 antagonists induce apoptosis and synergize with cisplatin
overcoming chemoresistance in TP53 wild-type ovarian cancer cells.
Int J Cancer. 132:1525–1536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kanayama H, Tanaka K, Aki M, et al:
Changes in expressions of proteasome and ubiquitin genes in human
renal cancer cells. Cancer Res. 51:6677–6685. 1991.PubMed/NCBI
|
|
59
|
Jiang RD, Zhang LX, Yue W, et al:
Establishment of a human nasopharyngeal carcinoma drug-resistant
cell line CNE2/DDP and screening of drug-resistant genes. Ai Zheng.
22:337–345. 2003.(In Chinese).
|
|
60
|
Jenssen TK, Laegreid A, Komorowski J and
Hovig E: A literature network of human genes for high-throughput
analysis of gene expression. Nat Genet. 28:21–28. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Casorelli I, Bossa C and Bignami M: DNA
damage and repair in human cancer: molecular mechanisms and
contribution to therapy-related leukemias. Int J Environ Res Public
Health. 9:2636–2657. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li J, Feng Q, Kim JM, et al: Human ovarian
cancer and cisplatin resistance: possible role of inhibitor of
apoptosis proteins. Endocrinology. 142:370–380. 2001.PubMed/NCBI
|
|
63
|
Shah MA and Schwartz GK: Cell
cycle-mediated drug resistance: an emerging concept in cancer
therapy. Clin Cancer Res. 7:2168–2181. 2001.PubMed/NCBI
|