Introduction
Head and neck cancer is among the 10 most common
types of cancer, with more than 600,000 new cases diagnosed
annually worldwide (1); the
squamous cell carcinoma subtype is the most frequent, corresponding
to 90% of all cases (2). This
disease originates at distinct head and neck topologies including
the oral cavity, the pharynx and the larynx. Due to the critical
location in the upper aerodigestive tract, these types of cancer
and their treatment significantly impair patient quality of life as
they affect breathing, swallowing, speech and even appearance. In
the past decades, a reduction in the mortality rates has been
observed in carcinomas of different anatomical sites; however, this
trend was not observed in the head and neck squamous cell carcinoma
(HNSCC) (3). Despite different
strategies used in the current treatment of HNSCC, a high
percentage of these patients progresses with locoregional and
distant recurrences (4). Therefore,
identifying molecular aberrations in HNSCC may improve our
understanding of head and neck carcinogenesis, allowing for the
identification of new strategies for subdividing patients into
biologically and clinically relevant subgroups, and highlighting
novel therapeutic options.
Cancer is a progressive genetic disease
characterized by the accumulation of genetic and epigenetic
modifications. Among the most common genetic alterations are the
mutations in coding and non-coding sequences (5) and the presence of mutations in the
phosphorylation domain of protein kinases that have been linked to
the development of various tumors. Therefore, protein kinase
inhibitors have demonstrated a significant efficacy in the
treatment of different types of cancer (6).
An important cellular signaling pathway is the
RAF/MEK/ERK (MAPK) pathway, which plays a role in mediating
cellular response to cell growth (7). Several studies have reported the
presence of hotspot mutations in KRAS (codons 12 and 13) and
BRAF (codons 469 and 600) which produce perpetually active
proteins resulting in increased proliferation and survival
signaling (8–13). There are few reports on the
frequency of mutations in KRAS and BRAF genes in
HNSCC cases (7,14–16).
KRAS downstream effector pathways also
include the PI3K-AKT molecular signaling cascade. The presence of
mutations in PIK3CA hotspots (codons 542, 545 and 1047) was
linked to increased cell survival by inhibiting apoptosis (17). Mutations in PIK3CA have been
found in different types of human cancer, including lung, gastric,
breast, ovarian, hepatocellular, pancreatic, esophageal and head
and neck (18–22).
Aside from the MAPK pathway, the JAK-STAT signaling
pathway also controls cell proliferation, differentiation and
survival by transmitting signals from the plasma membrane to the
cell nucleus (23). In cells,
cytokine receptors are associated through its cytosolic domain to
one of the four members of the JAK family of tyrosine kinases
(JAK1, JAK2, JAK3 and TYK2). Conformational changes in the receptor
enable JAKs to transphosphorylate and also phosphorylate the
receptor itself, activating them. Both are capable of recruiting
active substrates such as STAT proteins (signal transducer and
activator of transcription proteins), which when phosphorylated,
become active, translocate to the nucleus and regulate expression
of different genes (23). In recent
years, several studies have correlated the presence of mutations in
proteins of the JAK-STAT pathway with hematopoietic disorders. High
frequency of JAK2 V617F mutation is observed in myeloproliferative
cases, whereas JAK1 and JAK3 sequence alterations
have been found in cases of myeloid leukemia megakaryoblastic,
acute lymphoblastic leukemia and lymphomas (24–26).
In solid tumors, mutations in JAK1 have been detected in
lung and breast cancer and changes in the JAK3 gene were
identified in breast and gastric carcinomas (27).
Therapies targeting inhibition of multiple points
along the signal transduction pathways are potential new approaches
in the treatment of cancer. In non-small cell lung cancer (NSCLC),
the presence of activating mutations within the kinase domain of
epidermal growth factor receptor (EGFR) is common (28). Therapeutic approaches based on drugs
that inhibit the tyrosine kinase activity of this receptor
(gefitinib and erlotinib) have improved survival rates for NSCLC
patients (29).
Cetuximab, a chimeric monoclonal antibody against
epidermal growth factor receptor (EGFR), is used in treatment of
colorectal cancer (CRC). Recently, the presence of mutations in
KRAS was described as a marker for the resistance to therapy
with cetuximab in CRC (30). In
other words, CRC patients carrying mutations in this oncogene
cannot benefit from treatment with this drug, unlike patients whose
tumors have wild-type KRAS.
Cetuximab is the only kinase-targeted therapy
approved by the Food and Drug Administration (FDA) for the
treatment of locally and regionally advanced HNSCC in combination
with radiation. However, this agent generally presents only modest
efficacy, and efforts to identify the subset of responsive patients
who could benefit from this therapy have not been successful
(31).
The aim of the present study was to examine the
mutation status of KRAS, BRAF, PIK3CA,
JAK1 and JAK2 in primary head and neck cancer and to
verify if the mutation status could identify specific molecular
subgroups of HNSCC with clinical or therapeutic implications and if
KRAS mutations could be useful as a marker for the
resistance to therapy with cetuximab in HNSCC.
Materials and methods
Patients
This retrospective study involved 94 tumor specimens
obtained from patients diagnosed with HNSCC, who were treated at
the Department of Head and Neck Surgery of Barretos Cancer Hospital
(São Paulo, Brazil) between 2007 and 2010. These samples, available
at the tumor bank of the Hospital, were collected during surgery
for treatment of HNSCC. Tissue samples were snap-frozen in liquid
nitrogen within 30 min after resection and stored at −80°C. Only
patients diagnosed with primary HNSCC, not previously treated, that
were over 18 years of age, treated surgically with curative intent
and presenting with tumors at the oral cavity, the pharynx, or the
larynx were included in the study. The HNSCC diagnoses were
confirmed by a pathologist and all samples were examined
microscopically for the presence of neoplastic tissue and the
absence of contaminating normal mucosa.
The study protocol was reviewed and approved by the
Ethics Committee of Barretos Cancer Hospital and was performed in
accordance with the ethics guidelines of the 1975 Declaration of
Helsinki.
RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol®
reagent (Invitrogen, Carlsbad, CA, USA) following the
manufacturer's recommendations. The concentration of the resulting
RNA was measured using a spectrophotometer (NanoDrop 1000
Spectrophotometer, Thermo Fischer Scientific, Waltham, MA, USA) and
the quality of the RNAs was checked by electrophoresis on 1%
agarose gel stained with SYBR® Safe (Invitrogen).
Two micrograms of total RNA from each sample were
used to synthesize cDNA molecules using SuperScript® III
First-Strand Synthesis system (Invitrogen) according to the
conditions provided by the manufacturer. The cDNA obtained was
diluted 10× prior to use.
Sequencing analysis
PCR amplification of cDNA was performed and analyzed
for mutations at hotspots of KRAS (codons 12 and 13),
BRAF (codons 469 and 600) and PIK3CA (codons 542, 545
and 1047), as previously reported (32–34).
To search for mutations in JAK1 and JAK2, primers
were designed to generate seven fragments covering the entire
coding region of each gene. All primers used in this study are
presented in Table I.
 | Table IPrimers used for amplification of
fragments from BRAF, KRAS, PIK3CA, JAK1
and JAK2 genes. |
Table I
Primers used for amplification of
fragments from BRAF, KRAS, PIK3CA, JAK1
and JAK2 genes.
| Fragment | Sense | Sequence 5′-3′ | Annealing
temperature (°C) | Size (bp) |
|---|
| BRAF-11b | F |
GACGGGACTCGAGTGATGAT | 62 | 155 |
| R |
CTGCTGAGGTGTAGGTGCTG | | |
| BRAF-15b | F |
GCACAGGGCATGGATTACTT | 62 | 195 |
| R |
GATGACTTCTGGTGCCATCC | | |
| KRASc | F |
GGCCTGCTGAAAATGACTGAA | 62 | 253 |
| R |
CACAAAGAAAGCCCTCCCCA | | |
| PI3K-9d | Fa |
AATTGGTCTGTATCCCGAG | 60 | 199 |
| R |
CGGGGATAGTTACACAATAGT | | |
| PI3K-20d | Fa |
TGGAATGCCAGAACTACAATC | 60 | 175 |
| R |
ATGCTGTTTAATTGTGTGGAAG | | |
| JAK1-1 | F |
GCCCAGGCGCACACGGA | 62 | 680 |
| R |
GACAGCCATCCCTAGCACTCGTTC | | |
| JAK1-2 | F |
TTTGGTGAAATGCCTGGCTCCTAT | 62 | 594 |
| R |
CCACTCTTCCCGGATCTTGTTTTTC | | |
| JAK1-3 | F |
CTGGAAAATAAACACAAGAAGGATGAGGAG | 62 | 610 |
| R |
CGGGGCTTGGGCTGGC | | |
| JAK1-4 | F |
AGCCACCTCAAGAAGCAGATCCTG | 62 | 642 |
| R |
TGAGCTTGATGAATGGGCCACA | | |
| JAK1-5 | F |
GAAATGTGTGTACTAAAAACCTCCTCCTGG | 62 | 636 |
| R |
TCCTTTTTCAGATCAGCTATGTGGTTACC | | |
| JAK1-6 | F |
GGGGACAATACAGGGGAGCAGG | 62 | 408 |
| R |
GTGTAATACTCCTTATCGGTTTCAATTGCTTT | | |
| JAK1-7 | F |
TTCACCGGGACTTGGCAGCA | 62 | 525 |
| R |
CATTTGTTGCAGGAGAAGGACTTGATAA | | |
| JAK2-1 | F |
AGAAGCAGGCAACAGGAACAAGATG | 62 | 529 |
| R |
ACGTTCAGCACCTCGAGATATTCCAT | | |
| JAK2-2 | F |
TGAGTCAACCAGGCATAATGTACTCTACAG | 62 | 588 |
| R |
CCTCTTGACCACTGAATTCCACCG | | |
| JAK2-3 | F |
GGAAACTCTGCAGTCTGCCTTCTACAC | 62 | 638 |
| R |
AGTTCTTCTTTGTCCCACTGAGGTTGTAC | | |
| JAK2-4 | F |
TTTTTGACTTTTGCTGTCGAGCGA | 62 | 693 |
| R |
TTCTTCTAGAAAATGCATGGCCCA | | |
| JAK2-5 | F |
TGGAGTATGTGTCTGTGGAGACGAGAA | 62 | 693 |
| R |
AACTGTGTAGGATCCCGGTCTTCAAA | | |
| JAK2-6 | F |
GCCTTCTTTCAGAGCCATCATACGA | 62 | 703 |
| R |
CTCTCTGTCAGTGATTCTGGAGCATACC | | |
| JAK2-7 | F |
CGAGAAATATATTGGTGGAGAACGAGAAC | 62 | 659 |
| R |
AATGTCTTTTACTGGTGGCCTCATGAA | | |
Amplification reactions contained 1 μl of cDNA as
template in a 25 μl reaction mixture consisting of 1X reaction
buffer, 1 mmol/l deoxynucleotide triphosphate, 2 mmol/l magnesium,
200 nmol/l of each primer, and 1.2 unit Platinun Taq (Invitrogen).
Following purification using QIAquick® Multiwell PCR
Purification Kit (Qiagen, Hilden, Germany) and Gel Band
Purification (GE Healthcare, Little Chalfont, Buckinghamshire, UK),
the PCR products were subjected to sequencing (both directions)
using BigDye® Terminator v3.1 sequencing kit (Applied
Biosystems, Foster City, CA, USA), ethanol precipitation, and an
automated sequencer (ABI PRISM 3130XL Genetic Analyzer; Applied
Biosystems).
The sequences obtained were analyzed with the
BioEdit Sequence Alignment Editor software (35) and the Phred/Phrap/Consed Package
(36). The sequences were compared
to a human reference sequence, available at UCSC Genome
Bioinformatics (http://genome.ucsc.edu/). The annotation of the
sequence variations identified was performed using the Ensembl
database (http://www.ensembl.org/) and the NCBI
database (http://www.ncbi.nlm.nih.gov/).
Results
Patient characteristics
Table II summarizes
the clinical and histological characteristics of the patients
enrolled in the study. Ninety four primary tumor samples were
collected from untreated HNSCC patients (80 male and 14 female).
Median age was 57.8 years (range, 32–82 years). Tobacco or alcohol
consumption (current or former) was reported by 91.2 and 74.7%
patients, respectively. Primary tumor sites included: oral cavity
(70.2%), larynx (14.9%), oropharynx (12.8%) and hypopharynx (2.1%).
Regarding tumor stage, the majority was T3/T4 (81.7%). Forty two
patients presented positive lymph nodes, among those 15 (19.0%)
presented extracapsular spread. Perineural invasion was detected in
24 cases (27.0%) among 89 patients with data available, while
lymphovascular invasion was observed in 15 cases (17.4%) among 86
patients with data available.
 | Table IIDemographic and clinical
characteristics of the HNSCC patients included in this study
(n=94). |
Table II
Demographic and clinical
characteristics of the HNSCC patients included in this study
(n=94).
| Parameter | n | % |
|---|
| Age (years) |
| <55 | 33 | 35.1 |
| ≥55 | 61 | 64.9 |
| Mean | 57.8 | |
| Range | 32–82 | |
| Gender |
| Male | 80 | 85.1 |
| Female | 14 | 14.9 |
| Tobacco
consumption |
| Yes | 83 | 91.2 |
| No | 8 | 8.8 |
| Alcohol
consumption |
| Yes | 65 | 74.7 |
| No | 22 | 25.3 |
| Tumor site |
| Oral cavity | 66 | 70.2 |
| Larynx | 14 | 14.9 |
| Oropharynx | 12 | 12.8 |
| Hypopharynx | 2 | 2.1 |
| T stagea |
| T1/T2 | 17 | 18.3 |
| T3/T4 | 76 | 81.7 |
| N stage |
| N0 | 41 | 49.4 |
| N+ | 42 | 50.6 |
| Extracapsular
spread |
| Yes | 15 | 19.0 |
| No | 64 | 81.0 |
| Perineural
invasion |
| Yes | 24 | 27.0 |
| No | 65 | 73.0 |
| Lymphovascular
invasion |
| Yes | 15 | 17.4 |
| No | 71 | 82.6 |
Mutation analysis
The presence of sequence variations in the hotspots
previously described in the coding regions of KRAS (codons
12 and 13), BRAF (codons 469 and 600) and PIK3CA
(codons 542, 545 and 1047) was examined in cDNA molecules from 94
HNSCC specimens. The assembly of the consensus sequences, as well
as the evaluation of base qualities, were conducted with
Phred/Phrap/Consed Package (36).
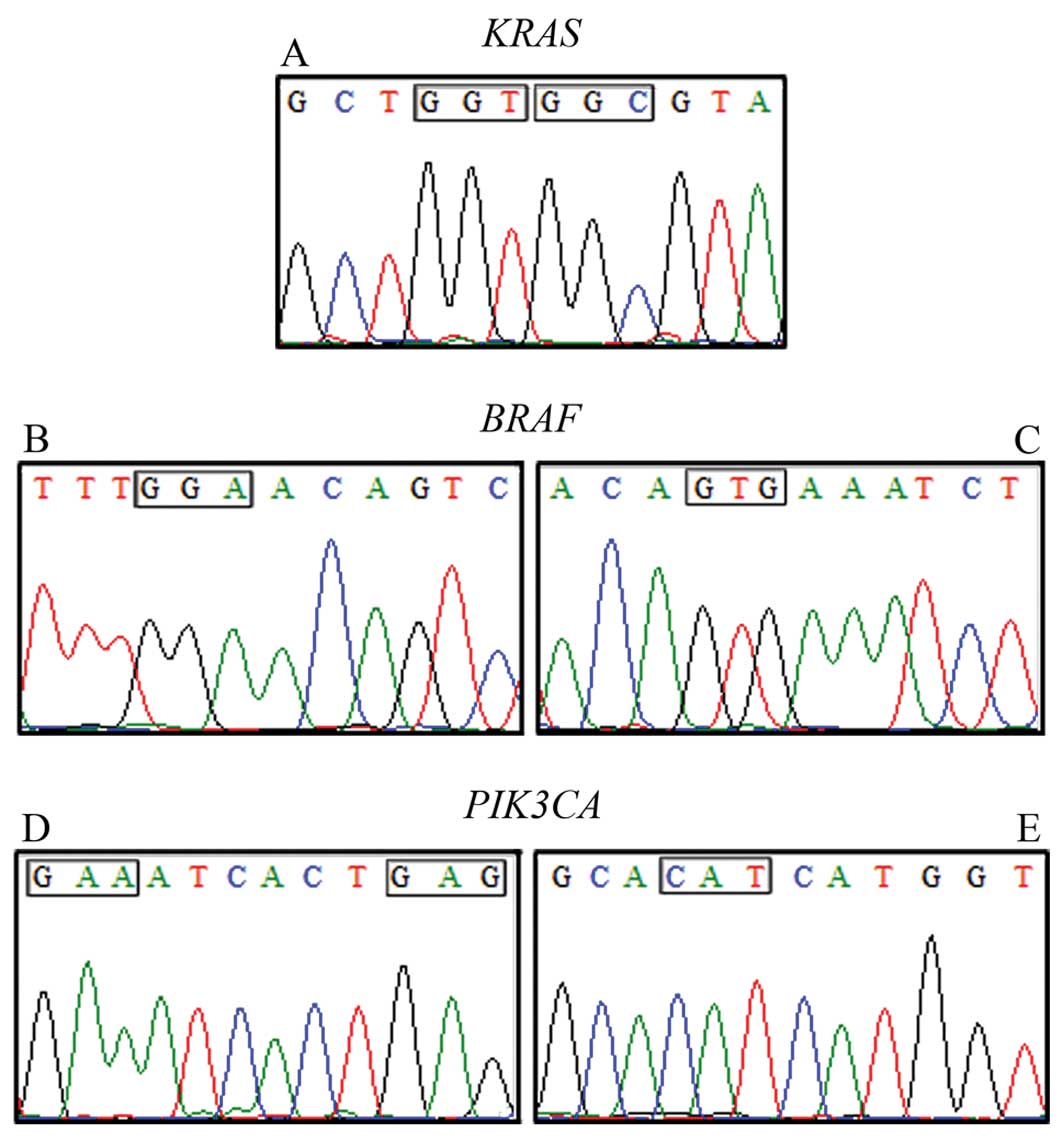
No somatic mutations were detected in any of these genes in all
cases examined. Representative cases are shown in Fig. 1.
Mutations in JAK1 and JAK2 genes have
been reported in different types of cancer, mainly in hematological
malignances; however, no hotspots were identified in the sequence
of these genes. Therefore, primers were designed to generate
overlapping fragments covering the entire coding region of
JAK1 (seven fragments, size 408 to 680 bp) and JAK2
(seven fragments, size 529 to 703 bp). We searched for sequence
variations in 20 HNSCC samples, but were not able to detect any
mutations within the JAK1 and JAK2 coding
sequences.
The analysis of the complete sequences from
JAK1 and JAK2 genes allowed the identification of
some sequence variations reported as polymorphisms in the Ensembl
database. JAK1 presented 6 different polymorphisms at
positions 579 T>C (codon 193), 1977 C>T (codon 659), 2049
C>T (codon 683), 2097 C>G (codon 699), 2199 A>G (codon
733) and 3096 G>A (codon 1032). Of note, none of these
alterations resulted in amino acid changes (Table III). For JAK2, we found
four different polymorphisms at positions 489 C>T (codon 163),
1177 C>G (codon 393), 2490 G>A (codon 830) and 3188 G>A
(codon 1063) (Table IV). The
alteration of the C nucleotide by a G at position 1177 promotes a
substitution of a Leucine residue by a Valine (codon 393), whereas
the replacement of G at position 3188 by an A induces the change of
an Arginine by a Histidine (codon 1063) (Table IV).
 | Table IIISingle nucleotide polymorphisms
(SNPs) of the JAK1 gene found in 20 HNSCC samples
analyzed. |
Table III
Single nucleotide polymorphisms
(SNPs) of the JAK1 gene found in 20 HNSCC samples
analyzed.
| SNP | Codon | Exon | Amino acid | No. of cases | Frequency observed
(%) |
|---|
| rs 45598436 | 193 GCT>GCC | 6 | Ala>Ala | 4 | 20 |
| rs 17127063 | 659 CGC>CGT | 14 | Arg>Arg | 1 | 5 |
| rs 2230587 | 683 AGC>AGT | 15 | Ser>Ser | 6 | 30 |
| rs 3737139 | 699 GCC>GCG | 15 | Ala>Ala | 1 | 5 |
| rs 2230588 | 733 CCA>CCG | 16 | Pro>Pro | 5 | 25 |
| rs 12129819 | 1032
AAG>AAA | 22 | Lys>Lys | 4 | 20 |
 | Table IVSingle nucleotide polymorphisms
(SNPs) of the JAK2 gene found in 20 HNSCC samples
analyzed. |
Table IV
Single nucleotide polymorphisms
(SNPs) of the JAK2 gene found in 20 HNSCC samples
analyzed.
| SNP | Codon | Exon | Amino acid | No. of cases | Frequency observed
(%) |
|---|
| rs 10429491 | 163 CAC>CAT | 7 | His>His | 13 | 65 |
| rs 2230723 | 393 CTT>GTT | 9 | Leu>Val | 1 | 5 |
| rs 2230724 | 830 CTG>CTA | 20 | Leu>Leu | 17 | 85 |
| rs 41316003 | 1063
CGT>CAT | 25 | Arg>His | 2 | 10 |
Discussion
The management of HNSCC patients has improved in
recent years with developments in surgical techniques, the
introduction of altered fractionation radiation, new
chemotherapeutic agents, and combined use of chemoradiotherapy;
however, long-term survival rates have improved only marginally,
and the 5-year overall survival rate remains approximately 50%
(3).
To date, two promising strategies tested for the
treatment of HNSCC are blocking growth factors signaling pathways
and interfering in pathways related to angiogenesis. EGFR has been
studied extensively as a therapeutic target due to its high
expression in different tumors and its influence in the regulation
of proliferation, apoptosis, metastasis, angiogenesis, and cell
differentiation. The extracellular portion of EGFR can be inhibited
by monoclonal antibodies and intracellular part by tyrosine kinase
inhibitors. The monoclonal antibody cetuximab, an EGFR
extracellular inhibitor, has been used in the treatment of
colorectal tumors (37). Therapies
targeting epidermal growth factor receptor (EGFR/HER-1/ERBB1)
pathways have also been used in HNSCC, but only a minority of
patients have benefited from these treatments, since small-molecule
tyrosine kinase inhibitors (gefitinib and erlotinib) and monoclonal
antibodies against EGFR (cetuximab) have shown limited efficacy
(38–40).
It is well known that cetuximab prolongs overall
survival of locally advanced HNSCC patients when delivered in
combination with radiation (41).
This drug has also shown activity in the first-line treatment of
recurrent and/or metastatic HNSCC in combination with cisplatin or
cisplatin/fluorouracil (42,43).
However, the response rates of cetuximab as a single agent are
approximately 13% (40). Thus, the
identification of clinically effective markers for selection of
HNSCC patients responsive to cetuximab therapy is of paramount
importance.
In colorectal cancer, there is a well-established
association between the presence of mutations in KRAS and
the response to cetuximab treatment (30). Considering all types of human
cancer, KRAS mutations are the most common type of Ras
mutation, with HRAS being the least common; however, in head
and neck cancer, Ras mutations appear to be exclusively
HRAS mutations (44). To
date, few studies have attempted to assess the KRAS mutation
status in HNSCC and their results are contradictory. In a recent
study, Smilek et al(14)
examined the KRAS mutation status of 27 HNSCC patients and
found alterations in 14.8% (4/27) of the cases. Weber et
al(7) found KRAS
mutations in 6% (3/89) of HNSCC samples examined, while Van Damme
et al(15) evaluated 22
HNSCC specimens and identified a mutation in one case (4.5%). On
the other hand, no KRAS mutation was detected among 183
HNSCC samples investigated from a Japanese cohort (45). Mutations in the KRAS gene
were also infrequent in oral squamous cell carcinomas (OSCCs)
(16,46,47).
In the present study, wild-type KRAS was detected in all 94
HNSCC samples evaluated. In light of these findings and in contrast
to colorectal cancer, our results together with others indicate
that KRAS mutations may not be considered a predictor of
sensitivity to cetuximab in HNSCC.
As well as the RAS family, another member of the
MAPK pathway is BRAF. Somatic point mutations of
BRAF, such as those that occur at hotspot V600E of its
kinase domain, can result in elevated BRAF kinase function
(48). Mutations in this gene have
been associated with various types of cancer, including non-Hodgkin
lymphoma, colorectal cancer, thyroid cancer and lung carcinoma
(10,11,49,50). A
German study found BRAF mutated in 3% (3/89) of HNSCC cases
evaluated (7), while a study
evaluating American HNSCC patients reported BRAF mutations
in 2.4% (1/42) of the cases (16).
In the present study, none of the 94 samples analyzed presented
alterations in BRAF sequence.
Mutations in PIK3CA, the catalytic subunit of
PI3-kinase, are reported to have higher oncogenic potential. Recent
exome sequencing analyses revealed PIK3CA activating
mutations in 6–8% of HNSCC (51,52).
Two independent studies, evaluating 38 HNSCC samples and 37 OSCC
specimens, identified PIK3CA mutations in approximately 11%
of the cases (47,53). Kozaki et al(54) detected mutations in PIK3CA in
7.4% (8/108) of OSCC, whereas Fenic et al(55) and Bruckman et al(16) found a lower frequency of mutations
in HNSCC (2.9 and 3%, respectively). A study analyzing South-Asian
HNSCC patients found PIK3CA mutations in 10.5% (2/19) of the
Indian cases, while only wild-type PIK3CA was detected in
Vietnamese tumors (0/18) (56). We
did not identify any PIK3CA mutations in all 94 HNSCC
patients evaluated. It should be noted that we are unable to
discuss the presence of mutations in other gene regions not within
hotspots previously described in KRAS, BRAF and
PIK3CA.
The MAPK and PI3K-AKT signaling pathways are
involved in HNSCC tumorigenesis, but the oncogenic activation of
these pathways may include additional mechanisms other than point
mutations, such as chromosomal amplification or gene
overexpression. Consistent with this, PIK3CA was found
frequently amplified in OSCC (57–59)
and Hoa et al(60) detected
increased expression of KRAS in HNSCC cell lines.
The JAK-STAT signaling pathway is essential for the
physiology of healthy individuals as it mediates cytokine responses
in hematopoietic cells. This pathway is also responsible for
regulating the hormone receptor signaling, including various growth
hormones and prolactin receptors. In recent years, several
mutations in JAK genes associated with constitutive
activation were identified and it is possible that mutations in
this pathway component are present in various types of cancer
(23). It is important to define
the role of this pathway in solid tumors, as new therapeutic agents
that have JAK as target are being developed and some JAK kinase
inhibitors are already being tested in clinical trials (61).
Mutations in JAK genes are commonly found in
hematological tumors (62–64); however, few mutations have been
described in solid tumors, such as breast, lung and gastric tumors
(27). To the best of our
knowledge, our study is the first to search for JAK
mutations in HNSCC. We analyzed the complete cDNA sequence of
JAK1 and JAK2 genes in 20 HNSCC samples and, although
some known polymorphisms have been evidenced, no mutation could be
detected.
In conclusion, the present study investigated the
presence of mutations in JAK1 and JAK2 genes in HNSCC
patients examined and also evaluated the presence of sequence
alterations in PIK3CA, BRAF and KRAS genes.
Overall, no mutation in these genes could be detected in the HNSCC
specimens examined. Also, KRAS mutation suggests that
alterations in this gene do not seem to be useful as a cetuximab
predictor of sensitivity in HNSCC.
Acknowledgements
The present study was supported by Fundação de
Amparo à Pesquisa do Estado de São Paulo-FAPESP grant
(2008/58460-9). T.G.C was the recipient of a scholarship from
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-CAPES.
A.C.C. was the recipient of a scholarship from Fundação de Amparo à
Pesquisa do Estado de São Paulo-FAPESP. J.K.O was the recipient of
scholarships from the National Counsel of Technological and
Scientific Development-CNPq. A.L.C. has a National Counsel of
Technological and Scientific Development-CNPq scholarship
(313181/2009-8). A.L.V. has a National Counsel of Technological and
Scientific Development-CNPq scholarship (302360/2008-5).
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wong DT, Todd R, Tsuji T and Donoff RB:
Molecular biology of human oral cancer. Crit Rev Oral Biol Med.
7:319–328. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carvalho AL, Nishimoto IN, Califano JA and
Kowalski LP: Trends in incidence and prognosis for head and neck
cancer in the United States: a site-specific analysis of the SEER
database. Int J Cancer. 114:806–816. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hinerman RW, Mendenhall WM, Morris CG,
Amdur RJ, Werning JW and Villaret DB: Postoperative irradiation for
squamous cell carcinoma of the oral cavity: 35-year experience.
Head Neck. 26:984–994. 2004.PubMed/NCBI
|
|
5
|
Aaltonen LA, Peltomaki P, Leach FS, et al:
Clues to the pathogenesis of familial colorectal cancer. Science.
260:812–816. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greenman C, Stephens P, Smith R, et al:
Patterns of somatic mutation in human cancer genomes. Nature.
446:153–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weber A, Langhanki L, Sommerer F,
Markwarth A, Wittekind C and Tannapfel A: Mutations of the BRAF
gene in squamous cell carcinoma of the head and neck. Oncogene.
22:4757–4759. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matthaios D, Zarogoulidis P, Balgouranidou
I, Chatzaki E and Kakolyris S: Molecular pathogenesis of pancreatic
cancer and clinical perspectives. Oncology. 81:259–272. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Santarpia L, El-Naggar AK, Cote GJ, Myers
JN and Sherman SI: Phosphatidylinositol 3-kinase/akt and
ras/raf-mitogen-activated protein kinase pathway mutations in
anaplastic thyroid cancer. J Clin Endocrinol Metab. 93:278–284.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rowe LR, Bentz BG and Bentz JS: Detection
of BRAF V600E activating mutation in papillary thyroid carcinoma
using PCR with allele-specific fluorescent probe melting curve
analysis. J Clin Pathol. 60:1211–1215. 2007. View Article : Google Scholar
|
|
11
|
Tie J, Gibbs P, Lipton L, et al:
Optimizing targeted therapeutic development: analysis of a
colorectal cancer patient population with the BRAF(V600E) mutation.
Int J Cancer. 128:2075–2084. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kullmann F, Hartmann A, Stohr R, et al:
KRAS mutation in metastatic pancreatic ductal adenocarcinoma:
results of a multicenter phase II study evaluating efficacy of
cetuximab plus gemcitabine/oxaliplatin (GEMOXCET) in first-line
therapy. Oncology. 81:3–8. 2011. View Article : Google Scholar
|
|
13
|
Rako I, Jakic-Razumovic J, Caban D, et al:
The role of KRAS gene mutation testing in colorectal cancer - a
predictive biomarker of response to EGFR inhibitors therapy. Lijec
Vjesn. 133:403–407. 2011.(In Croatian).
|
|
14
|
Smilek P, Neuwirthova J, Jarkovsky J, et
al: Epidermal growth factor receptor (EGFR) expression and
mutations in the EGFR signaling pathway in correlation with
anti-EGFR therapy in head and neck squamous cell carcinomas.
Neoplasma. 59:508–515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van Damme N, Deron P, Van Roy N, et al:
Epidermal growth factor receptor and K-RAS status in two cohorts of
squamous cell carcinomas. BMC Cancer. 10:1892010.PubMed/NCBI
|
|
16
|
Bruckman KC, Schonleben F, Qiu W, Woo VL
and Su GH: Mutational analyses of the BRAF, KRAS, and PIK3CA genes
in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol
Oral Radiol Endod. 110:632–637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rameh LE and Cantley LC: The role of
phosphoinositide 3-kinase lipid products in cell function. J Biol
Chem. 274:8347–8350. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee JW, Soung YH, Kim SY, et al: PIK3CA
gene is frequently mutated in breast carcinomas and hepatocellular
carcinomas. Oncogene. 24:1477–1480. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Levine DA, Bogomolniy F, Yee CJ, et al:
Frequent mutation of the PIK3CA gene in ovarian and breast cancers.
Clin Cancer Res. 11:2875–2878. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Phillips WA, Russell SE, Ciavarella ML, et
al: Mutation analysis of PIK3CA and PIK3CB in esophageal cancer and
Barrett's esophagus. Int J Cancer. 118:2644–2646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schonleben F, Qiu W, Ciau NT, et al:
PIK3CA mutations in intraductal papillary mucinous
neoplasm/carcinoma of the pancreas. Clin Cancer Res. 12:3851–3855.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu G, Mambo E, Guo Z, et al: Uncommon
mutation, but common amplifications, of the PIK3CA gene in thyroid
tumors. J Clin Endocrinol Metab. 90:4688–4693. 2005. View Article : Google Scholar
|
|
23
|
Constantinescu SN, Girardot M and Pecquet
C: Mining for JAK-STAT mutations in cancer. Trends Biochem Sci.
33:122–131. 2008. View Article : Google Scholar
|
|
24
|
Flex E, Petrangeli V, Stella L, et al:
Somatically acquired JAK1 mutations in adult acute lymphoblastic
leukemia. J Exp Med. 205:751–758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Walters DK, Mercher T, Gu TL, et al:
Activating alleles of JAK3 in acute megakaryoblastic leukemia.
Cancer Cell. 10:65–75. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Melzner I, Bucur AJ, Bruderlein S, et al:
Biallelic mutation of SOCS-1 impairs JAK2 degradation and sustains
phospho-JAK2 action in the MedB-1 mediastinal lymphoma line. Blood.
105:2535–2542. 2005. View Article : Google Scholar
|
|
27
|
Jeong EG, Kim MS, Nam HK, et al: Somatic
mutations of JAK1 and JAK3 in acute leukemias and solid cancers.
Clin Cancer Res. 14:3716–3721. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tokumo M, Toyooka S, Kiura K, et al: The
relationship between epidermal growth factor receptor mutations and
clinicopathologic features in non-small cell lung cancers. Clin
Cancer Res. 11:1167–1173. 2005.PubMed/NCBI
|
|
29
|
Rosell R, Moran T, Carcereny E, et al:
Non-small-cell lung cancer harbouring mutations in the EGFR kinase
domain. Clin Transl Oncol. 12:75–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Siddiqui AD and Piperdi B: KRAS mutation
in colon cancer: a marker of resistance to EGFR-I therapy. Ann Surg
Oncol. 17:1168–1176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Licitra L, Mesia R, Rivera F, et al:
Evaluation of EGFR gene copy number as a predictive biomarker for
the efficacy of cetuximab in combination with chemotherapy in the
first-line treatment of recurrent and/or metastatic squamous cell
carcinoma of the head and neck: EXTREME study. Ann Oncol.
22:1078–1087. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nikiforova MN, Kimura ET, Gandhi M, et al:
BRAF mutations in thyroid tumors are restricted to papillary
carcinomas and anaplastic or poorly differentiated carcinomas
arising from papillary carcinomas. J Clin Endocrinol Metab.
88:5399–5404. 2003. View Article : Google Scholar
|
|
33
|
Mathur A, Moses W, Rahbari R, et al:
Higher rate of BRAF mutation in papillary thyroid cancer over time:
a single-institution study. Cancer. 117:4390–4395. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mori R, Ishiguro H, Kimura M, et al:
PIK3CA mutation status in Japanese esophageal squamous cell
carcinoma. J Surg Res. 145:320–326. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hall TA: BioEdit: a user-friendly
biological sequence alignment editor and analysis program for
Windows 95/98/NT. Nucleic Acids Symposium Series. 41:95–98.
1999.
|
|
36
|
Ewing B, Hillier L, Wendl M and Green P:
Base-calling of automated sequencer traces using phred. I. Accuracy
assessment. Genome Res. 8:175–185. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bianchini C, Ciorba A, Pelucchi S, Piva R
and Pastore A: Targeted therapy in head and neck cancer. Tumori.
97:137–141. 2011.PubMed/NCBI
|
|
38
|
Cohen EE, Kane MA, List MA, et al: Phase
II trial of gefitinib 250 mg daily in patients with recurrent
and/or metastatic squamous cell carcinoma of the head and neck.
Clin Cancer Res. 11:8418–8424. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Soulieres D, Senzer NN, Vokes EE, Hidalgo
M, Agarwala SS and Siu LL: Multicenter phase II study of erlotinib,
an oral epidermal growth factor receptor tyrosine kinase inhibitor,
in patients with recurrent or metastatic squamous cell cancer of
the head and neck. J Clin Oncol. 22:77–85. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vermorken JB, Trigo J, Hitt R, et al:
Open-label, uncontrolled, multicenter phase II study to evaluate
the efficacy and toxicity of cetuximab as a single agent in
patients with recurrent and/or metastatic squamous cell carcinoma
of the head and neck who failed to respond to platinum-based
therapy. J Clin Oncol. 25:2171–2177. 2007.
|
|
41
|
Bonner JA, Harari PM, Giralt J, et al:
Radiotherapy plus cetuximab for squamous-cell carcinoma of the head
and neck. N Engl J Med. 354:567–578. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Burtness B, Goldwasser MA, Flood W, Mattar
B and Forastiere AA: Phase III randomized trial of cisplatin plus
placebo compared with cisplatin plus cetuximab in
metastatic/recurrent head and neck cancer: an Eastern Cooperative
Oncology Group study. J Clin Oncol. 23:8646–8654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bourhis J, Rivera F, Mesia R, et al: Phase
I/II study of cetuximab in combination with cisplatin or
carboplatin and fluorouracil in patients with recurrent or
metastatic squamous cell carcinoma of the head and neck. J Clin
Oncol. 24:2866–2872. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Loyo M, Li RJ, Bettegowda C, et al:
Lessons learned from next-generation sequencing in head and neck
cancer. Head Neck. 35:454–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fujii S, Uryu H, Akashi K, et al: Clinical
significance of KRAS gene mutation and epidermal growth factor
receptor expression in Japanese patients with squamous cell
carcinoma of the larynx, oropharynx and hypopharynx. Int J Clin
Oncol. Mar 24–2012.(Epub ahead of print).
|
|
46
|
Chang SE, Bhatia P, Johnson NW, et al: Ras
mutations in United Kingdom examples of oral malignancies are
infrequent. Int J Cancer. 48:409–412. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cohen Y, Goldenberg-Cohen N, Shalmon B, et
al: Mutational analysis of PTEN/PIK3CA/AKT pathway in oral squamous
cell carcinoma. Oral Oncol. 47:946–950. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mercer KE and Pritchard CA: Raf proteins
and cancer: B-Raf is identified as a mutational target. Biochim
Biophys Acta. 1653:25–40. 2003.PubMed/NCBI
|
|
49
|
Lee JW, Yoo NJ, Soung YH, et al: BRAF
mutations in non-Hodgkin's lymphoma. Br J Cancer. 89:1958–1960.
2003. View Article : Google Scholar
|
|
50
|
Kobayashi M, Sonobe M, Takahashi T, et al:
Clinical significance of BRAF gene mutations in patients with
non-small cell lung cancer. Anticancer Res. 31:4619–4623.
2011.PubMed/NCBI
|
|
51
|
Agrawal N, Frederick MJ, Pickering CR, et
al: Exome sequencing of head and neck squamous cell carcinoma
reveals inactivating mutations in NOTCH1. Science. 333:1154–1157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Stransky N, Egloff AM, Tward AD, et al:
The mutational landscape of head and neck squamous cell carcinoma.
Science. 333:1157–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qiu W, Schonleben F, Li X, et al: PIK3CA
mutations in head and neck squamous cell carcinoma. Clin Cancer
Res. 12:1441–1446. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kozaki K, Imoto I, Pimkhaokham A, et al:
PIK3CA mutation is an oncogenic aberration at advanced stages of
oral squamous cell carcinoma. Cancer Sci. 97:1351–1358. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fenic I, Steger K, Gruber C, Arens C and
Woenckhaus J: Analysis of PIK3CA and Akt/protein kinase B in head
and neck squamous cell carcinoma. Oncol Rep. 18:253–259.
2007.PubMed/NCBI
|
|
56
|
Murugan AK, Hong NT, Fukui Y, Munirajan AK
and Tsuchida N: Oncogenic mutations of the PIK3CA gene in head and
neck squamous cell carcinomas. Int J Oncol. 32:101–111.
2008.PubMed/NCBI
|
|
57
|
Estilo CL, POC, Ngai I, et al: The role of
novel oncogenes squamous cell carcinoma-related oncogene and
phosphatidylinositol 3-kinase p110alpha in squamous cell carcinoma
of the oral tongue. Clin Cancer Res. 9:2300–2306. 2003.PubMed/NCBI
|
|
58
|
Freier K, Schwaenen C, Sticht C, et al:
Recurrent FGFR1 amplification and high FGFR1 protein expression in
oral squamous cell carcinoma (OSCC). Oral Oncol. 43:60–66. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu CJ, Lin SC, Chen YJ, Chang KM and
Chang KW: Array-comparative genomic hybridization to detect
genomewide changes in microdissected primary and metastatic oral
squamous cell carcinomas. Mol Carcinog. 45:721–731. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hoa M, Davis SL, Ames SJ and Spanjaard RA:
Amplification of wild-type K-ras promotes growth of head and neck
squamous cell carcinoma. Cancer Res. 62:7154–7156. 2002.PubMed/NCBI
|
|
61
|
Lai SY and Johnson FM: Defining the role
of the JAK-STAT pathway in head and neck and thoracic malignancies:
implications for future therapeutic approaches. Drug Resist Updat.
13:67–78. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Baxter EJ, Scott LM, Campbell PJ, et al:
Acquired mutation of the tyrosine kinase JAK2 in human
myeloproliferative disorders. Lancet. 365:1054–1061. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Levine RL, Wadleigh M, Cools J, et al:
Activating mutation in the tyrosine kinase JAK2 in polycythemia
vera, essential thrombocythemia, and myeloid metaplasia with
myelofibrosis. Cancer Cell. 7:387–397. 2005. View Article : Google Scholar
|
|
64
|
Colaizzo D, Amitrano L, Tiscia GL,
Grandone E, Guardascione MA and Margaglione M: A new JAK2 gene
mutation in patients with polycythemia vera and splanchnic vein
thrombosis. Blood. 110:2768–2769. 2007. View Article : Google Scholar : PubMed/NCBI
|