Introduction
The most common malignant primary brain tumors are
gliomas. Despite aggressive surgery, radiation and chemotherapy,
the median survival is only 12–15 months for glioblastoma
multiforme (GBM) (1). It is
critical to explore the mechanism involved in the development and
progression of glioma and to find new therapeutic targets. Few
biomarkers have thus far been integrated into clinical
practice.
Nanog is a stem cell transcription factor that is
essential for embryonic development, reprogramming normal adult
cells and malignant transformation and progression (2). Oncogenesis has long been considered an
abnormal embryogenesis and tumor cells share a few biological
properties with ESCs (3). Several
tumor cell types have previously been reported to express Nanog
(4,5). Downregulation of Nanog by histone
deacetylase inhibitor apicidin could lead to cell cycle arrest,
differentiation and apoptosis in human embryonic carcinoma NCCIT
cells (6). Our previous research
demonstrated the overexpression of Nanog in glioma tissues and
brain tumor stem cells (BTSCs) compared with normal brain tissues,
indicating that Nanog may contribute to the existence of BTSCs and
may be related to tumorigenesis of the cerebrum by maintaining the
undifferentiated state of glioma cells (7).
Phosphorylation on serine or threonine residue
preceding proline (Ser/Thr-Pro) is a major intracellular signaling
mechanism. The conformation of certain phosphorylated Ser/Thr-Pro
bonds is regulated specifically by the prolyl isomerase Pinl. Pin1
is the only one of the prolyl isomerase family that can recognize
the phosphorylated Ser/Thr-Pro motif (pS/pT-P motif) and induce the
cis/trans conversion of the proline bond (8,9). It
has been reported that Pin1 is markedly overexpressed in several
types of human cancer (10–12). Pinl might amplify and translate
multiple oncogene signal mechanisms during oncogenesis and function
as a pivotal catalyst for multiple oncogenic pathways.
Nanog is phosphorylated at several Ser/Thr-Pro
motifs, which promotes the interaction between Nanog and the prolyl
isomerase Pin1 (13). The
interaction is important for Nanog stabilization by suppressing its
ubiquitin dependent degradation. Disruption of Pin1-Nanog
interaction in ESCs suppresses their capability to self-renew and
to form teratomas in immunodeficient mice (13). In human colorectal cancer, it has
been found that both Pin1 and Nanog are located in the perinuclear
space in the cytoplasm where they may interact to affect cell
proliferation and maintain the stemness of human colorectal cancer
(2).
In the present study, we first investigated the
expressions of Pin1 and Nanog in gliomas, as well as the
correlation between them. For both Pin1 and Nanog, their mRNA and
protein expressions were detected and found highly expressed in
human gliomas and positively correlated with pathological grade of
patients with gliomas. Furthermore, we frequently observed a
positive relationship between Pin1 and Nanog in gliomas. We also
confirmed that the co-location of Pin1 and Nanog was mainly in the
perinuclear space in the cytoplasm of glioma cells. However,
further study is required to determine the precise role of the
Pin1-Nanog pathway, and the mechanism of Pin1-Nanog transcriptional
regulation in gliomas.
Materials and methods
Clinical sample collection
The patients had received no treatment prior to the
craniotomy. Human glioma tissues (n=120) were obtained from
patients with newly diagnosed glioma who had received no therapy
before sample collection and had undergone resection at the Anhui
Provincial Hospital Affiliated to Anhui Medical University between
2007 and 2010. Normal brain specimens were acquired from 7 trauma
patients for whom partial resection of normal brain tissue was
required. All specimens were collected in the operating room
immediately (≤15 min) after tumor resection and were then snap
frozen in liquid nitrogen and stored at −80°C. The enrollment
criteria for the glioma patients in the present study were: glioma
diagnosis by pathology based on World Health Organization (WHO)
grading; no prior antiglioma treatment; suitable formalin fixed,
paraffin-embedded tissues and frozen tissues were available. All
glioma samples were verified by pathological analysis and
classified according to the WHO 2007 classification standard. There
were 22 low-grade (WHO grade II) and 98 high-grade tumors (WHO
grades III 42 and IV 56). None of the patients had received
chemotherapy, immunotherapy and radiotherapy prior to specimen
collection. Ethics approval for human subjects was obtained from
the Research Ethics Committee of the Anhui Provincial Hospital
Affiliated to Anhui Medical University and informed consent was
obtained from each patient.
RT-PCR
Reverse transcription-polymerase chain reaction
(RT-PCR) was performed as previously described (7). Total RNA was extracted from the human
glioma samples with TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
and treated with DNase (Fermentas, Vilnius, Lithuania) to remove
DNA contamination. RNA (200 ng to 1 μg) and M-MLV (Takara, Shiga,
Japan) and oligo-dT (Takara) were used for cDNA synthesis. PCR was
performed with 2X Taq Plus PCR Master Mix (Tiangin, China). The
primer sequences and the size of the amplified product were: Pin1
(427 bp), 5′-TCAGGCC GAGTGTACTAC-3′ (forward) and 5′-CGGAGGATGAT
GTGGATG-3′ (reverse); Nanog (403 bp), 5′-ATGCCTGT GATTTGTGGGCC-3′
(forward) and 5′-GCCAGTTGTTT TTCTGCCAC-3′ (reverse); β-actin (252
bp), 5′-ATGGATGA TGATATCGCCGCGCTC-3′ (forward), and 5′-TTTCTCCAT
GTCGTCCCAGTTGG-3′ (reverse).
β-actin was used as the internal control. In
semi-quantitative RT-PCR, standardized template amounts were used
to amplify cDNA for 30–35 cycles. The PCR products were separated
on 1.5% agarose gels by electrophoresis. The intensity of the bands
was determined using the Quantity One software (Bio-Rad
Laboratories).
Western blotting
Western blotting was performed as previously
described (7). Membranes were
probed with mouse anti-human Nanog polyclonal antibody (1:100;
Abcam) or rabbit anti-human Pin1 polyclonal antibody (1:500; Abcam)
at 4°C overnight or mouse monoclonal anti-β-actin antibody (1:1,000
dilution) (Beyotime Institute of Biotechnology, Nanjing, China) for
1 h at room temperature followed by the horseradish peroxidase
(HRP)-conjugated goat anti-mouse or goat anti-rabbit IgG antibody
(ZSGB-BIO, Beijing, China). Immunoblots were visualized by
chemiluminescence using an ECL Detection system (BeyoECL Plus;
Beyotime Institute of Biotechnology). The intensity of the bands
was determined using the Image-Pro Plus 6.0 software.
Immunohistochemical analysis
Immunohistochemical analysis was performed as
previously described (7,14). Slides were deparaffinized and
rehydrated following standard methods. A microwave antigen
retrieval procedure was carried out for 20 min in citrate buffer
(pH 6.0). Hydrogen peroxide was used to block non-specific
peroxidase reaction. Sections were blocked with normal goat serum
(20 min), then incubated with rabbit anti-human Pin1 polyclonal
antibody (1:200; Abcam, Cambridge, UK) or mouse anti-human
monoclonal antibody Nanog (1:100; Abcam) for 12 h at 4°C followed
by treatment with biotinylated secondary antibody; color reactions
were performed with diaminobenzidine (DAB) (Sigma). The sections
were lightly counterstained with hematoxylin. Negative control
sections were incubated with PBS instead of the primary
antibody.
In the present study, positive cells were scored
based on nucleus and cytoplasm staining of Nanog protein. The
number of positive immunostained cells out of 100 in 10 random
high-power fields (Olympus BX51; Tokyo, Japan) was scored (7,15).
Nanog expression was classified semi-quantitatively according to
the following criteria: 0 when <5% of glioma cells discretely
expressed Nanog in their nucleus and cytoplasm; 1+ when >5 to
<25% of glioma cells discretely expressed Nanog in their nucleus
and cytoplasm; 2+ when >25% to <50% of tumor cells are
immunopositive; 3+ when >50% of morphologically unequivocal
neoplastic cells discretely expressed Nanog in the nucleus and
cytoplasm. We considered samples scored as 2+ and 3+ as high
expression, while 0 and 1+ as low expression. The Pin1 expression
was evaluated visually and semi-quantified according to previous
studies (16–18). The scoring system was based on both
the intensity and the labeling frequency percentage. Cases with
Pin1 score 0–2 were assigned to the low Pin1 expression group, and
cases with Pin1 score 3–6 to the high Pin1 expression group.
Sections were scored by two independent pathologists with no
knowledge of the associated clinical data. In cases of occasional
scoring discrepancy, consensus was always achieved after a
discussion of the findings.
Immunofluorescence staining
The U87 human glioma cell line used in the present
study was purchased from the Chinese Academy of Sciences Type
Culture Collection. The cells were routinely maintained in high
glucose DMEM supplemented with 10% FBS, 100 U/ml penicillin, and
100 mg/ml streptomycin at 37°C in a humidified incubator with a 5%
CO2 atmosphere.
Immunofluorescence staining studies were performed
as previously described (7,18). U87 cells were grown on coverslips
for 24–48 h. They were then fixed with 4% paraformaldehyde for 20
min at room temperature and washed three times with 0.2% Triton
X-100/PBS for 15 min for permeabilization. The coverslips were
blocked with 10% normal goat serum for 30 min and then incubated at
4°C overnight with primary antibody Nanog (1:100 dilution) or Pin1
(1:200 dilution), followed by FITC-or TRITC-conjugated secondary
antibodies. The cells were counterstained with
4′,6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO, USA).
The images were acquired using an Olympus BX51 fluorescence
microscope.
Statistical analysis
All statistical analyses were performed by SPSS 17.0
software package for Windows. Data in the text and figures are
expressed as the means ± SD. The independent Student's t-test or
one-way analysis of variance (ANOVA) was used to compare the
expression level of Pin1 or Nanog between groups. Correlation
analysis of the expression levels of Pin1 and Nanog was performed
using the Spearman rank-sum test. P<0.05 was considered to
indicate a statistically significant difference in all tests.
Results
Pin1 is highly expressed in human gliomas
and is positively correlated with pathological grade
We initially analyzed the expression profiles of 120
gliomas to examine whether Pin1 was enriched in glioma tissues. We
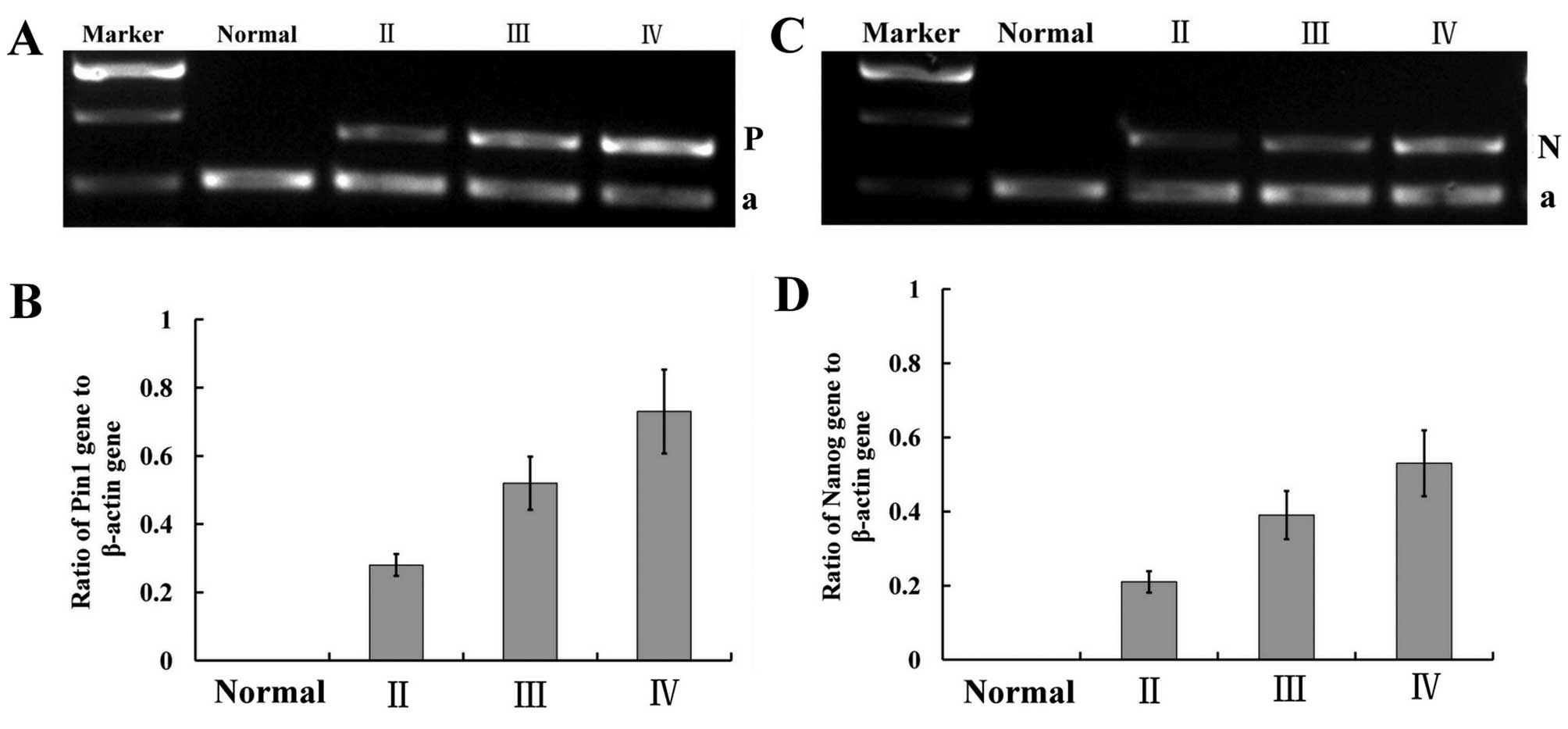
used the primers described above to investigate Pin1 mRNA
expression levels in the glioma tissues of different pathological
grade (Fig. 1). Densitometric
evaluation of the relative expression showed that the mRNA level of
Pin1 in the high-grade primary gliomas was significantly higher
than that in the low-grade gliomas (F=21.814, P<0.01) (Fig. 1). When observed by H&E staining
and excluding necrotic and hemorrhagic tissues, the glioma cells
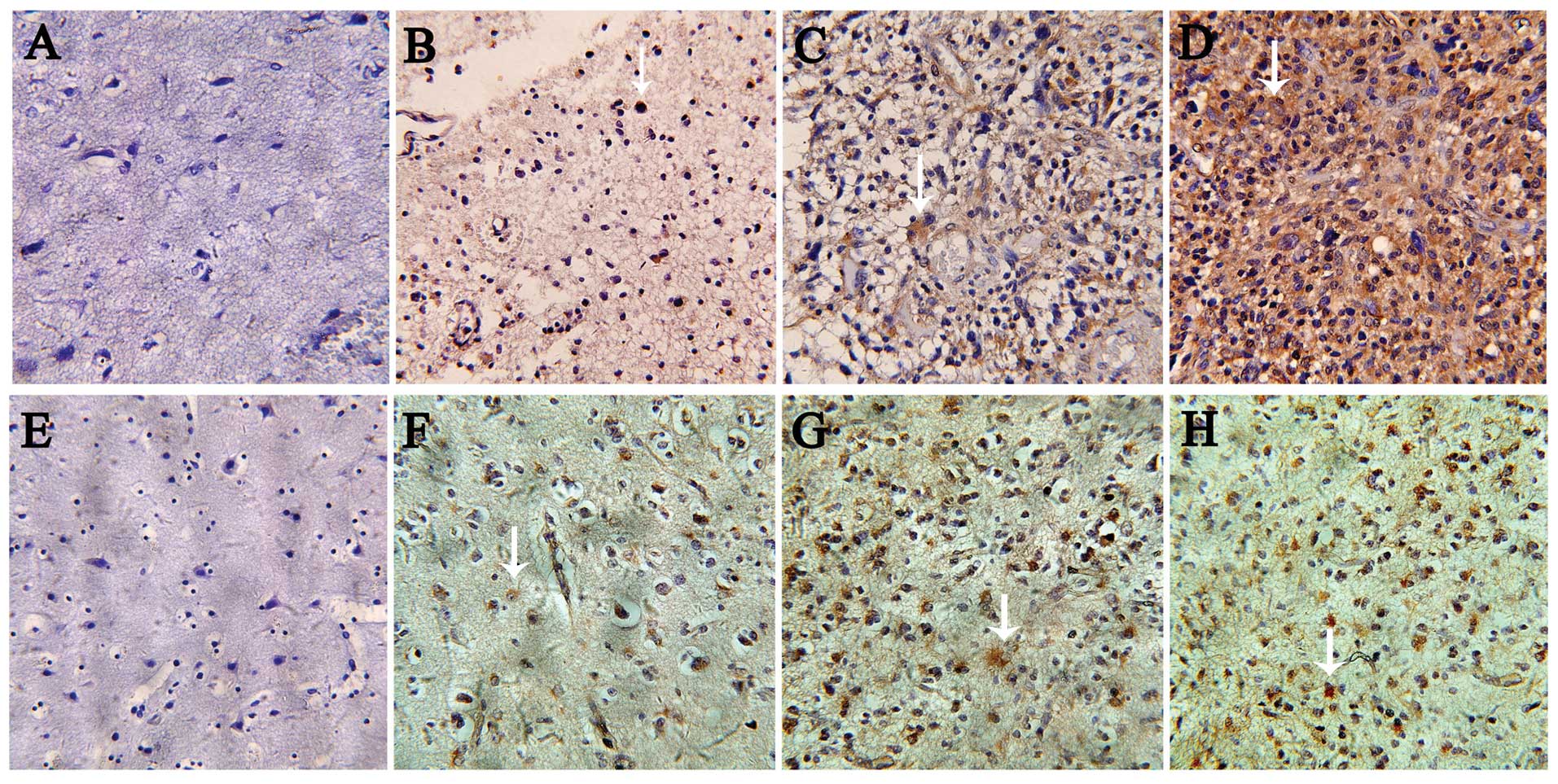
within gliomas were relatively homogeneous. The immunohistochemical
staining results showed that 95 (79.17%) glioma samples were
positively stained and 25 (20.83%) glioma samples were negatively
stained. Among the normal brain specimens, all 7 (100%) specimens
were negatively stained. In addition, Pin1 expression was mainly
confined to the nuclei in low grade glioma in a lower degree of
enrichment and weak expression, but exhibited enhanced expression
in both the cytoplasm and nuclei of high grade glioma. Moreover, a
marked positive correlation was noted between the expression of
Pin1 and pathological grade (r=0.279, P<0.01) (Fig. 2 and Table I). By contrast, no evident
expression was observed in the 7 normal brain samples. Following
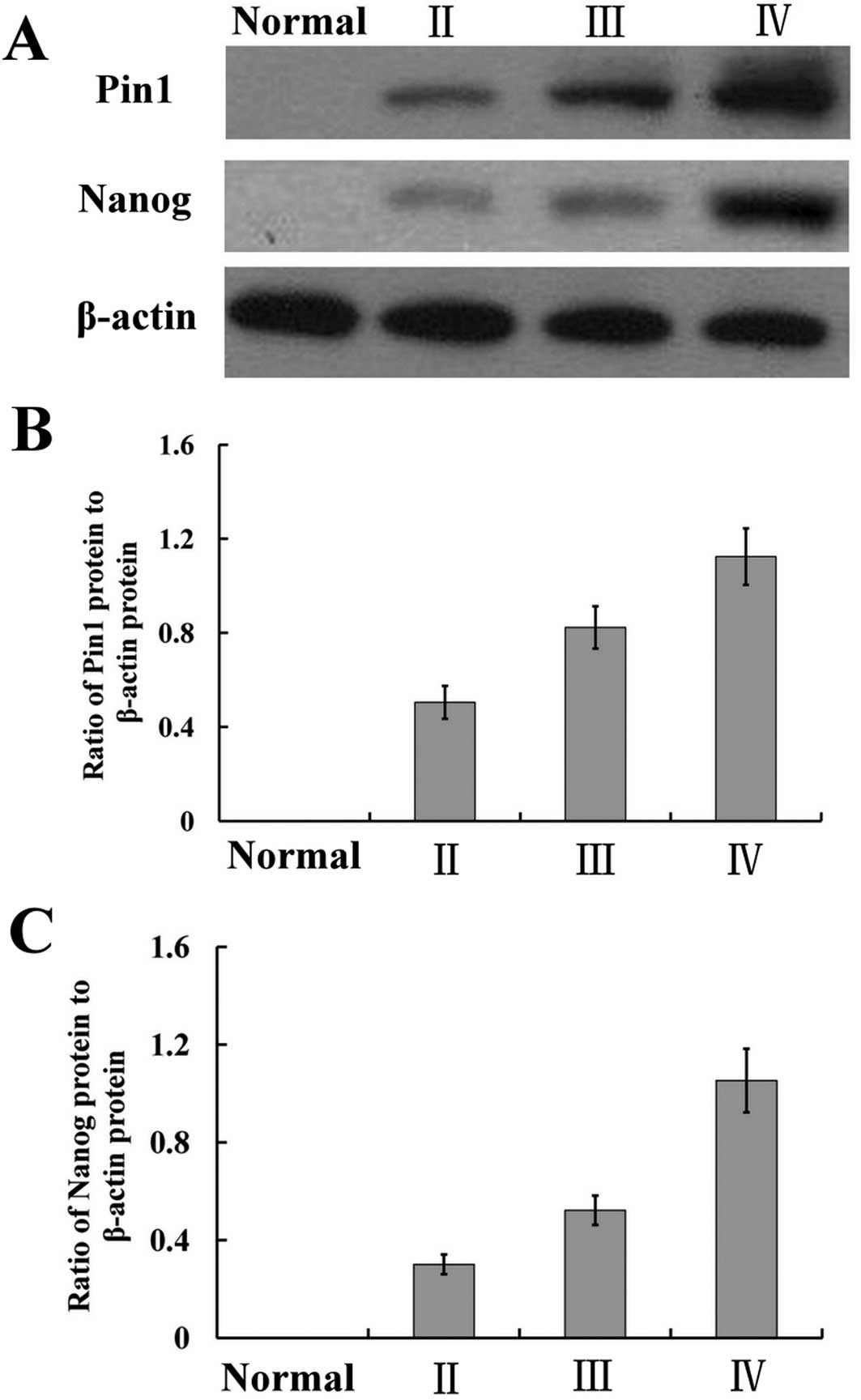
the above observations, we carried out western blot analysis to
confirm the relationship between the Pin1 expression and
pathological grade. As expected, a similar differential expression
pattern was observed; the higher expression of Pin1 correlated with
a more highly malignant glioma (F=22.962, P<0.01) (Fig. 3).
 | Table IPositive correlation between
Pin1-Nanog expression and pathological grade in human glioma
tissues. |
Table I
Positive correlation between
Pin1-Nanog expression and pathological grade in human glioma
tissues.
| | Pin1
expression | | Nanog
expression | |
|---|
| |
| |
| |
|---|
| WHO | Cases | Low | High | P-value | Low | High | P-value |
|---|
| II | 22 | 13 | 9 | | 17 | 5 | |
| III | 42 | 20 | 22 | 0.002 | 20 | 22 | 0.020 |
| IV | 56 | 14 | 42 | | 24 | 32 | |
Nanog is highly expressed in human
gliomas and is positively correlated with pathological grade
Our previous research demonstrated the
overexpression of Nanog in glioma tissues and BTSCs compared with
normal brain tissues (7). Our
current data also reveal that Nanog showed predominantly nuclear or
perinuclear staining with some cytoplasmic localization in glioma
cells. The immunohistochemical staining results showed that 88
(73.33%) glioma specimens were positively stained and 32 (26.67%)
glioma specimens were negatively stained. The normal brain tissues
were all negatively stained. Nanog mRNA and protein expressions
were highly expressed in gliomas, particularly WHO IV glioma
samples (F=18.381, P<0.01, ANOVA; F=42.691, P<0.01, ANOVA).
Moreover, the protein expression levels of Nanog were positively
correlated with pathological grade (r=0.211, P<0.05) (Figs. 1–3
and Table I).
Correlation between Pin1 and Nanog
expression in human gliomas
Positive immunostaining of Pin1 and Nanog was
observed in glioma cells. Based on the hierarchical scores of the
immunohistochemical staining described above, we proceeded to
analyze the correlation between Pin1 and Nanog in gliomas. The
results indicated that the expression levels of Pin1 and Nanog were
positively correlated in gliomas (r=0.209, P<0.05) (Table II), which suggest a high
correlation between the levels of Pin1 and Nanog in glioma
development.
 | Table IICorrelation between Pin1 and Nanog
expression in human glioma tissues. |
Table II
Correlation between Pin1 and Nanog
expression in human glioma tissues.
| Low Pin1 | High Pin1 | P-value |
|---|
| Low Nanog | 30 | 31 | 0.022 |
| High Nanog | 17 | 42 | |
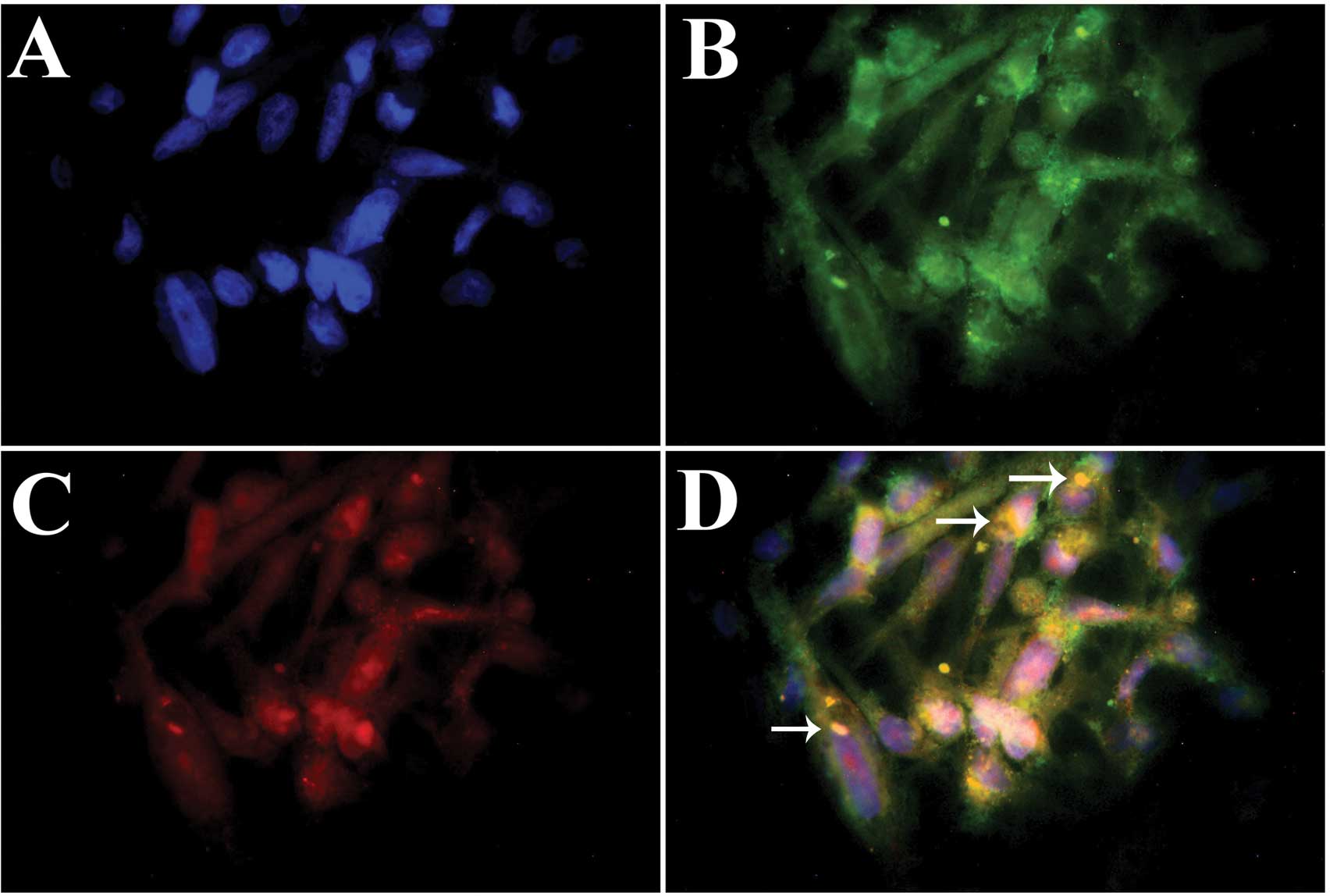
Subcellular localization and coexpression
of Pin1 and Nanog in glioma cells
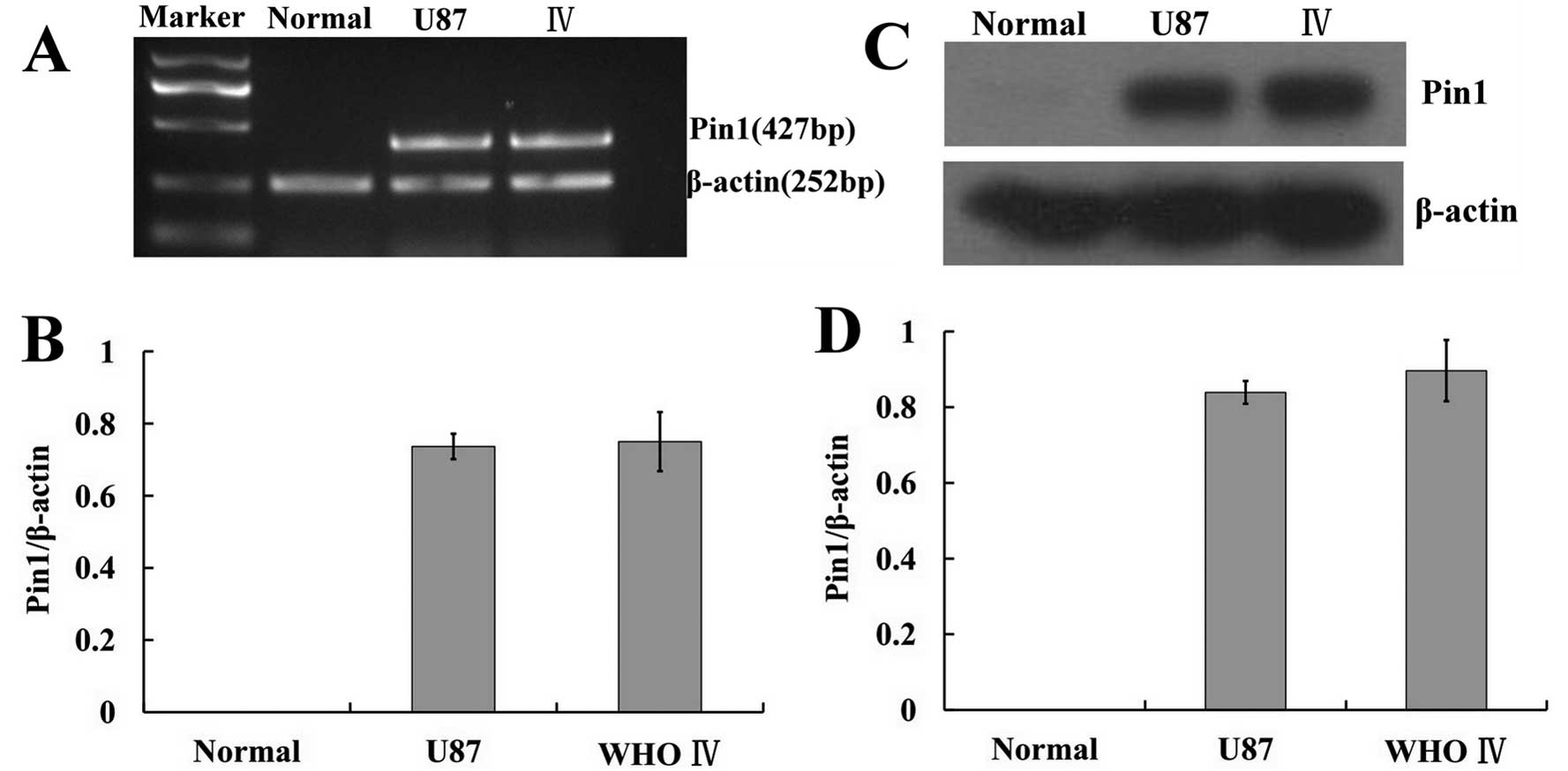
In previous research, Nanog mRNA and protein
expression in U87 glioma cells was confirmed (7). In the present study, Pin1 mRNA and
protein expression in U87 cells was examined using RT-PCR and
western blotting, respectively (Fig.
4). RT-PCR analysis of cells revealed the expected 427-bp Pin1
band in both the U87 cells and WHO IV glioma tissues (t=0.259,
P>0.05). No obvious band was observed in the normal brain
tissues. Western blot analysis confirmed that Pin1 was highly
expressed in both U87 cells and WHO IV glioma tissues (t=1.138,
P>0.05). For further analysis, immunofluorescence staining was
performed to detect the subcellular localization and coexpression
of Pin1 and Nanog. Pin1 was expressed in both the cytoplasm and
nuclei of U87 glioma cells. At the same time, Nanog showed mainly
nuclear and perinuclear staining with some cytoplasmic
localization. The majority of glioma cells coexpressed Pin1 and
Nanog (Fig. 5B and C). Furthermore,
Pin1 and Nanog were co-located in the perinuclear space in the
cytoplasm of glioma cells (Fig.
5D), where they may interact and have a cytoplasmic function to
affect glioma cells.
Discussion
Gliomas are the most common primary tumors in the
central nervous system (CNS). Malignant gliomas are the most lethal
tumors originating in the CNS, which account for 70% of gliomas
with a high recurrence and mortality rate (1). The most biologically aggressive
subtype of gliomas is glioblastoma multiforme (GBM), a tumor
associated with a rather poor prognosis. Although major advances
have been made in surgery, chemotherapy and radiotherapy for
gliomas, the life expectancy of patients with GBM and anaplastic
astrocytoma (WHO grade III) remains short, with a median survival
of approximately only 14–16 months and 2–5 years, respectively
(19). Advances in the treatment of
malignant gliomas require improved understanding of the biology and
the molecular mechanisms of glioma development and progression, as
well as the elucidation of novel molecular markers and signaling
pathways. Identification of the sets of genes that are
differentially expressed in different grade glioma specimens and
normal brain tissues is important to understand the molecular basis
of glioma, to predict patient prognosis and to develop novel
therapeutic strategies.
Pin1, which catalyzes cis-to-trans conformational
switches of target proteins presenting the phospho Ser/Thr-Pro
(pS/T-Pro) motif, has received considerable attention as a cofactor
that regulates the phosphorylation of several target proteins
(20). Previous studies showed that
Pin1 is overexpressed in a number of common tumors (10–12),
and several of its target proteins have an altered phosphorylation
profile (20,21). Such Pin1 activity is correlated with
a change in target protein stability through a ubiquitin-mediated
mechanism (22–24). Pin1 dependent conformational changes
are a unique signaling mechanism essential in regulating numerous
cellular functions. For instance, functional inactivation of RUNX3,
a tumor suppressor, is frequently observed in various types of
cancer, including glioma and breast cancer (25–27).
Expression of Pin1 inversely correlates with the expression of
RUNX3 in human breast cancer samples. Of note, Pin1 recognizes four
phosphorylated Ser/Thr-Pro motifs in RUNX3 via its WW domain.
Binding of Pin1 to RUNX3 could suppress the transcriptional
activity of RUNX3 by inducing the ubiquitination and proteasomal
degradation of RUNX3 (25).
Similarly, Fbw7, a well-characterized major tumor suppressor, is
the substrate recognition component of the Skp1-Cullin-F-box
(SCF)-type E3 ligase complex, which is frequently inactivated by
mutation or genetic deletion in various types of human cancer
(28–30). Min et al(31) found that Pin1 directly interacts
with Fbw7 to disrupt Fbw7 dimerization. As a result, Pin1 blocks
the ability of Fbw7 to mediate substrate degradation, but promotes
Fbw7 self-ubiqutination instead. These results support the idea
that Pin1 promotes the progression of cancer (32). In our research, we found
significantly higher Pin1 mRNA and protein expression in glioma
samples as compared with the normal brain tissue samples. An
association between higher Pin1 expression and aggressive grades of
gliomas was also demonstrated, which suggests that Pin1 may
participate in the pathogenesis of gliomas, which are defined as
poorly differentiated according to purely histopathological
criteria (7). However, whether
these findings are associated with Pin1 dependent conformational
changes, which change target protein stability through a
ubiquitin-mediated mechanism, remains to be further analyzed.
Nanog, a core transcription factor reported by
Mitsui et al(33), plays a
critical role in maintaining self-renewal and pluripotency of ESCs
by regulating cell fate of pluripotent inner cell mass (ICM)
(34–36). Apart from controlling such
‘stemness’ properties, the role of Nanog in tumorigenesis has
attracted significant attention. Increasing evidence suggests that
most tumors are heterogeneous. Of these, a small subset of cells,
known as cancer stem cells, arise from mutated adult
stem/progenitor cells possessing stem cell-like properties, which
are responsible for tumor growth, metastasis, chemoresistance and,
thus, cancer recurrence (37). Only
by targeting these populations of cells, which share several key
biological properties with normal stem cells, can the disease be
cured (38,39). Nanog overexpression has already been
detected in a number of human tumors, including glioma cells, and
is involved in some oncogenic pathways, suggesting that Nanog plays
a critical role in tumor genesis and progression (2,4–7,37,40).
Moretto-Zita et al(13)
reported that Pin1 could induce conformational change in Nanog by
isomerizing the pS/T-Pro bonds, leading to the inhibition of
ubiquitin-proteasome-dependent degradation of Nanog. The disruption
of the interaction between Pin1 and Nanog suppressed ESC
self-renewal. Pin1 plays critical roles in various types of cancer
by changing target protein stability through a ubiquitin-mediated
mechanism. However, why Pin1 facilitates ubiquitin-mediated
degradation of tumor suppressors but also stabilizes Nanog, a novel
oncogene, by suppressing its ubiquitination has yet to be fully
clarified and requires further research.
In the present study, the association between Pin1
and Nanog in human gliomas, the subcellular localization and
coexpression of Pin1 and Nanog in glioma cells were investigated.
We have shown that high Pin1 and Nanog expressions were detected in
glioma specimens by RT-PCR, western blotting and
immunohistochemical analysis. We also confirmed that Pin1 was
expressed in both the cytoplasm and nuclei of glioma cells, which
was consistent with Ryo et al(12) who reported that Pin1 expression was
found to be confined to the nuclei in low grade astrocytoma at
relatively low expression levels but exhibited enhanced expression
in both the cytoplasm and nuclei of anaplastic astrocytoma and
glioblastoma. Furthermore, Nanog showed mainly nuclear and
perinuclear staining with some cytoplasmic localization in glioma
cells. This finding was consistent with previous results which
found the Nanog protein was located in both the nuclei and in the
cytoplasm of breast carcinoma, prostate cancer and glioma cells
(4,41,42).
We further confirmed that majority of glioma cells coexpressed Pin1
and Nanog, which were co-located in the perinuclear space in the
cytoplasm of glioma cells, where they may interact and have a
cytoplasmic function to affect glioma cells. Additionally, Pin1 and
Nanog expression were positively correlated in glioma tissues,
indicating they may interact to affect cell proliferation and
maintain the cell viability and stemness of glioma. On the basis of
these findings, we hypothesize that the Pin1-Nanog pathway may be
important in the tumorigenesis of the gliomas. Targeting the
Pin1-Nanog pathway may be an approach to improve the therapeutic
intervention for poorly differentiated gliomas.
In conclusion, we have shown that Pin1 and Nanog
expression in human gliomas appears to be associated with the
pathogenesis of gliomas. Furthermore, Pin1 expression is positively
correlated and co-located with Nanog expression in glioma. Pin1 and
Nanog may play an important role in glioma tumorigenesis through
interaction. Further research is required to elucidate the
difference in response and control of expression of Pin1 and Nanog
in glioma, and to explore if Pin1 could act as a ubiquitination
switch in regulating glioma cellular functions through Nanog
conformational changes.
Acknowledgements
Grant support for this study was provided by the
National Natural Science Foundation of China (#81172407). The
authors thank the surgeons at the Neurosurgery Department of Anhui
Provincial Hospital Affiliated to Anhui Medical University for
assisting in the collection of the tumor samples.
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang J, Espinoza LA, Kinders RJ, et al:
NANOG modulates stemness in human colorectal cancer. Oncogene.
October 22–2012.(Epub ahead of print). View Article : Google Scholar
|
|
3
|
Linderholm BK, Gruvberger-Saal S, Ferno M,
et al: Vascular endothelial growth factor is a strong predictor of
early distant recurrences in a prospective study of premenopausal
women with lymph-node negative breast cancer. Breast. 17:484–491.
2008. View Article : Google Scholar
|
|
4
|
Ezeh UI, Turek PJ, Reijo RA, et al: Human
embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are
expressed in both seminoma and breast carcinoma. Cancer.
104:2255–2265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bourguignon LY, Peyrollier K, Xia W and
Gilad E: Hyaluronan-CD44 interaction activates stem cell marker
Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated
multidrug efflux in breast and ovarian tumor cells. J Biol Chem.
283:17635–17651. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
You JS, Kang JK, Seo DW, et al: Depletion
of embryonic stem cell signature by histone deacetylase inhibitor
in NCCIT cells: involvement of Nanog suppression. Cancer Res.
69:5716–5725. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Niu CS, Li DX, Liu YH, et al: Expression
of NANOG in human gliomas and its relationship with
undifferentiated glioma cells. Oncol Rep. 26:593–601.
2011.PubMed/NCBI
|
|
8
|
Ranganathan R, Lu KP, Hunter T and Noel
JP: Structural and functional analysis of the mitotic rotamase Pin1
suggests substrate recognition is phosphorylation dependent. Cell.
89:875–886. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen M, Stukenberg PT, Kirschner MW and Lu
KP: The essential mitotic peptidyl-prolyl isomerase Pin1 binds and
regulates mitosis-specific phosphoproteins. Genes Dev. 12:706–720.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lam PB, Burga LN and Wu BP: Prolyl
isomerase Pin1 is highly expressed in Her2-positive breast cancer
and regulates erbB2 protein stability. Mol Cancer. 7:912008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuramochi J, Arai T and Ikeda S: High Pin1
expression is associated with tumor progression in colorectal
cancer. J Surg Oncol. 94:155–160. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ryo A, Hirai A and Nishi M: A suppressive
role of the prolyl isomerase Pin1 in cellular apoptosis mediated by
the death-associated protein Daxx. J Biol Chem. 282:36671–36681.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moretto-Zita M, Jin H and Shen Z:
Phosphorylation stabilizes Nanog by promoting its interaction with
Pin1. Proc Natl Acad Sci USA. 107:13312–13317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He H, Niu CS and Li MW: Correlation
between glioblastoma stem-like cells and tumor vascularization.
Oncol Rep. 27:45–50. 2012.PubMed/NCBI
|
|
15
|
Sotomayor P, Godoy A, Smith GJ and Huss
WJ: Oct4A is expressed by a subpopulation of prostate
neuroendocrine cells. Prostate. 69:401–410. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ayala G, Wang D, Wulf G, et al: The prolyl
isomerase Pinl is a novel prognostic marker in human prostate
cancer. Cancer Res. 63:6244–6251. 2003.PubMed/NCBI
|
|
17
|
Bao L, Kimzey A, Sauter G, et al:
Prevalent overexpression of prolyl isomerase Pinl in human cancers.
Am J Pathol. 164:1727–1737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sasaki T, Ryo A, Uemura H, et al: An
immunohistochemical scoring system of prolyl isomerase Pin1 for
predicting relapse of prostate carcinoma after radical
prostatectomy. Pathol Res Pract. 202:357–364. 2006. View Article : Google Scholar
|
|
19
|
Van Meir EG, Hadjipanayis CG, Norden AD,
et al: Exciting new advances in neuro-oncology: the avenue to a
cure for malignant glioma. CA Cancer J Clin. 60:166–193.
2010.PubMed/NCBI
|
|
20
|
Lu KP and Zhou XZ: The prolyl isomerase
PIN1: a pivotal new twist in phosphorylation signalling and
disease. Nat Rev Mol Cell Biol. 8:904–916. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saegusa M, Hashimura M and Kuwata T: Pin1
acts as a modulator of cell proliferation through alteration in
NF-kappaB but not β-catenin/TCF4 signalling in a subset of
endometrial carcinoma cells. J Pathol. 222:410–420. 2010.PubMed/NCBI
|
|
22
|
Luo Z, Wijeweera A, Oh Y, et al: Pin1
facilitates the phosphorylation-dependent ubiquitination of SF-1 to
regulate gonadotropin β-subunit gene transcription. Mol Cell Biol.
30:745–763. 2010.PubMed/NCBI
|
|
23
|
Nakano A, Koinuma D, Miyazawa K, et al:
Pin1 down-regulates transforming growth factor-β (TGF-β) signaling
by inducing degradation of Smad proteins. J Biol Chem.
284:6109–6115. 2009.PubMed/NCBI
|
|
24
|
Siepe D and Jentsch S: Prolyl isomerase
Pin1 acts as a switch to control the degree of substrate
ubiquitylation. Nat Cell Biol. 11:967–972. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nicole Tsang YH, Wu XW, Lim JS, et al:
Prolyl isomerase Pin1 downregulates tumor suppressor RUNX3 in
breast cancer. Oncogene. 32:1488–1496. 2012.PubMed/NCBI
|
|
26
|
Mei PJ, Bai J, Liu H, et al: RUNX3
expression is lost in glioma and its restoration causes drastic
suppression of tumor invasion and migration. J Cancer Res Clin
Oncol. 137:1823–1830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mueller W, Nutt CL, Ehrich M, et al:
Downregulation of RUNX3 and TES by hypermethylation in
glioblastoma. Oncogene. 26:583–593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Akhoondi S, Sun D, von der Lehr N, et al:
FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer
Res. 67:9006–9012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maser RS, Choudhury B, Campbell PJ, et al:
Chromosomally unstable mouse tumours have genomic alterations
similar to diverse human cancers. Nature. 447:966–971. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Strohmaier H, Spruck CH, Kaiser P, et al:
Human F-box protein hCdc4 targets cyclin E for proteolysis and is
mutated in a breast cancer cell line. Nature. 413:316–322. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Min SH, Lau AW, Lee TH, et al: Negative
regulation of the stability and tumor suppressor function of Fbw7
by the Pin1 prolyl isomerase. Mol Cell. 46:771–783. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ryo A, Liou YC, Lu KP, et al: Prolyl
isomerase Pin1: a catalyst for oncogenesis and a potential
therapeutic target in cancer. J Cell Sci. 116:773–783. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mitsui K, Tokuzawa Y, Itoh H, et al: The
homeoprotein Nanog is required for maintenance of pluripotency in
mouse epiblast and ES cells. Cell. 113:631–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pan G and Thomson JA: Nanog and
transcriptional networks in embryonic stem cell pluripotency. Cell
Res. 17:42–49. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pereira L, Yi F and Merrill BJ: Repression
of Nanog gene transcription by Tcf3 limits embryonic stem cell
self-renewal. Mol Cell Biol. 26:7479–7491. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suzuki A, Raya A, Kawakami Y, et al:
Maintenance of embryonic stem cell pluripotency by Nanog-mediated
reversal of mesoderm specification. Nat Clin Pract Cardiovasc Med.
3:114–122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Siu MK, Wong ES, Kong DS, et al: Stem cell
transcription factor NANOG controls cell migration and invasion via
dysregulation of E-cadherin and FoxJ1 and contributes to adverse
clinical outcome in ovarian cancers. Oncogene. September
3–2012.(Epub ahead of print). View Article : Google Scholar
|
|
38
|
Singh SK, Clarke ID, Terasaki M, et al:
Identification of a cancer stem cell in human brain tumors. Cancer
Res. 63:5821–5828. 2003.PubMed/NCBI
|
|
39
|
Huff CA, Matsui W, Smith BD and Jones RJ:
The paradox of response and survivalin cancer therapeutics. Blood.
107:431–434. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang L, Zhang X, Zhang M, et al: Increased
nanog expression promotes tumor development and Cisplatin
resistance in human esophageal cancer cells. Cell Physiol Biochem.
30:943–952. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ye F, Zhou C, Cheng Q, et al:
Stem-cell-abundant proteins Nanog, Nucleostemin and Musashi1 are
highly expressed in malignant cervical epithelial cells. BMC
Cancer. 8:1082008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo Y, Liu S, Wang P, et al: Expression
profile of embryonic stem cell-associated genes Oct4, Sox2 and
Nanog in human gliomas. Histopathology. 59:763–775. 2011.
View Article : Google Scholar : PubMed/NCBI
|