Introduction
Glioblastoma multiforme (GBM), a grade IV and
malignant glioma, is the most common, aggressive and proliferative
among all gliomas (1). Due to its
nature, and following high recurrence rate, as well as strong
resistance to treatment, the prognosis of GBM patients remains poor
even after surgical resection and adjuvant radiotherapy or
chemotherapy. In 2001, Sullivan reported that the median survival
time for GBM patients was approximately 12 months, compared with 5
years in patients with low-grade glioma (2). Despite decades of concerted efforts
and the advances in surgery, radiotherapy and chemotherapy, the
overall 5-year survival rate of GBM remains less than 5% and is
even poorer for elderly patients (3). Therefore, it is important to elucidate
the mechanism of glioma tumorigenesis and to find and develop the
key molecular targets for effective therapy.
NF-E2-related factor 2 (Nrf2) is considered a
critical regulator of intracellular antioxidants and phase II
detoxification enzymes. Nrf2 belongs to the Keap1-Nrf2-ARE
(antioxidant response element) signaling pathway and upregulates
several ARE-containing genes, including heme oxygenase-1 (HO-1) and
NAD(P)H:quinone oxidoreductase-1 (NQO-1) (4). Nrf2 was formerly considered to help
protect cells in normal tissues from harmful stimulus, including
inflammation, trauma, ischemia, hemorrhage and cancer (5–9).
However, recent findings suggest that Nrf2 might play the dark role
in tumors. A number of studies showed constitutively high levels of
Nrf2 promote cancer formation and contribute to chemoresistance
(4,10–12).
Further investigation demonstrated that Nrf2 plays a pivotal role
in cell proliferation by activating its target genes, including
various detoxification enzymes, glutathione-related enzymes and
cell-cycle regulatory proteins (13–15).
Kim et al(16) showed that
Nrf2 as a candidate molecular target might control colon tumor
angiogenesis by imposing a blockade to HIF-1α (hypoxia-inducible
factor-1α) signaling. Recently, we also demonstrated that Nrf2 was
involved in migration and invasion of U251MG glioma cells (17). Collectively, these results
demonstrated that Nrf2 contributes to proliferation, migration,
invasion, angiogenesis and chemoresistance observed in several
types of malignant tumors. However, there are few reports on the
function of Nrf2 in glioma. In the present pilot study, we further
investigated the role of Nrf2 in the proliferation and tumor growth
of human U251MG glioma cells in the nude mouse xenograft model
following Nrf2 knockdown.
Materials and methods
Patients and tissue samples
The present study was approved by the Research
Ethics Committee of Jinling Hospital, School of Medicine, Nanjing
University, China. Written informed consent was obtained from all
patients. Surgically removed human GBM tissue samples were
continuously obtained paraffin-embedded from 49 patients (34 males
and 15 females (2.23:1), average age 52 years (27–82) attended at
Jinling Hospital from 2008 to 2010, according to the ethical and
legal standards. All the slides were re-evaluated according to WHO
classifications by two pathologists, with differences resolved by
careful discussion (18). None of
the patients had received chemotherapy or radiotherapy prior to the
surgery. Human normal brain tissues (mostly from the cortex) were
obtained from 5 male and 5 female patients in the pathway during
surgical removal of deep benign tumor from 2010 to 2011.
A total of 37 patients completed the follow-up until
mortality and the survival time was censored in August 2012, while
12 patients dropped out of the study. No patients succumbed to a
disease not directly related to their gliomas or due to unexpected
events. Follow-up was conducted every 6 months by telephone. The
adjuvant chemotherapy and radiotherapy were fully discussed with
the patients prior to the treatment and all patients received the
standard chemotherapy and radiotherapy. Overall survival (OS) was
defined as the time from the date of surgery to the date of death
from all cases. Progression-free survival (PFS) was defined as the
time from surgery to the time of tumor progression on MRI, or
mortality due to GB.
Cell culture, plasmid transfection and
establishment of Nrf2 knockdown cells
Human U251MG glioma cells were obtained from the
American Type Culture Collection (ATCC, Manassas, VA, USA). The
cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco,
Los Angeles, CA, USA) supplemented with 10% fetal bovine serum
(Gibco) and 1% penicillin/streptomycin (Gibco), and were incubated
at 37°C in a 5% CO2 incubator. The lentiviral particles
with Nrf2 short hairpin RNA (shRNA) were purchased from GenePharma
(Shanghai, China) and the target sequence was
GCAGTTCAATGAAGCTCAACT. The new plasmid was termed Si-Nrf2. Random
sequence, TTCTCCGAACGTGTCACGT, was used as the negative control,
and was termed Si-control. Cell transfection was performed in
6-well plates with lentiviral particles containing either
Si-control RNA or Nrf2 shRNA expression plasmid using Polybrene
(GenePharma) according to the instructions of the manufacturer.
Transfection was continued for 24 h and followed by a 24-h recovery
in the complete medium. For the selection of cells with target
plasmids, cells were grown in the medium containing 1.5 μg/ml
puromycin (Sigma-Aldrich, St. Louis, MO, USA) for up to 2 weeks.
Then, the positive clonal cells were selected and cultured in
96-well plates with the medium containing 1.0 μg/ml puromycin.
Total RNA extraction and RT-PCR
analysis
Total RNA was isolated from cells using the TRIzol
reagent (Invitrogen) following the manufacturer’s recommendations
and subjected to DNase (Promega, Madison, WI, USA) treatment.
Reverse transcription (RT) reaction was performed by incubating 400
ng of total RNA with the first-strand cDNA synthesis kit (Takara,
Dalian, China) following the manufacturer’s recommendations. The
concentration and purity of total RNA were determined by
spectrophotometer analysis (OD260/280:1.8–2.2) and agarose gel
electrophoresis. Obtained cDNA was amplified immediately using the
following primers: for Nrf2, 5′-TCAGCGACGGAAAGAGTATGA-3′ and
5′-CCACTGGTTTCTGACTGGATGT-3′; for NQO-1, 5′-ATGGTCGGCAGAAGAGC-3′
and 5′-GGAAATGATGGGATTGAAGT-3′; for HO-1,
5′-TCTCCGATGGGTCCTTACACTC-3′ and 5′-GGCATAAAGCCCTACAGCAACT-3′; and
for GAPDH, 5′-GAAATCCCATCACCATCTTC-3′ and
5′-GGACTCCACGACGTACTCA-3′. The amplification and data acquisition
were carried out on a real-time PCR system (Agilent, Anaheim, CA,
USA) using FastStart Universal SYBR Green Master (Roche, Mannheim,
Germany). The conditions were predenaturated at 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. All
samples were analyzed in triplicates in three independent
experiments. Reaction without cDNA was used as no-template control,
and no-RT controls were also set up to rule out genomic DNA
contamination. Relative quantification of mRNA expression was
determined using the 2−ΔΔCq method. PCR amplification
efficiency of each gene was established by means of calibration
curves. All the real-time PCR experiments were performed in
accordance to the minimum information for publication of
quantitative real-time PCR experiments (19).
Western blot analysis
Total protein was separated by 8% SDS-polyacrylamide
gel electrophoresis using the Criterion system (Bio-Rad
Laboratories, Hercules, CA, USA) at a constant voltage of 90 V.
Proteins were subsequently transferred to PVDF membranes
(Millipore, Billerica, MA, USA) at a constant voltage of 15 V for
30 min. Non-specific binding sites were blocked with 3% skim milk
for 2 h at room temperature and incubated overnight with primary
antibodies at 4°C. The following antibodies were used: 1:500
anti-Nrf2 (Abcam, Cambridge, MA, USA), 1:1,000 anti-HO-1 (Abcam),
1:2,000 anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA,
USA). The membranes were then incubated with the appropriate
secondary antibodies (Cell Signaling Technology, Danvers, MA, USA;
1:2,000) for 2 h at room temperature. After washing, protein bands
were visualized with Chemiluminescent HRP Substrate (Millipore) for
5 min at room temperature and exposed to X-ray film (Fuji
Hyperfilm). Relative changes in protein expression were estimated
from the mean pixel density using Quantity One software 4.6.2
(Bio-Rad Laboratories), normalized to β-actin and presented as
relative density units.
Cell proliferation assay
Cell proliferation was measured by Cell-Counting
kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) and cell proliferation
ELISA kit (Roche) according to the manufacturer’s instruction.
Cells were plated in 96-well plates at a density of
2×103 cells/well. After the incubation for 24 and 48 h,
10 μl CCK-8 were added and cells were further incubated for 1 h.
The absorbance was then measured at 450 nm using an enzyme-linked
immunosorbent assay (ELISA) microplate reader (Bio-Rad
Laboratories). DNA synthesis of cells was measured using the ELISA
kit, which is a colorimetric immunoassay based on the assessment of
bromodeoxyuridine (BrdU) incorporation during DNA synthesis. Cells
were plated in 96-well plates at a density of 2×103
cells/well and grown for 24 h. Then, cells were labeled with 10 μM
BrdU for 2 h and incorporated BrdU was quantified using a plate
reader at 492 nm using the ELISA microplate reader (Bio-Rad
Laboratories).
Tumor xenograft study
Animal studies were performed according to the
institutional Committee for Animal Research and in conformity with
national guidelines for the care and use of laboratory animals.
Si-Nrf2 and Si-control-stable-transfected cells and U251MG cells
(5.0×106) were suspended in 100 μl phosphate-buffered
saline (PBS) and then injected subcutaneously into either side of
the posterior flank of the male BALB/c athymic nude mice (Charles
River Breeding Laboratories, Wilmington, MA, USA) at 5–6 weeks of
age. Tumor growth was examined weekly for at least 5 weeks. Tumors
were then stripped and weighed before paraffin-embedding. Tumor
volumes were determined by external measurements and calculated
according to V= [L × W2] × 0.52, where V is the volume,
L is the length and W is the width.
Immunohistochemical staining
For the immunohistochemical analysis of human brain
tissues and xenograft tumors, 3-μm serial sections were dewaxed,
and endogenous peroxidase was quenched with 3%
H2O2 in methanol for 30 min (20). Before staining, non-specific binding
was blocked by incubation with 10% bovine serum albumin (BSA) in
PBS at 37°C for 1 h. Then, all incubations with 1:50 anti-Nrf2
(Abcam), 1:200 anti-Ki-67 (Abcam), 1:50 anti-caspase-3 (Abcam) and
1:100 anti-CD31 (Abcam) antibodies in PBS containing 1% BSA were
carried out at 4°C overnight. All sections were briefly washed in
PBS and incubated at room temperature with the anti-rabbit antibody
and avidin-biotin peroxidase (Vector Laboratories Inc., Burlingame,
CA, USA). Color was then developed by incubation with the
diaminobenzidine solution (Dako Corp., Carpinteria, CA, USA).
Nuclei were counterstained blue with Meyer’s hematoxylin
(Sigma-Aldrich). Non-neoplastic brain tissues were used as control
and non-immune IgG was used as negative control antibody for the
staining. The slides were scored by two independent observers. The
number of positive-staining cells was counted from two areas of
each specimen and the percentage of positive cells was calculated.
The staining intensity was estimated and stratified as 0
(negative), 1 (weak), 2 (moderate) and 3 (strong). The
immunohistochemistry positive results for Nrf2 were recorded as
previously described (20). Then we
scored the percentage of immunoreactive tumor cells as 0 (0 %), 1
(1–10%), 2 (11–50%) and 3 (>50%). Final immunoreactivity scores
(IRS) were obtained for each specimen by multiplying the percentage
and the intensity score. We then estimated the protein expression
levels by classifying IRS values as low (based on an IRS value
<5) and as high (based on an IRS value >5).
Microvessel density
Microvessel density (MVD) of the tumor tissues was
assessed through the endothelial marker CD31 immunohistochemical
analysis and determined according to the method previously
described (21). The immunostained
sections were initially screened at low magnification (x50) to
identify hot spots of the neovascularization. Any yellow-brown
stained endothelial cell or endothelial cell cluster clearly
separate from the adjacent microvessels, tumor cells and other
connective tissue elements was considered a single, countable
microvessel. Within the hot spot area, the stained microvessels
were counted in a single high-power (x200) field, and the average
vessel count in three hot spots was considered the value of MVD.
All counts were performed by three investigators in a blinded
manner. Microvessel counts were compared between the observers and
discrepant results were reassessed. The consensus was used as the
final score for analysis (22).
Statistical analysis
SPSS 19.0 software was used for statistical
analysis. OS was reported in months and defined as the interval
between the date of the surgery and the date of death. OS curves
were estimated by the Kaplan-Meier method, and the difference in
survival was evaluated using the log-rank test. Statistical data
are presented as the means ± SD. One-way analysis of variance
(ANOVA) followed by Tukey post-hoc comparison tests was used to
compare the levels of different experimental groups. P-values of
<0.05 and <0.01 were considered to indicate statistically
significant differences.
Results
Overexpression of Nrf2 in human GBM
tissues and the higher expression levels of Nrf2 correlate with
shorter survival
Nrf2 protein expression levels were investigated by
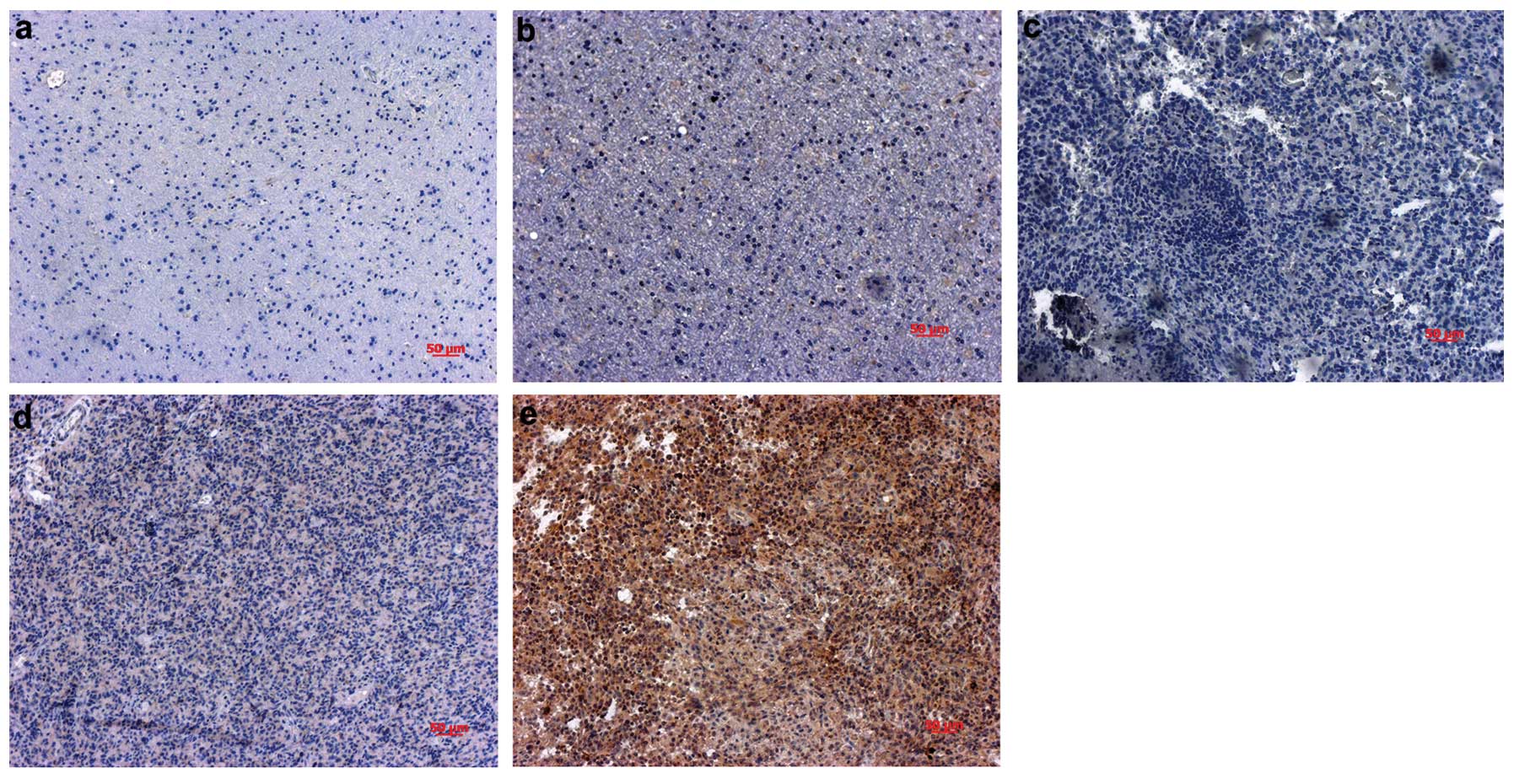
immunohistochemical analysis in 49 paraffin-embedded GBM samples,
15 adjacent normal tissues and 10 normal tissues. Immunoreactivity
for the Nrf2 antigen was seen in 43/49 (87.78%) of the tumor
samples and in 1/15 (6.67%) adjacent normal tissues, and the 10
normal brain tissues were negative for Nrf2 (Fig. 1). The Nrf2-positive label was
confined mainly to the cytoplasm, with some positive labels in the
nuclei. Of the 37 tumor samples for the follow-up patients, high
expression levels of Nrf2 were detected in 40.5% (15/37) and 59.5%
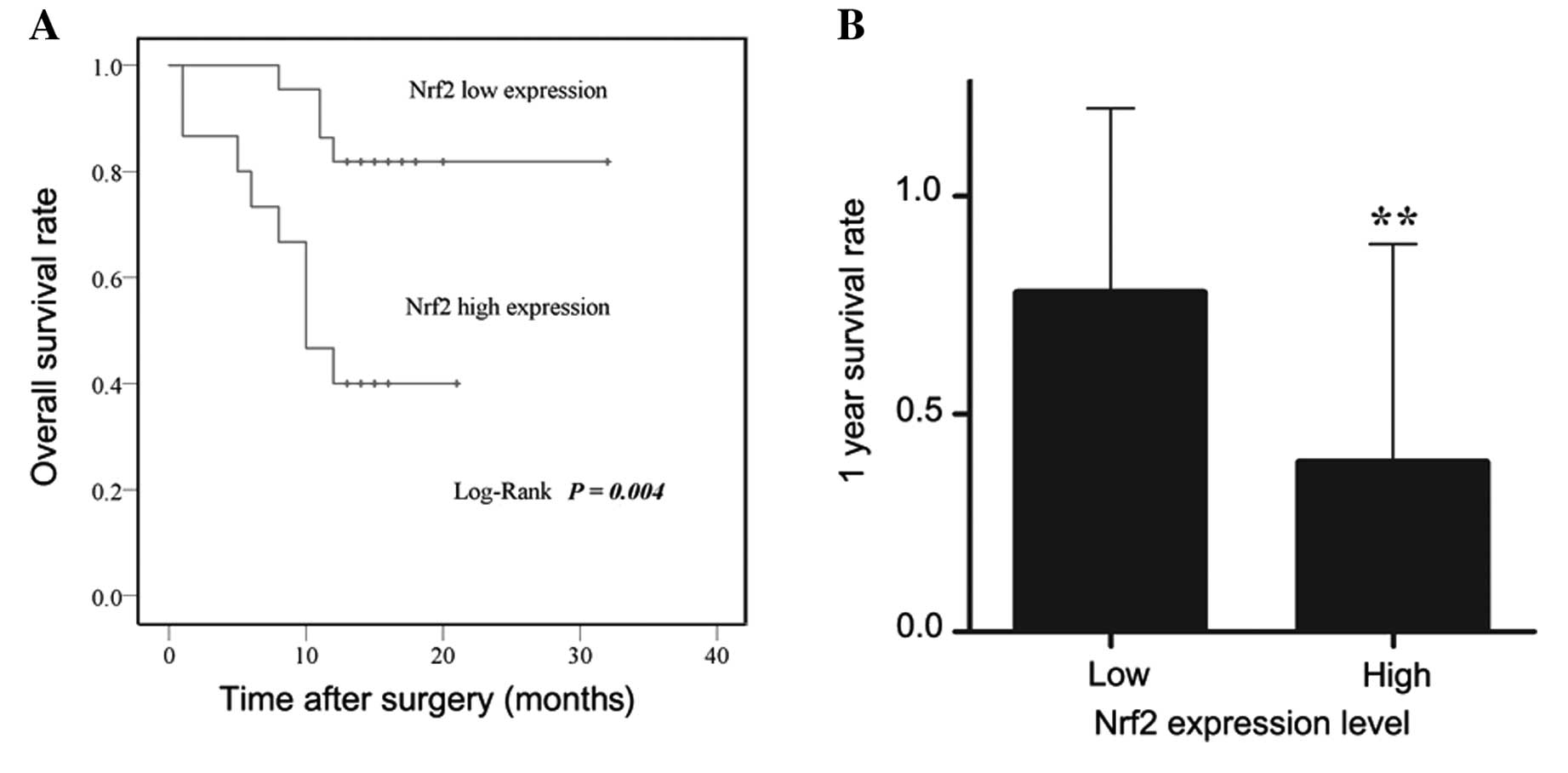
(22/37) of patients, respectively. The log-rank test showed that
the expression levels of Nrf2 were significantly correlated with
the OS of GBM patients (P=0.004, Fig.
2A). Thus, patients with high expression levels of Nrf2 (IRS
value >5) had shorter OS than the patients with low expression
levels (IRS value <5) (28.09±1.77 vs. 12.60±1.93 months,
respectively). We further evaluated whether the Nrf2
immunoreactivity was correlated with 1-year survival rates, and
found the 1-year survival rates to be 40.0% (18/22) for patients
with high Nrf2 expression and 81.8% (6/15) for patients with low
Nrf2 expression (P=0.0079, Fig.
2B). These results highlight the clinical significance of Nrf2
in determining the prognosis for patients with GBM.
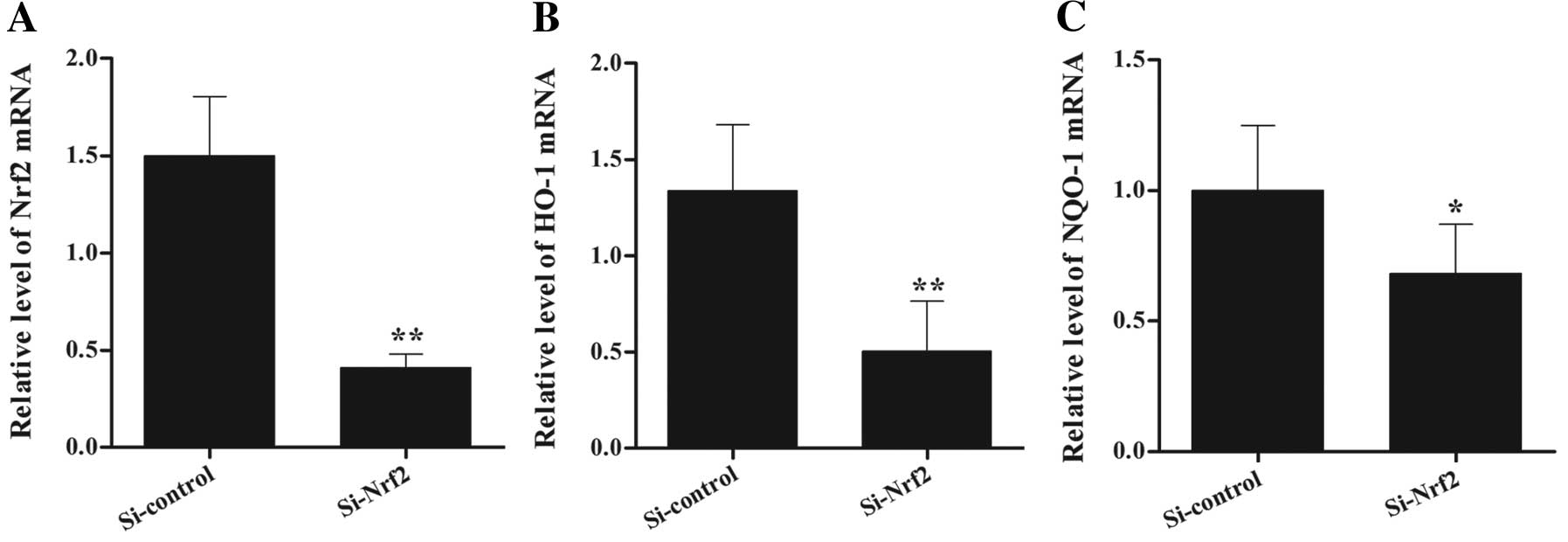
Stable transfection effect on the mRNA
and protein levels of Nrf2
To further investigate the role of Nrf2 in glioma
cell proliferation and tumor growth, we established an Nrf2
knockdown U251MG cell line. We transduced cells with plasmids
encoding non-specific Si-control RNA and Nrf2-targeting shRNA with
lentiviral particles. The stable cell line was obtained following
puromycin selection. Compared to the Si-control, cells with stable
Si-Nrf2 expression showed repressed transcript levels of Nrf2 and
its target genes, such as the catalytic subunit of HO-1 and NQO-1
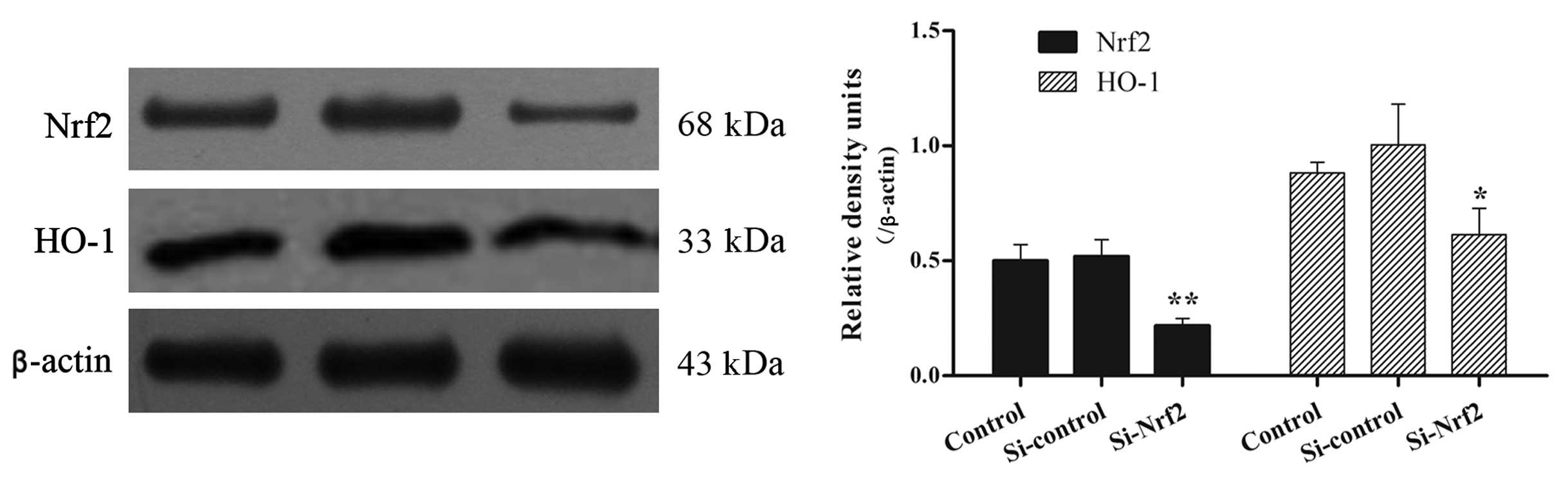
(Fig. 3A–C). Similar patterns were
measured by Nrf2 and HO-1 immunoblot analysis. The total protein
levels were much lower in Si-Nrf2 cells in comparison to the
Si-control (Fig. 4). In total,
stable Si-Nrf2 transfection reduced the Nrf2 mRNA and protein level
to 27 and 42% in comparison to the Si-control, respectively
(P<0.01).
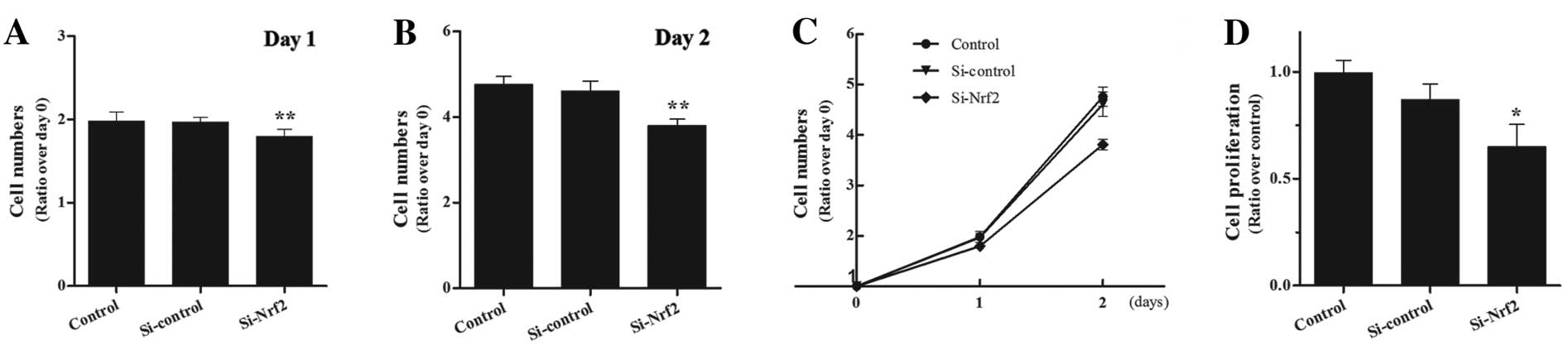
Suppressed cell proliferation in vitro
and tumor growth in Nrf2 knockdown xenograft
To investigate the role of Nrf2 in in vitro
proliferation, we performed the CCK-8 assay and the BrdU
incorporation assessment. First, we performed the CCK-8 assay at 24
and 48 h and found that the Si-Nrf2 cells were suppressed in cell
proliferation rate, particularly at 48 h (Fig. 5A–C). Similarly, the assessment of
BrdU incorporation confirmed the CCK-8 result (Fig. 5D). To further examine the role of
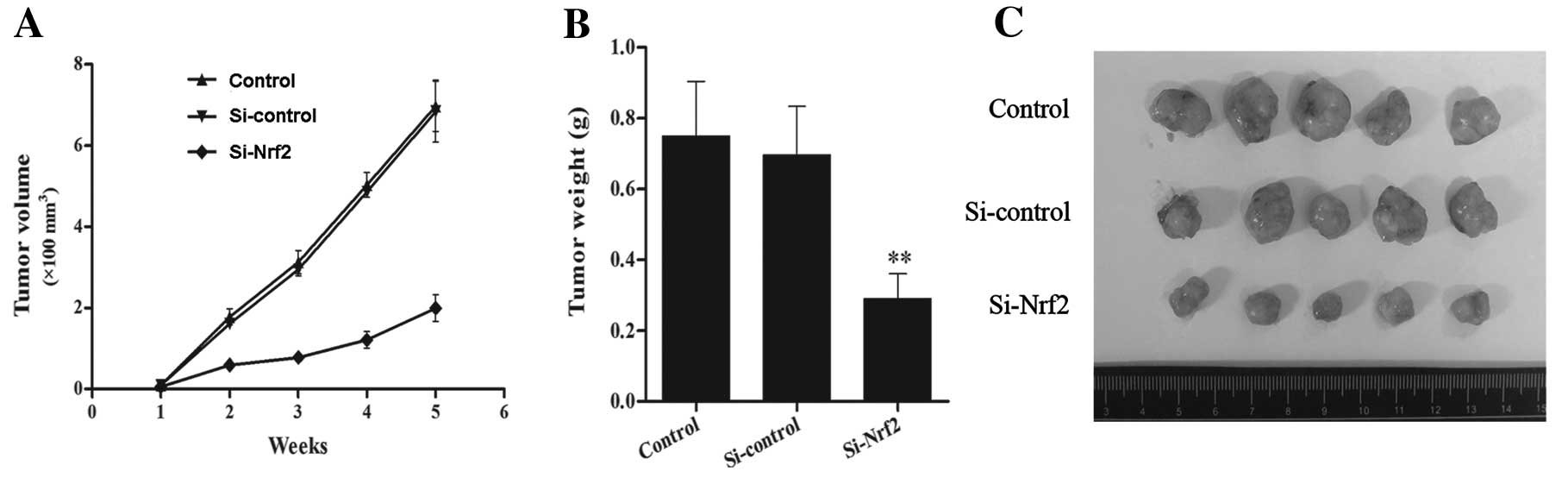
Nrf2 in tumorigenesis, we implanted the stable transfected U251MG
cells into nude mice and assessed the tumor growth. We measured the
volume changes during tumor growth and the tumor weight in the end,
and observed that tumor size of Si-Nrf2 cells increased more slowly
than the size of Si-control cells (Fig.
6A). A similar result was observed in the tumor weight
(Fig. 6B and C). Thus, the tumor
growth of the Si-Nrf2 cells was markedly suppressed in comparison
to the Si-control in the xenograft model.
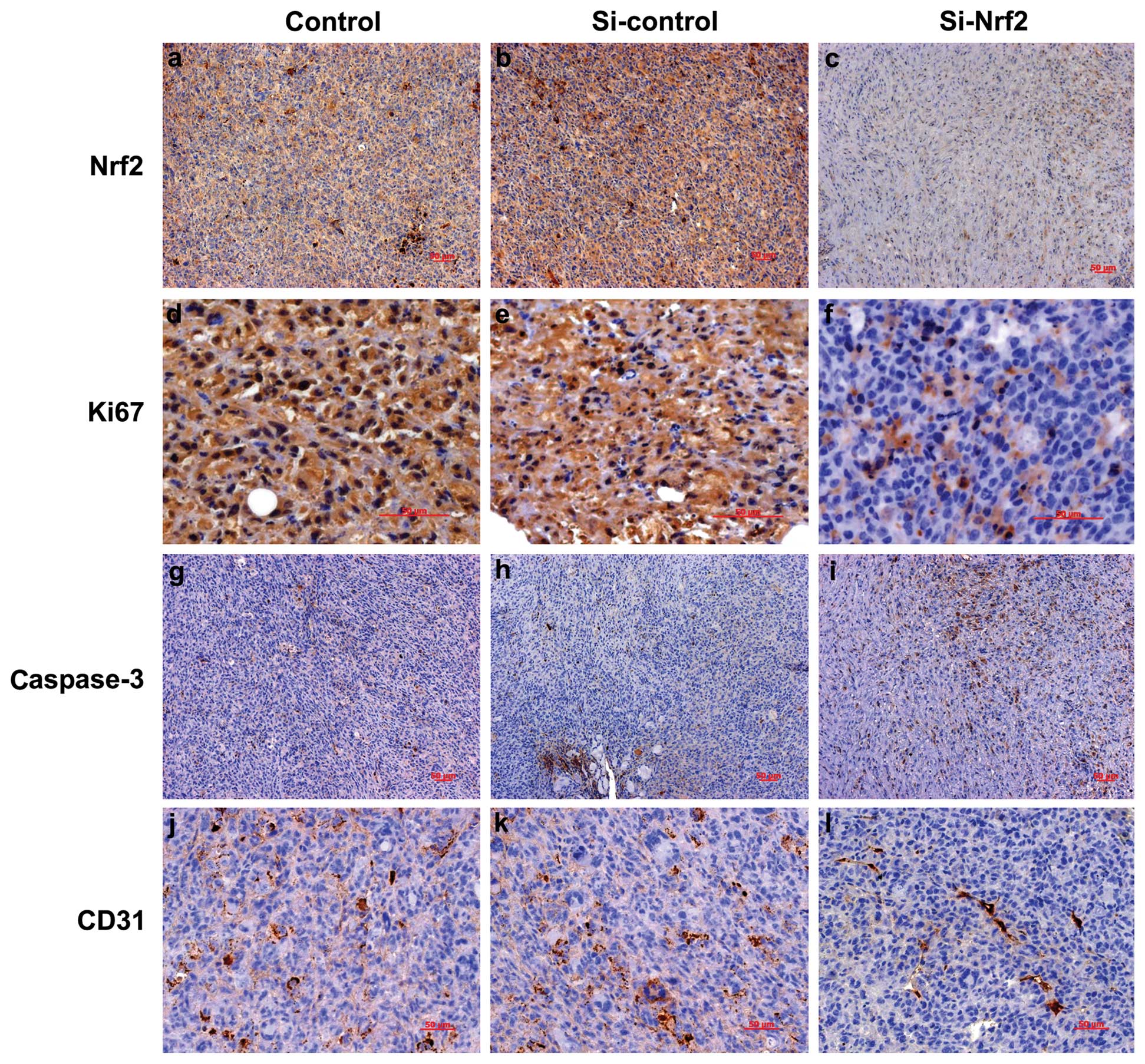
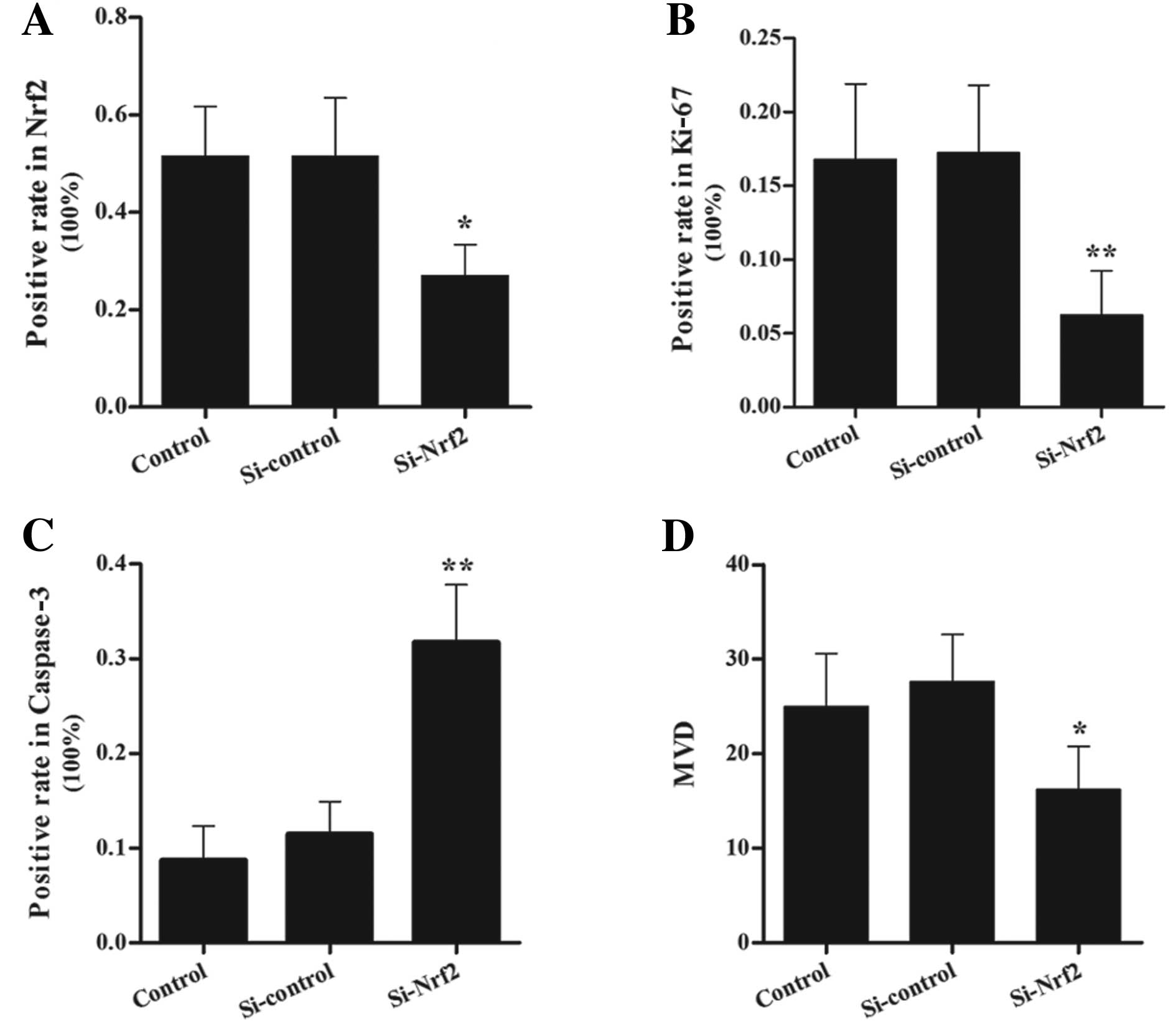
Analysis of the immunohistochemical
staining in the xenograft tumors
We performed immunohistochemistry analysis to detect
the protein levels of Nrf2, Ki-67, caspase-3 and CD31 in the
xenograft tumors (Figs. 7 and
8). Nrf2 was mainly detected in the
cytoplasm with various expression levels and a few in the nuclei.
Immunoreactivities in the tumor sections of Si-Nrf2 cells ranged
from undetectable to low, whereas the immunoreactivities of
Si-control cells were much higher (Fig.
7a-c and Fig. 8A). Ki-67,
located in the nucleus of eukaryocytes, is a sensitive biological
marker for cell proliferation (23). The expression level of Ki-67 in the
sections of Si-Nrf2 cells was lower in comparison to the Si-control
group (Fig. 7d-f and Fig. 8B). This observation confirmed the
results of cell proliferation assay in vitro. In order to
further explore the mechanism of the tumor suppression, we detected
the cell apoptosis and angiogenesis in the xenograft model.
Coincident with reduced tumor mass of the Si-Nrf2 group, the number
of caspase-3 positive cells was significantly increased in
Nrf2-inhibited tumors (Fig. 7g-i
and Fig. 8C). Subsequently, we
performed immunohistochemical analysis of endothelial marker CD31
to assess the MVD in the tumor tissues. Compared with the
Si-control group, MVD values in the Si-Nrf2 group decreased
significantly (Fig. 7j-l and
Fig. 8D).
Discussion
The present study focused on human GBM, which is an
aggressive tumor with heterogeneous tumor biology, high
invasiveness, rapid tumor cell proliferation and poor prognosis.
The results of our analysis showed that the protein level of Nrf2
in human GBM tissues was markedly upregulated in comparison to
non-neoplastic brain tissues, and GBM patients with high Nrf2
expression tended to have poorer OS. Furthermore, stable
downregulation of Nrf2 in GBM U251MG suppressed the cell
proliferation in vitro and the tumor growth in a nude mice
xenograft model. These findings suggest that Nrf2 is a valuable
transcription factor in cell proliferation, tumor growth and
clinical prognosis.
Although the cytoprotective roles of Nrf2 have been
confirmed by extensive studies since the first report by Moi et
al in 1994 (24), there are
increasing concerns regarding the deleterious effects of the Nrf2
signal in cancer cell biology. Research on Nrf2 in cancer have been
promoted by reports that Nrf2 is often overactivated in various
cancer cells. Studies revealed that mutations of the Keap1 protein
were identified from lung cancer cell lines and the tumor tissues
of lung cancer patients (25,26).
These mutations caused the loss of Keap1 repression of Nrf2, and
then led to the constitutive Nrf2 activation. Aside from Keap1,
mutations of Nrf2 were also identified, and, due to their
mutations, Nrf2 was overexpressed in other types of tumor tissues
and cell lines (27–30). Merikallio et al(31) found that Nrf2 and its stabilizing
protein DJ1 affected the prognosis of patients with lung cancer.
Further studies indicated that Nrf2 played a pivotal role in tumor
cell proliferation and chemoresistance (13,15).
These findings led to the hypothesis that inhibition of Nrf2
expression could reverse the phenotypic characteristics of cancer
cells, such as rapid proliferation, unresponsiveness to apoptosis,
drug resistance and angiogenesis. Studies have demonstrated that
RNAi-mediated inhibition of Nrf2 expression in lung cancer cells
could induce generation of reactive oxygen species, attenuate tumor
growth, and circumvent chemoresistance in vitro and in
vivo(13,14,32).
Our study focused on glioma and the results suggested that Nrf2 is
involved in the cell proliferation and tumor growth in GBM. Thus,
Nrf2 is critical in cancer cell pathobiology and targeting Nrf2
activity in cancer, particularly in cancer with Keap1 mutations,
may be a promising clinical strategy to inhibit tumor growth.
Ki-67, as a widely used proliferation marker, is
associated with cell cycle activity and is expressed at various
levels during the G1, S, G2, and M phases, with the exception of
the G0 phase (23). Thus, we could
perform immunohistochemical staining to detect Ki-67 during the
peak level in mitosis. The protein level of Ki-67 shows a good
correlation with the growth fraction in several model systems, and
its expression has therefore been investigated in various types of
human cancer, including glioma (33). Herein, our immunohistochemical
results of tumor tissues indicated inhibition of Nrf2 repressed
Ki-67 immunoreactivity, indicating that the effects of Nrf2 on
tumor growth could be attributed to Nrf2-mediated active and rapid
tumor cell division. Regarding the molecular mechanisms, tumor cell
proliferation might be dually regulated by epidermal growth factor
receptor (EGFR) signaling and the Nrf2 repressor protein Keap1
(14).
Tumor growth is considered the consequence of
balancing tumor cell proliferation and spontaneous cell death
(34). Apoptosis is an important
cell death form and a critical process in cancer cells. It is
deregulated resulting in tumorigenesis and drug resistance. In
general, the rate of apoptosis in malignant gliomas is very low,
and induction of apoptosis may be an important therapeutic strategy
for malignant glioma patients. A study recently demonstrated Nrf2
upregulated anti-apoptotic protein Bcl-2, leading to a decrease in
Bax, cytochrome c release from mitochondria, activation of
caspases, decreased DNA fragmentation and preventing
etoposide-induced apoptotic cell death (35). The present study indicated
inhibition of Nrf2 upregulated caspase-3 expression, which
confirmed the Nrf2-mediated anti-apoptotic effect in cancer. As a
result, increased apoptosis by inhibiting Nrf2 expression cannot be
excluded from the cause of tumor growth.
It is well established that the growth, invasion and
metastasis of most solid malignant tumors are
angiogenesis-dependent. Kim et al(16) performed a study in colon cancer
cells using stable RNAi-mediated knockdown of Nrf2. Their study
showed retarded tumor growth, diminished vascular formation, lower
VEGF expression and suppressed angiogenesis, and these may all be
contributed to Nrf2-mediated HIF-1α inhibition through the adaptive
reduction in mitochondrial O2 consumption. Another study
demonstrated that carbon monoxide produced by HO-1 increased HIF-1α
stability and enhanced VEGF expression (36). As one of the downstream genes of
Nrf2, HO-1 plays a role in the control of mitochondrial function.
As a result, suppressed HO-1 activity by the inhibition of Nrf2 may
contribute to reduced mitochondrial function and consequent HIF-1α
defect (16). These may explain our
observation of decreased MVD value in the Si-Nrf2 group xenograft
tumors.
The current study demonstrated the overexpression of
Nrf2 in GBM samples and correlated it with the clinical prognosis.
We further showed the role of Nrf2 in cell proliferation and tumor
growth in vitro and in vivo. Our findings support the
potential use of Nrf2 siRNA gene therapy for GBM patients. However,
the study needs to be repeated in more cell lines and in
vivo. In addition, the underlying molecular mechanisms of Nrf2
in cancer remain to be determined by further studies.
Acknowledgements
The present study was supported partly by the
Natural Science Foundation of China (81271377 to L.Z. and 81070974
to H.W.). We are grateful to Bo Yu, Juehua Zhu, Yanwen Yao and
Genbao Feng for their technical assistance.
Abbreviations:
|
ARE
|
antioxidant response element
|
|
BrdU
|
bromo- deoxyuridine
|
|
BSA
|
bovine serum albumin
|
|
kDa
|
kilodalton
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
GBM
|
glioblastoma multiforme
|
|
HIF-1α
|
hypoxia-inducible factor-1α
|
|
HO-1
|
heme oxygenase-1
|
|
IRS
|
immunoreactivity scores
|
|
MVD
|
microvessel density
|
|
Nrf2
|
NF-E2-related factor 2
|
|
NQO-1
|
NAD(P)H:quinone oxidoreductase-1
|
|
PBS
|
phosphate-buffered saline
|
|
PCR
|
polymerase chain reaction
|
|
RT
|
reverse transcription
|
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sullivan PR: Brain tumors. N Engl J Med.
344:1478author reply 1479. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen J, McKay RM and Parada LF: Malignant
glioma: lessons from genomics, mouse models, and stem cells. Cell.
149:36–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lau A, Villeneuve NF, Sun Z, Wong PK and
Zhang DD: Dual roles of Nrf2 in cancer. Pharmacol Res. 58:262–270.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dinkova-Kostova AT, Liby KT, Stephenson
KK, et al: Extremely potent triterpenoid inducers of the phase 2
response: correlations of protection against oxidant and
inflammatory stress. Proc Natl Acad Sci USA. 102:4584–4589. 2005.
View Article : Google Scholar
|
|
6
|
Kwak MK, Itoh K, Yamamoto M, Sutter TR and
Kensler TW: Role of transcription factor Nrf2 in the induction of
hepatic phase 2 and antioxidative enzymes in vivo by the cancer
chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol Med.
7:135–145. 2001.PubMed/NCBI
|
|
7
|
Shih AY, Li P and Murphy TH: A
small-molecule-inducible Nrf2-mediated antioxidant response
provides effective prophylaxis against cerebral ischemia in vivo. J
Neurosci. 25:10321–10335. 2005. View Article : Google Scholar
|
|
8
|
Yan W, Wang HD, Feng XM, Ding YS, Jin W
and Tang K: The expression of NF-E2-related factor 2 in the rat
brain after traumatic brain injury. J Trauma. 66:1431–1435. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Fields J, Zhao C, et al: Role of
Nrf2 in protection against intracerebral hemorrhage injury in mice.
Free Radic Biol Med. 43:408–414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hayes JD and McMahon M: NRF2 and KEAP1
mutations: permanent activation of an adaptive response in cancer.
Trends Biochem Sci. 34:176–188. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayes JD and McMahon M: The double-edged
sword of Nrf2: subversion of redox homeostasis during the evolution
of cancer. Mol Cell. 21:732–734. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kensler TW and Wakabayashi N: Nrf2: friend
or foe for chemoprevention? Carcinogenesis. 31:90–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Homma S, Ishii Y, Morishima Y, et al: Nrf2
enhances cell proliferation and resistance to anticancer drugs in
human lung cancer. Clin Cancer Res. 15:3423–3432. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamadori T, Ishii Y, Homma S, et al:
Molecular mechanisms for the regulation of Nrf2-mediated cell
proliferation in non-small-cell lung cancers. Oncogene.
45:4768–4777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lister A, Nedjadi T, Kitteringham NR, et
al: Nrf2 is overexpressed in pancreatic cancer: implications for
cell proliferation and therapy. Mol Cancer. 10:372011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim TH, Hur EG, Kang SJ, et al: NRF2
blockade suppresses colon tumor angiogenesis by inhibiting
hypoxia-induced activation of HIF-1α. Cancer Res. 71:2260–2275.
2011.PubMed/NCBI
|
|
17
|
Pan H, Wang H, Zhu L, Mao L, Qiao L and Su
X: The role of Nrf2 in migration and invasion of human glioma cell
U251. World Neurosurg. Nov 7–2011.(Epub ahead of print).
|
|
18
|
Rousseau A, Mokhtari K and Duyckaerts C:
The 2007 WHO classification of tumors of the central nervous system
- what has changed? Curr Opin Neurol. 21:720–727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bustin SA, Benes V, Garson JA, et al: The
MIQE guidelines: minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu Z, Lin D, Yuan J, et al: Overexpression
of osteopontin is associated with more aggressive phenotypes in
human non-small cell lung cancer. Clin Cancer Res. 11:4646–4652.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weidner N, Semple JP, Welch WR and Folkman
J: Tumor angiogenesis and metastasis--correlation in invasive
breast carcinoma. N Engl J Med. 324:1–8. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang M, Tang J, Liu S, Yoshida D and
Teramoto A: Expression of cathepsin B and microvascular density
increases with higher grade of astrocytomas. J Neurooncol. 71:3–7.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yerushalmi R, Woods R, Ravdin PM, Hayes MM
and Gelmon KA: Ki67 in breast cancer: prognostic and predictive
potential. Lancet Oncol. 11:174–183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moi P, Chan K, Asunis I, Cao A and Kan YW:
Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic
leucine zipper transcriptional activator that binds to the tandem
NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl
Acad Sci USA. 91:9926–9930. 1994. View Article : Google Scholar
|
|
25
|
Padmanabhan B, Tong KI, Ohta T, et al:
Structural basis for defects of Keap1 activity provoked by its
point mutations in lung cancer. Mol Cell. 21:689–700. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Singh A, Misra V, Thimmulappa RK, et al:
Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer.
PLoS Med. 3:e4202006. View Article : Google Scholar
|
|
27
|
Shibata T, Kokubu A, Gotoh M, et al:
Genetic alteration of Keap1 confers constitutive Nrf2 activation
and resistance to chemotherapy in gallbladder cancer.
Gastroenterology. 135:1358–1368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nioi P and Nguyen T: A mutation of Keap1
found in breast cancer impairs its ability to repress Nrf2
activity. Biochem Biophys Res Commun. 362:816–821. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shibata T, Ohta T, Tong KI, et al: Cancer
related mutations in NRF2 impair its recognition by Keap1-Cul3 E3
ligase and promote malignancy. Proc Natl Acad Sci USA.
105:13568–13573. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Konstantinopoulos PA, Spentzos D,
Fountzilas E, et al: Keap1 mutations and nrf2 pathway activation in
epithelial ovarian cancer. Cancer Res. 71:5081–5089. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Merikallio H, Paakko P, Kinnula VL, Harju
T and Soini Y: Nuclear factor erythroid-derived 2-like 2 (Nrf2) and
DJ1 are prognostic factors in lung cancer. Hum Pathol. 43:577–584.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singh A, Boldin-Adamsky S, Thimmulappa RK,
et al: RNAi-mediated silencing of nuclear factor
erythroid-2-related factor 2 gene expression in non-small cell lung
cancer inhibits tumor growth and increases efficacy of
chemotherapy. Cancer Res. 68:7975–7984. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wiesner FG, Magener A, Fasching PA, et al:
Ki-67 as a prognostic molecular marker in routine clinical use in
breast cancer patients. Breast. 18:135–141. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thompson CB: Apoptosis in the pathogenesis
and treatment of disease. Science. 267:1456–1462. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niture SK and Jaiswal AK: Nrf2
up-regulates anti-apoptotic protein Bcl-2 and prevents cellular
apoptosis. J Biol Chem. 287:9873–9886. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choi YK, Kim CK, Lee H, et al: Carbon
monoxide promotes VEGF expression by increasing HIF-1α protein
level via two distinct mechanisms, translational activation and
stabilization of HIF-1α protein. J Biol Chem. 285:32116–32125.
2010.PubMed/NCBI
|