1. Introduction
Cancer is currently the most lethal human disease.
Lung and colorectal cancers are the first and third most common
types of cancers and the leading causes of cancer-related mortality
worldwide (1). Bladder cancer, a
urological cancer, is the second most common malignancy that
involves the urinary system, and the clinical outcome is often poor
once the tumor becomes invasive (2). Although much progress has been made in
the prevention, early diagnosis and treatment of cancer, survival
rates are still not optimistic, indicating that a more powerful
method to detect cancer in the early stages is needed.
Early detection of cancer has been reported to
greatly improve both the survival rate and prognosis, suggesting
that the key to oncotherapy may lie in early diagnosis (3–7). Thus,
developing a method for the early detection of cancer is both
important and necessary. Ideally, an early detection method would
have high sensitivity, specificity and repeatability, and would be
safe, affordable and acceptable to the patient as well.
Traditional methods, such as colonoscopy (8), bronchoscopy (9) and cystoscopy (10), are used to detect colon cancer,
non-small cell lung cancer (NSCLC) and bladder cancer,
respectively. These methods have greatly benefited many individuals
in the past and they are still used to diagnose cancer. However,
their use has been hampered by their invasive nature, the manpower
resources they require, their high cost and the discomfort they
cause patients (11–13).
Biological screening methods, including the fecal
occult blood test (FOBT) for colon cancer; sputum cytology for
NSCLC (14); and the bladder tumor
antigen (BTA test), BTA stat test, nuclear matrix protein 22
(NMP22), and urinary cytology for bladder cancer, have also been
applied in recent years. However, these methods each have a
significant sensitivity (15,16) or
specificity (13), but not
both.
As previously mentioned, these methods have
drawbacks that prevent their wide application. However, miRNA
research in recent years has shed new light on early stage cancer
detection. miRNAs can function as oncogenes and tumor suppressors
(17). Many studies have reported
that miRNA levels are altered during cancer (18–20),
suggesting that miRNA dysregulation may be the perfect tool for the
early diagnosis of cancer.
miRNAs are short, non-coding RNA sequences of 20–22
nucleotides that are involved in crucial biological processes, such
as development, differentiation, apoptosis and proliferation
(21–23). Each miRNA has numerous gene targets,
and miRNAs mainly function by pairing with the 3′-untranslated
regions of target mRNAs (24).
Nevertheless, a recent study reported that miR-34a modulates MDM4
expression via a target site in the open reading frame (25). According to existing data, miRNAs
regulate at least 30% of protein-coding genes (26) suggesting that miRNAs may control
cellular processes in this manner.
This review discusses the possibility of detecting
miRNAs in feces, sputum, pleural effusion and urine in order to
screen for certain types of cancer, such as colon, lung and bladder
cancers, respectively. These three body fluids and stool have been
widely used to detect diseases in the clinic for many years, and
the results of their biochemical indices have high diagnostic
value. These materials have the advantages of reproducibility,
abundant content and tissue-specificity.
With the development of genetic sequencing tools,
many researchers have realized that traditional clinical detection
methods cannot make full use of the genetic value of these
materials. Notably, we found that many studies have focused on the
potential use of miRNAs in these materials as biomarkers to detect
cancer. microRNAs in these materials are useful candidates for
cancer detection for the following reason: miRNAs in body fluids
and stool are stable under extreme conditions, including a range of
temperatures and pH values, after extended storage and after
multiple freeze-thaw cycles (27–32),
indicating that miRNAs in these materials are stable enough to
detect even after the time of collection. However, synthetic miRNAs
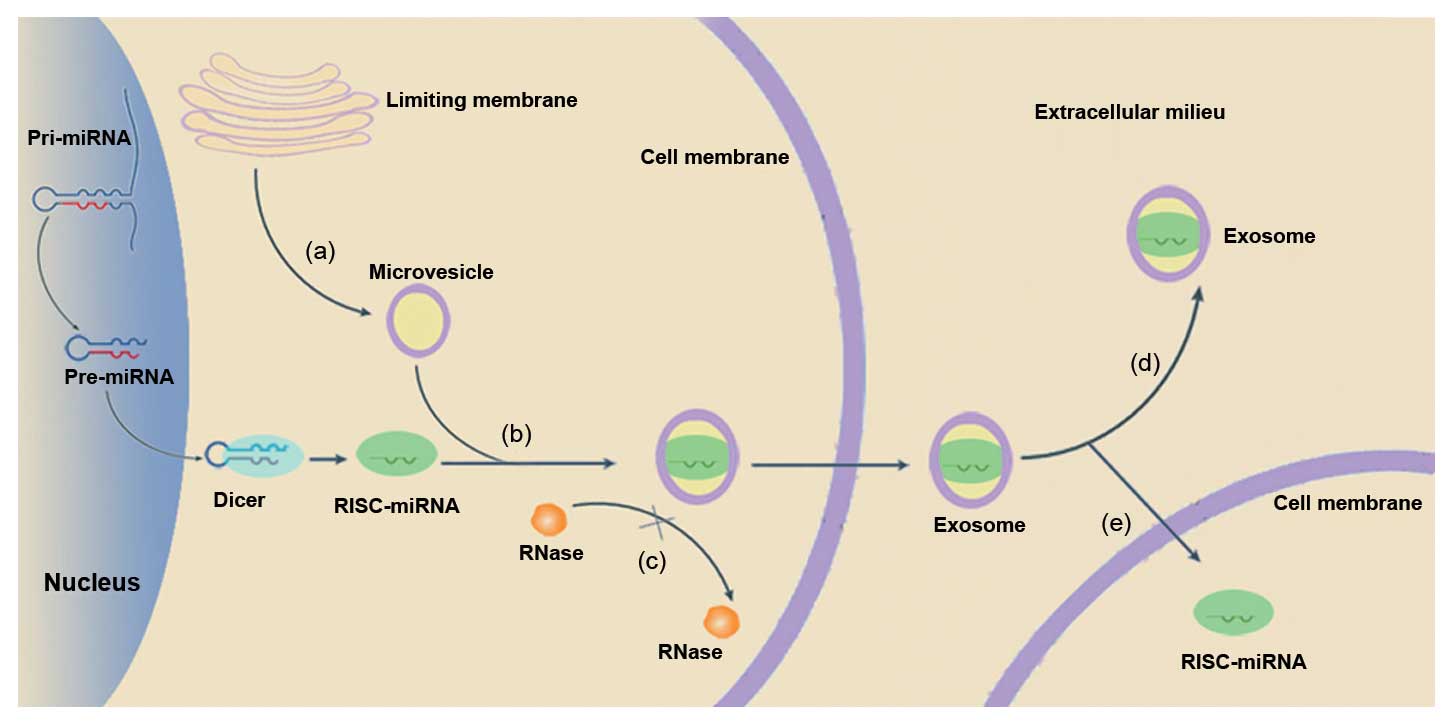
can be quickly degraded by RNase in the plasma (28). Brase et al(33) hypothesized that miRNAs hide in
microvesicles, which protect them against RNase activity, resulting
in their stability (Fig. 1).
Microvesicles are small particles that are released into the
cellular space and blood stream from cell membranes (34,35).
Evidence indicates that mRNAs and miRNAs can be transported through
microvesicles between cells (36).
Furthermore, these encapsulated miRNAs have been found to be
involved in the regulation of hematopoiesis and cellular
differentiation (37).
Microvesicles, also known as exosomes, have been correlated with
both cancer stage and miRNA levels in primary cancers when secreted
into the extracellular milieu (29,38),
suggesting that exosomes can be used to transport genetic
information, such as miRNAs, to support tumor growth and
progression (39). Additionally,
there is another mechanism that can explain miRNA stability.
Certain miRNAs have been reported to bind to a specific
DNA/RNA-binding protein to avoid degradation (40). miRNAs have been abundantly detected
in the stool, sputum, pleural effusion and urine, suggesting that
changes in the expression levels of miRNAs can be easily detected.
Xie et al(41) showed that
miRNAs were more stable than RNA molecules, despite significant
miRNA deposition. Evidence has shown that miRNAs can act as
oncogenes and tumor suppressors. Therefore, changes in miRNA
content may indicate that cancer is present.
In the present study, we summarize the value of
miRNAs in three body fluids and stool for the early diagnosis and
prognosis of tumors.
2. Fecal miRNA detection in colon cancer
screening
Colorectal cancer (CRC) is the third most common
cancer worldwide and the leading cause of cancer-related mortality.
Approximately 50% of patients will die from the development of
distant metastases, and the survival rate over a 5-year period is
~40% after diagnosis and treatment (1). However, early detection of such
neoplasms leads to a better prognosis. There are several methods
for detecting CRC, but their drawbacks have limited their wide
application and dissemination worldwide. Colonoscopy is the gold
standard for CRC diagnosis. However, the limitation outlined
previously (including the invasiveness of the procedure, the high
cost of the equipment and the manpower required), have restricted
the wide application of this procedure. Furthermore, clinical
guidelines suggest that colonoscopic screening should begin at the
age of 50. However, over 80% of these individuals could potentially
be spared the procedure as no relevant lesions are found (42,43).
The FOBT is one of the most commonly used biological methods, but
its effects are undesirable. The sensitivity of a single FOBT to
detect CRC is only 30–50% (44),
indicating that a substantial number of neoplasms may be missed
(45). One meta-analysis also
reported that FOBT screening reduces the relative risk of
CRC-related mortality by ~16% (46), suggesting that FOBT may not be an
ideal method for the diagnosis of CRC. Compared with colonoscopy,
CT colonography (CTC) has the advantage of reducing the side
effects and drawbacks of colonoscopy, including bleeding and
cardiorespiratory events. CTC also has a high sensitivity and
specificity of 55–90% and 86–96%, respectively (47–50).
However, the sensitivity of CTC decreases as the size of the polyps
decrease (50). In short, CTC is a
useful method for colon cancer screening, aside from the low
sensitivity in the detection of small polyps and the high cost.
Another promising approach for the early detection
of CRC is the analysis of molecular biomarkers, such as mRNA and
DNA in stool. One study showed that COX-2 mRNA could be detected in
26 out of 29 CRC cases (90% sensitivity) (51). In fecal DNA-based testing, which was
developed in the early 1990s, a number of genes in the stool,
including APC, p53 and K-Ras, are used as targets for CRC
identification (45). The
diagnostic sensitivity of this test ranged from 52 to 94% for CRC
detection (52), and the
specificity ranged from 93 to 97% (53,54).
However, fecal mRNAs and DNA degrade easily due to the activity of
RNase and DNase, limiting the wide application of this test
(55). In addition, the cost of
sDNA (stool DNA) screening can be as high as $800 (56), which is another factor limiting the
widespread use of the test.
miRNAs are short non-coding RNA sequences that play
an important role in the regulation of gene expression. Aberrant
gene expression can alter miRNA expression in cancer cells
(21). Changes in miRNA expression
can be observed in many types of cancers, including CRC (57). Many studies have reported that
miRNAs are detectable in the stool (Table I). Stool-based miRNAs are
continuously released and well mixed with the stool, leading to
high repeatability of tests on the same stool sample (31). In addition, the miRNA content is
very high in stool samples and is detectable in CRC patients
(58). miRNAs are the result of
cell exfoliation and easily accumulate in the stool, which makes
miRNAs detectable in stool samples (59). miRNAs have also been reported to
remain stable in stool samples (31,60).
The high content and stability make it possible to detect miRNAs in
stool samples (61,62).
 | Table ISummary of the characteristics of
miRNAs in the stool. |
Table I
Summary of the characteristics of
miRNAs in the stool.
| miRNA | Refs. | Dysregulation
(stool) | Specificity
(%) | Sensitivity
(%) | Samples | Normalization |
|---|
| miR-144* | (57) | Upregulated | 87 | 74 | 75 | miR-378 |
| miR-92a | (30) | Upregulated | 73.3 | 71.6 | 246 | RNU6B |
| miR-21 | (30) | Upregulated | 73.3 | 55.7 | 246 | RNU6B |
| miR-21 | (21) | Upregulated | -- | -- | 37 | miR-16 and
miR-26b |
| miR-135 | (31) | Upregulated | 95 | 46.2 | 340 | U6 snRNA |
| miR-17–92 | (31) | Upregulated | 81.5 | 69.5 | 340 | U6 snRNA |
| miR-34b/c | (59) | Upregulated | 87.2 | 75 | 67 | RNU19 and
RNU6B |
| miR-148a | (59) | Upregulated | -- | -- | 67 | RNU19 and
RNU6B |
| miR-106a | (21) | Upregulated | -- | -- | 37 | miR-16 and
miR-26b |
| miR-145 | (58) | Downregulated | -- | -- | 51 | miR-16 |
| miR-143 | (58) | Downregulated | -- | -- | 51 | miR-16 |
In addition to the advantages mentioned above,
stool-based miRNA detection also has high sensitivity and
specificity (Table I). miR-144* was
found to be overexpressed in the feces of CRC patients, indicating
that it could be a potential diagnostic marker for CRC detection,
with a sensitivity of 74% and a specificity of 87% (n=75, P=0.0001)
(60). miR-92a and miR-21 were also
reported to have these two advantages. miR-92a was found to have a
sensitivity of 71.6% and a specificity of 73.3%, whereas miR-21 had
a sensitivity of 55.7% and a specificity of 73.3% for CRC (31). Compared with miR-21, miR-92a was
able to detect polyps to a great extent and is likely to be a
relevant precancerous polyp marker. The level of miR-92a decreased
significantly after the removal of the tumor or advanced adenoma,
whereas the level of miR-21 decreased only after the removal of the
tumor (31). Link et
al(59) reported increased
expression of miR-21 and miR-106a in CRC stool samples, compared
with normal ones. They used a newly developed DMA (direct microRNA
analysis) methodology that easily detected miRNAs in the stool.
Kalimutho et al(63) found
that promoter methylation of miR-34b/c and miR-148a was detected in
the feces of CRC patients, suggesting that miR-34b/c and miR-148a
may be involved in colorectal tumorigenesis and metastasis.
3. Sputum and pleural effusion miRNA
detection in lung cancer
Non-small cell lung cancer (NSCLC) is the most
common type of lung cancer. Lung cancer is the leading cause of
cancer-related mortality worldwide (6,64).
Therefore, we focused on the application of new miRNA techniques in
lung cancer detection. NSCLC can be histologically subdivided into
four subtypes: adenocarcinoma, squamous cell carcinoma, large cell
carcinoma and ‘other’ (neuroendocrine cancers, carcinoid tumors)
(6). The disease is often diagnosed
during the advanced stages and carries a poor prognosis, with a
5-year survival rate of 13% (6,32,65).
However, the survival rate of NSCLC increases to 83% when detected
during stage I. Many methods are currently used to detect and
diagnose lung cancer, including computed tomography, magnetic
resonance imaging and bronchoscopy (66). Even though the sensitivities of
computed tomography (CT) were reported to be as high as 100%
(67–69), the cumulative frequency of subjects
with suspicious lesions is high, especially in silicosis patients,
in which CT generates a considerable number of false-positive
results due to high detection of many non-calcified nodules, which
have the potential to be confused with lung cancer (70). These results suggest that although
CT is widely used to detect NSCLC, it is plagued by false-positive
results at the cost of improved sensitivity (71). Similar to colonoscopy, bronchoscopy
is also invasive (12,72). Although sputum cytology is gentle,
the low sensitivity limits its wide application. The levels of
bronchial epithelial cells, which are detected by sputum cytology,
are very low in the sputum (14).
Aside from traditional methods, many studies have
reported the use of biological methods, such as molecular genetics,
to screen for lung cancer that may meet the standards for an ideal
diagnostic method. It has been reported that tissue-based
biomarkers can distinguish the tumors which originate in the lung
from metastases that originated in other sites in the body by
detecting significant biomarkers, such as tumor-suppressor genes,
regions of chromosomal amplification, differential miRNA expression
and variable miRNA expression (73–78).
However, this method is limited by the accessibility of the
specimens and the stability of the assessment offered. Blood-based
biomarkers are another biomarker method that can be used to detect
lung cancer. Indeed, blood is an ideal material due to the
abundance of cancer-specific biomarkers, such as DNA methylation
(79), gene expression (80), blood-miRNA (81), CTC (82) and cell-free DNA. Unfortunately,
there are still some drawbacks preventing blood-based biomarkers
from being successful clinical biomarkers of cancer, such as low
sensitivity, scarce quantities of any given marker, the complex
nature of the blood matrix and lack of reproducibility (73). One study attempted to detect
specific DNA in the sputum to screen for lung cancer but did not
detect any differences in either the free DNA or cellular DNA
concentrations in the sputum of lung cancer patients compared with
that of healthy controls (83),
indicating that DNA in the sputum may not be an effective biomarker
for lung cancer.
Many miRNAs have been proven to be abnormally
expressed in cancer tissue (57).
For this reason, miRNAs are potentially a useful tool for
diagnosing and screening human malignancies, including lung cancer
(78). Sputum in particular has
been considered to be a potential surrogate material for the
non-invasive diagnosis of lung cancer. Taken together, these
results indicate that miRNAs in the sputum may be used to screen
for lung cancer. miRNAs in the sputum can be detected using
real-time RT-PCR with TaqMan miRNA assays (Applied Biosystems)
(6,32). Sputum miRNAs are very stable
(6,32), similar to the stool miRNAs mentioned
above. The combination of sputum miRNAs have shown promising
results (Table II). miR-21 has
been found to be overexpressed in many types of cancer and this
finding has been demonstrated in many studies (84). In two recent studies, miR-21 was
reported to have a sensitivity of 72.6% and a specificity of 79.5%
(32) and a specificity of 69.66%
(95% CI, 0.46–0.86) and specificity of 100.00% (95% CI, 0.77–1.00)
(32). Therefore, examination of
miR-21 expression had higher sensitivity than that of sputum
cytology [47.82% (95% CI, 0.27–0.69) sensitivity and 100.00% (95%
CI, 0.77–1.00) specificity (32)]
for the diagnosis and early detection of lung cancer in patients.
In addition, it has been reported that increased miR-21 expression
is not significantly associated with length of smoking exposure in
both cancer patients and controls, suggesting that dysregulation of
miR-21 in lung cancer might not be caused by tobacco
smoking-related damage (32). Even
though overexpression of miR-155 may not distinguish lung cancer
patients from controls (32), it is
correlated with shortened survival of patients after resection
(78). In other words, elevated
miR-155 may indicate poor prognosis in lung cancer. Yu et
al(6) reported that detection
of a combination of different miRNAs (miR-486, miR-21, miR-200b and
miR-375) may be a better predictor, with a sensitivity and
specificity of 80.6 and 91.7%, when compared with that of a single
miRNA (as shown in Table II). Xing
et al(65) also showed that
detection of a combination of miRNAs (miR-205, miR-210 and miR-708)
greatly improve sensitivity and specificity. These two studies
indicate that the future detection of miRNAs may involve the
detection of a combination rather than a single miRNA. In addition,
Yu et al(6) found that miRNA
markers had higher diagnostic efficiency for adenocarcinomas than
for squamous cell carcinomas of the lung.
 | Table IISummary of the characteristics of
miRNAs in sputum and pleural effusion. |
Table II
Summary of the characteristics of
miRNAs in sputum and pleural effusion.
| Materials | miRNA | Refs. | Dysregulation | Specificity
(%) | Sensitivity
(%) | Samples | Normalization |
|---|
| Sputum | miR-21 | (31) | Upregulated | 100 | 69.7 | 40 | RNU6B |
| miR-155 | (31) | Upregulated | -- | -- | 40 | RNU6B |
| miR-486 | (6) | Downregulated | 79.4 | 66.9 | 72 | RNU6B |
| miR-126 | (6) | Downregulated | 73.8 | 67.2 | 72 | RNU6B |
| miR-145 | (6) | Downregulated | 82.9 | 59.5 | 72 | RNU6B |
| miR-21 | (6) | Upregulated | 79.2 | 72.6 | 72 | RNU6B |
| miR-182 | (6) | Upregulated | 79.5 | 64.3 | 72 | RNU6B |
| miR-200b | (6) | Upregulated | 78.5 | 62.9 | 72 | RNU6B |
| miR-375 | (6) | Upregulated | 80.6 | 63.9 | 72 | RNU6B |
| Malignant
effusion | miR-93 | (75) | Downregulated | -- | -- | 184 | ath-miR159a |
| miR-100 | (75) | Upregulated | -- | -- | 184 | ath-miR159a |
| miR-134 | (75) | Downregulated | -- | -- | 184 | ath-miR159a |
| miR-151 | (75) | Downregulated | -- | -- | 184 | ath-miR159a |
| miR-345 | (75) | Downregulated | -- | -- | 184 | ath-miR159a |
| miR-24 | (40) | Upregulated | 80.5 | 53.6 | 110 | ath-miR156a |
| miR-30d | (40) | Upregulated | 67.1 | 71.4 | 110 | ath-miR156a |
| miR-26a | (40) | Upregulated | -- | -- | 29 | ath-miR156a |
Pleural effusion is tightly correlated with NSCLC.
Approximately 15% of cancer patients are diagnosed with malignant
pleural effusions (MPEs) during early diagnosis (85). MPEs are an important route of
proliferation of tumor cells and are a frequent cause of morbidity
in NSCLC in lung cancer (85). MPEs
are very crucial for the treatment of NSCLC. Not all patients
benefit from chemotherapy, particularly those with short overall
survival times (86). There are
many methods that can be used to detect MPEs, including cytology,
needle biopsy and medical thoracoscopy. Cytology is the standard
diagnostic method for malignant effusions. Malignant cells are used
as a diagnostic sign, but the quantity of malignant cells may be
rather low, limiting the rate of positives (~50–70%), even with
repeated testing (87). Although
needle biopsy and medical thoracoscopy can improve the sensitivity
of diagnosis, their invasiveness and high cost limit their wide use
(88). Many studies have reported
using biomarkers to detect pleural effusion, such as marker
proteins (89), DNA methylation
status (90) and cell-free mRNA
levels (91), but these methods are
limited by their diagnostic accuracy. Research personnel have
noticed the close relationship between miRNAs and cancer, thus,
they attempted to find evidence that could demonstrate that pleural
effusion miRNAs are novel biomarkers for lung cancer diagnosis and
early detection (92). However, to
the best of our knowledge, few studies have been conducted in this
new field of interest. Xie et al(41) demonstrated that the levels of
miR-30d, miR-24, miR-26a are higher in malignant effusions compared
with normal effusions. miR-152 was first found to be a potential
diagnostic biomarker for drug sensitivity since the amounts of
miR-152 in tumor cells that were resistant to docetaxel were lower
than those of chemosensitive tumor cells (41). Wang et al(86) reported five miRNA expression
signatures (high expression levels of miR-100 and low expression
levels of miR-134, miR-345, miR-151 and miR-93) that were an
independent prognostic marker of poor survival. This was the first
report of miRNA expression signatures in MPEs that predicted NSCLC
patient prognosis. It seems that using miRNAs as a biomarker to
screen for MPE is a promising strategy, yet the mechanism remains
unknown. Thus, further study is warranted. In conclusion, detection
of miRNAs in the sputum and pleural effusion is a promising method
that may be used to prevent lung cancer, both by early detection
and accurate prognosis.
4. Urine miRNA detection in urological
cancer
Bladder cancer is the second most common malignancy
of the urinary system. These tumors are often invasive at the time
of diagnosis (2). Urothelial
carcinoma (UC) is among the five most common malignancies
worldwide, and it is also the second leading cause of mortality in
patients with genitourinary tract malignancies (93). UCs are the most common histological
type of bladder cancer. Ninety-five percent of primary urothelial
cell cancers arise from the bladder.
There are several clinical methods that are used to
detect bladder cancer. Cystoscopy is currently the standard
diagnostic tool, but it is difficult for cystoscopy to detect flat
lesions or carcinoma in situ. In addition, the invasive
nature and high expense of the procedure restrict it from being
widely used (94,95). Urinary cytology may be a useful
method for the detection of bladder cancer, due to the non-invasive
nature and high specificity of the procedure (90–95%); however, it
has a rather low sensitivity (30–40%) (96). Therefore, many alternative methods
have been presented to diagnose bladder cancer, such as the BTA
test, BTA stat test and NMP22 (97–99).
These methods have a higher sensitivity (50–70%) than cystoscopy,
but the increased sensitivity comes at the cost of specificity
(60–80%) (95). Even though many
achievements have been made in prevention and treatment in recent
years, the rates of morbidity and mortality remain high (100). A new biomarker for bladder cancer
detection is urgently needed.
As previously discussed, miRNAs are aberrantly
expressed or mutated in many types of cancers, suggesting that
detection of aberrant miRNA in the urine may be a useful method for
bladder cancer screening. In addition, miRNAs have been reported to
be stable in the urine and also show high specificity and
sensitivity (95,101–103). These characteristics indicate that
urinary miRNAs are a potential biomarker for bladder cancer and UC
screening. It has been reported that patients with bladder cancer
have lower expression of miR-200 family members (miR-200a, miR-200b
and miR-200c), miR-192 and miR-155 in the urinary sediment, and
higher expression of miR-155 in the urinary supernatant (101). It was also shown that the levels
of these miRNAs were altered after surgery. The postsurgical levels
of miR-200a, miR-200b, miR-200c, miR-141, miR-429, miR-205, miR-192
and miR-146a increased significantly, whereas the level of miR-155
remained similar (101). Taken
together, these results suggest that bladder cancer is the direct
cause of depressed urinary miRNA levels, but the mechanism of this
suppression is unknown. This study also revealed reverse
correlations between the expression of miR-200 family members and
EMT markers (ZEB1, vimentin, TGF-β1 and RhoA) (101). Downregulation of miR-200 family
members facilitates EMT of the transitional epithelium and promotes
cancer progression (104). This
may explain the mechanism, but further study is required. In other
studies, miR-452 and miR-222 were reported to play an oncogenic
role, while miR-143 was able to function as a tumor suppressor
(103,105–107). The present study also revealed
that miR-452 may contribute to tumorigenesis and aid in bladder
cancer diagnostics, whereas miR-143 and miR-222 may be related to
tumor progression and may be used for clinical outcome assessment
(103,107). In addition, expression levels of
of miR-222 and miR-452 were inversely correlated with ERBB4
expression, while ERBB4 was localized to several cellular
counterparts, including the membrane (108), cytoplasm (108) and nucleus (109). miR-222 was correlated with ERBB3
protein expression (103), which
is also related to tumor stage, grade, size, growth pattern,
recurrence, disease-specificity and overall survival (103). Although the study did not reveal
the translocation mechanisms of ERBB3 and ERBB4 in bladder cancer
progression, it did reveal the clinical relevance of subcellular
protein localization (103),
providing new insight into the relationship between miRNA, protein
and bladder cancer.
In UC, miR-96 and miR-183 levels in patient urine
samples were found to be significantly higher than those of the
control group, with 71.0% sensitivity and 89.2% specificity, and
74.0% sensitivity and 77.3% specificity, respectively. However,
more false-positive cases were found in miR-183 detection compared
with miR-96 detection, suggesting that miR-183 may be useful as a
staging marker but not as a diagnostic marker. miR-183 is
upregulated and functions in UTI as well as UC, and miR-96, which
has a high sensitivity and specificity, seems to be a tumor
biomarker that can be used to distinguish UC patients from non-UC
patients (95). The present study
also showed that 9 genes involved in activating apoptosis were
commonly downregulated in both miR-96 and miR-183 transfectants
(95). The characteristics of the
miRNAs in the urine are summarized in Table III.
 | Table IIISummary of the characteristics of
miRNAs in the urine. |
Table III
Summary of the characteristics of
miRNAs in the urine.
| miRNA | Refs. | Dysregulation | Specificity
(%) | Sensitivity
(%) | Samples | Normalization |
|---|
| miR-143 | (93) | Downregulated | -- | -- | 37 | miR-16 |
| miR-222 | (93) | Upregulated | -- | -- | 37 | miR-16 |
| miR-452 | (93) | Upregulated | -- | -- | 37 | miR-16 |
| miR-96 | (85) | Upregulated | 89.2 | 71 | 149 | RNAU6B |
| miR-183 | (85) | Upregulated | 77.3 | 74 | 149 | RNAU6B |
| miR-200a-b-c | (91) | Downregulated
(urinary sediment) | 52.6
(mir-200a) | 100 (mir-200a) | 75 | β-glucuronidase and
RNU48 |
| miR-192 | (91) | Downregulated
(urinary sediment) | -- | -- | 75 | β-glucuronidase and
RNU48 |
| miR-192 | (91) | Downregulated
(urinary supernatant) | -- | -- | 75 | β-glucuronidase and
RNU48 |
| miR-155 | (91) | Downregulated
(urinary sediment) | -- | -- | 75 | β-glucuronidase and
RNU48 |
| miR-155 | (91) | Upregulated
(urinary sediment) | -- | -- | 75 | β-glucuronidase and
RNU48 |
5. Common methods used in miRNA
diagnosis
The potential of miRNAs in four materials (stool,
sputum, pleural effusion and urine) to serve as biomarkers for
cancer screening was discussed above. miRNA diagnostic methods are
varied (110). In this section of
the study, we will focus on the steps that need to be taken to
obtain miRNAs and the methods that are used to detect them.
To obtain miRNA profiles, the following steps need
to be taken: sample collection, miRNA extraction, miRNA detection,
data processing and statistical analysis. Each step is important to
the final result. Sample collection is particularly important as it
determines the reliability of the results. In this step,
researchers should consider many factors, such as age, ethnic
group, gender and prior treatments (111). Concerning miRNA extraction, miRNAs
can be isolated from samples using three pre-methods: miRNeasy,
TRIzol and mirVANA (112). Even
though all three methods are suitable for profiling miRNAs from
total RNA, researchers still need to be prudent in choosing a
method, since small differences exist among the methods and may be
a source of bias. miRNA detection is based on the expression levels
of miRNAs that have been demonstrated to play a role in disease
(111). Researchers should choose
the appropriate technology to detect miRNAs using the various
available methods. In addition to the detection itself, the
stability and reproducibility of the method should also be taken
into consideration, to reduce the deviation (113). Data processing mainly refers to
the pre-processing of miRNAs for detection and normalization
(111), which is necessary to
minimize systematic experimental or technical variations.
Statistical analysis is the last step and mainly focuses on
comparing the differences between groups and indicating the
probability that the differences are clinically relevant, using the
Student's t-test as many studies have reported (113,114).
Although miRNA detection methods vary, the vast
majority of them rely on Watson-Crick base-pairing between
complementary chains of nucleotides and hybridization between a
strand of nucleic acid and its target miRNA (111). We summarized the advantages,
disadvantages and improvements of 6 of the most widely used
methods: Northern blot analysis, bioluminescence, RT-PCR,
fluorescence correlation spectroscopy, in situ hybridization
and microarray (115–132) (Table
IV).
 | Table IVFeatures of the common methods used
in miRNA diagnosis. |
Table IV
Features of the common methods used
in miRNA diagnosis.
| Method | Refs. | Advantage | Limitation | Improvement |
|---|
| Northern blot
analysis | (105–108) | Gold standard for
miRNA expression profiling
High specificity | Poor
sensitivity
Time-consuming
Not practical in a large amount | The use of locked
nucleic acid (LNA)-modified oligonucleotide probes (107) |
|
Bioluminescence | (109) | Rapid and
high-sensitivity
Suitable for application in clinical diagnostic | Complex steps | |
| RT-PCR | (110,111) | High sensitivity
and accuracy
Easy to operate | Expensive
Low throughput | Use of LNA-modified
primers (112)
Use of quantitative stem-loop
RT-PCR for the detection of mature miRNAs (110) |
| Fluorescence
correlation spectroscopy | (113) | High
sensitivity
Low detectable concentration | Special equipment
is needed | Use of a dual probe
labeling system (113) |
| In situ
hybridization | (114–116) | Specific to the
type of cell
Semiquantitative analysis | Low quantification
power
Low throughput | LNA miRNA oligo
probe (116)
Use of RNA molecules act as a primer (117) |
| Microarray | (118–120) | High
throughput
Widely used | Lack of
quantitative data
Expensive equipment | Probe
design
Sample labeling
Immobilization chemistry
Microarray chip signal-detection method (121,122) |
6. Conclusions and prospects
In the present review, we discussed the possibility
of screening miRNAs in the stool, sputum, pleural effusion and
urine to distinguish colon cancer, NSCLC and bladder cancer. Other
body fluids contain miRNAs as well, such as amniotic fluid,
cerebrospinal fluid, colostrums, peritoneal fluid, plasma, saliva,
seminal fluid and tears (133).
Few studies have reported the relationship between cancer and
miRNAs in amniotic fluid, cerebrospinal fluid, colostrums, saliva,
peritoneal fluid, seminal fluid and tears. Although many studies
have reported the close relationship between plasma miRNAs and
cancer, one plasma miRNA has been shown to be altered in multiple
types of cancers [e.g. changes in miR-21 in the plasma can
potentially indicate colorectal cancer (134) and gastric cancer (135)], which makes the diagnostic value
lower than the four materials we listed previously. We did not put
much emphasis on the blood biomarker limitations we discussed (low
sensitivity, scarce quantity, complex nature and lack of
reproducibility). In addition to miRNAs in the blood, a recent
study also reported that miR-421 in gastric fluid could be used as
a biomarker to screen for gastric cancer, with a sensitivity and
specificity that were equal to 71.4 and 71.7%, respectively
(136). This study also
demonstrated that miRNAs in gastric fluid had superior purity to
miRNAs in the plasma. However, it is not easy to obtain gastric
fluid in clinical practice and more research on miRNAs in gastric
fluid is still needed. Thus, we did not focus on the application of
miRNA detection in gastric fluid.
Biomarkers are important for the early detection and
prevention of malignancies as they are altered before histological
and morphological changes occur. The ideal biomarker must be
non-invasive, inexpensive, specific and sensitive to the disease
state and a reliable early indication of disease before clinical
symptoms appear (133). Even
though many biomarkers (most of them are protein) have been used to
screen for cancer, they do not function as expected. Improving the
diagnostic specificity and sensitivity of proteins is expensive,
time-consuming and difficult (133). Meanwhile, miRNA detection is much
easier due to PCR or other DNA amplification methods, which can
compensate for the low content limits, indicating that miRNAs may
be promising biomarkers for screening cancer.
miRNAs are excellent biomarker. The specificity and
sensitivity of miRNAs for screening cancer are higher than other
biomarkers, particularly the combination of specific miRNAs, as we
have previously discussed (Tables
I–III). Detection of miRNAs
is also an inexpensive and rapid method that costs ~$10 (US) and
takes ~3.5 h to obtain the results (95), indicating its possible use to detect
tumor biomarkers. In addition, it is a non-invasive method for
screening cancer and the materials are easy to obtain.
However, even with all of these advantages, more
effort is required to clarify the usefulness of miRNAs. Recent
research has focused on the phenomenon of aberrantly expressed
miRNAs in related body fluids and feces. The mechanisms of action
remain unknown, suggesting that the mechanisms require future
investigation. In addition, it is not easy to identify miRNA target
genes. A single miRNA may regulate the transcription of more than
one mRNA, and one specific mRNA may be regulated by several miRNAs
(103,137), making it difficult to determine a
particular miRNA. In many studies, RNU6B was reported to be an
endogenous control that could be used to normalize the expression
of miRNAs in tissue specimens, but to the best of our knowledge, it
may not be the ideal endogenous control for miRNAs due to its rapid
degradation in samples (such as stool) (138) and uncertain changes in the content
(95). Therefore, it is important
to find a stable powerful endogenous control. Although several
studies have proposed solutions (59,95),
further examination is required.
In conclusion, miRNA detection is a promising method
for cancer screening. Many opportunities and challenges lie ahead.
We believe that miRNA-based detection will be used for cancer
screening in the near future.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81272689), and
the Chongqing Science Fund for Distinguished Young Scholars (CSTC,
2009BA5045).
Abbreviations:
|
microRNA
|
miRNA
|
|
NSCLC
|
non-small cell lung cancer
|
|
FOBT
|
fecal occult blood test
|
|
BTA test
|
bladder tumor antigen test
|
|
NMP22
|
nuclear matrix protein 22
|
|
BC
|
bladder cancer
|
|
CRC
|
colorectal cancer
|
|
CTC
|
CT colonography
|
|
CT
|
computed tomography
|
|
MPE
|
malignant pleural effusion
|
|
UC
|
urothelial carcinoma
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ERBB4
|
epidermal growth factor receptor
|
|
COX-2
|
cyclooxygenase-2
|
|
APC
|
adenomatous polyposis coli
|
|
UTI
|
urinary tract infection
|
References
|
1
|
Corte H, Manceau G, Blons H and
Laurent-Puig P: MicroRNA and colorectal cancer. Digest Liver Dis.
44:195–200. 2012. View Article : Google Scholar
|
|
2
|
Jemal A, Siegel R, Ward E, Hao YP, Xu JQ
and Thun MJ: Cancer Statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
3
|
Frost JK, Ball WC Jr, Levin ML, et al:
Early lung cancer detection: results of the initial (prevalence)
radiologic and cytologic screening in the Johns Hopkins study. Am
Rev Respir Dis. 130:549–554. 1984.PubMed/NCBI
|
|
4
|
Flehinger BJ, Kimmel M and Melamed MR: The
effect of surgical treatment on survival from early lung cancer.
Implications for screening. Chest. 101:1013–1018. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Patz EF Jr, Rossi S, Harpole DH Jr,
Herndon JE and Goodman PC: Correlation of tumor size and survival
in patients with stage IA non-small cell lung cancer. Chest.
117:1568–1571. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu L, Todd NW, Xing LX, et al: Early
detection of lung adenocarcinoma in sputum by a panel of microRNA
markers. Int J Cancer. 127:2870–2878. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Subramanian J and Simon R: Gene
expression-based prognostic signatures in lung cancer: ready for
clinical use? J Natl Cancer Inst. 102:464–474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Greer KB and Cooper GS: Receipt of
colonoscopy is key to reduction of colorectal cancer mortality.
Gastrointest Endosc. 76:365–366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang KC and Yew WW: What is the role of
autofluorescence bronchoscopy in screening lung cancer among
silicotic subjects? reply. Int J Tuberc Lung Dis. 15:1277–1278.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Blick CG, Nazir SA, Mallett S, et al:
Evaluation of diagnostic strategies for bladder cancer using
computed tomography (CT) urography, flexible cystoscopy and voided
urine cytology: results for 778 patients from a hospital haematuria
clinic. BJU Int. 110:84–94. 2012. View Article : Google Scholar
|
|
11
|
Ladabaum U and Song K: Projected national
impact of colorectal cancer screening on clinical and economic
outcomes and health services demand. Gastroenterology.
129:1151–1162. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toloza EM, Harpole L and McCrory DC:
Noninvasive staging of non-small cell lung cancer: a review of the
current evidence. Chest. 123:137S–146S. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van Rhijn BW, van der Poel HG and van der
Kwast TH: Urine markers for bladder cancer surveillance: a
systematic review. Eur Urol. 47:736–748. 2005.
|
|
14
|
Thunnissen FB: Sputum examination for
early detection of lung cancer. J Clin Pathol. 56:805–810. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Simon MA, Lokeshwar VB and Soloway MS:
Current bladder cancer tests: unnecessary or beneficial? Crit Rev
Oncol Hematol. 47:91–107. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bassi P, De Marco V, De Lisa A, et al:
Non-invasive diagnostic tests for bladder cancer: a review of the
literature. Urol Int. 75:193–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
18
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Patnaik SK, Kannisto E, Mallick R and
Yendamuri S: Overexpression of the lung cancer-prognostic
miR-146b microRNAs has a minimal and negative effect on the
malignant phenotype of A549 lung cancer cells. PLoS One.
6:e223792011.PubMed/NCBI
|
|
20
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:E249–E258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Harfe BD: MicroRNAs in vertebrate
development. Curr Opin Genet Dev. 15:410–415. 2005. View Article : Google Scholar
|
|
24
|
Kloosterman WP and Plasterk RHA: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mandke P, Wyatt N, Fraser J, Bates B,
Berberich SJ and Markey MP: MicroRNA-34a modulates MDM4 expression
via a target site in the open reading frame. PLoS One.
7:e420342012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ho AS, Huang X, Cao HB, et al: Circulating
miR-210 as a novel hypoxia marker in pancreatic cancer. Transl
Oncol. 3:109–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu CW, Ng SSM, Dong YJ, et al: Detection
of miR-92a and miR-21 in stool samples as potential screening
biomarkers for colorectal cancer and polyps. Gut. 61:739–745. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie Y, Todd NW, Liu ZQ, et al: Altered
miRNA expression in sputum for diagnosis of non-small cell lung
cancer. Lung Cancer. 67:170–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brase JC, Wuttig D, Kuner R and Sultmann
H: Serum microRNAs as non-invasive biomarkers for cancer. Mol
Cancer. 9:3062010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
van Niel G, Porto-Carreiro I, Simoes S and
Raposo G: Exosomes: a common pathway for a specialized function. J
Biochem. 140:13–21. 2006.PubMed/NCBI
|
|
36
|
Valadi H, Ekstrom K, Bossios A, Sjostrand
M, Lee JJ and Lotvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–672. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hunter MP, Ismail N, Zhang X, et al:
Detection of microRNA expression in human peripheral blood
microvesicles. PLoS One. 3:e36942008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rabinowits G, Gercel-Taylor C, Day JM,
Taylor DD and Kloecker GH: Exosomal microRNA: a diagnostic marker
for lung cancer. Clin Lung Cancer. 10:42–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Skog J, Wurdinger T, van Rijn S, et al:
Glioblastoma microvesicles transport RNA and proteins that promote
tumour growth and provide diagnostic biomarkers. Nat Cell Biol.
10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu Z and Hecht NB: The DNA/RNA-binding
protein, translin, binds microRNA122a and increases its in vivo
stability. J Androl. 29:572–579. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie L, Chen X, Wang L, et al: Cell-free
miRNAs may indicate diagnosis and docetaxel sensitivity of tumor
cells in malignant effusions. BMC Cancer. 10:5912010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Levine JS and Ahnen DJ: Clinical practice.
Adenomatous polyps of the colon. N Engl J Med. 355:2551–2557. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Levin B, Lieberman DA, McFarland B, et al:
Screening and surveillance for the early detection of colorectal
cancer and adenomatous polyps, 2008: a joint guideline from the
American Cancer Society, the US Multi-Society Task Force on
Colorectal Cancer, and the American College of Radiology.
Gastroenterology. 134:1570–1595. 2008. View Article : Google Scholar
|
|
44
|
Lieberman DA and Weiss DG: One-time
screening for colorectal cancer with combined fecal occult-blood
testing and examination of the distal colon. N Engl J Med.
345:555–560. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Imperiale TF, Ransohoff DF, Itzkowitz SH,
Turnbull BA and Ross ME: Fecal DNA versus fecal occult blood for
colorectal-cancer screening in an average-risk population. N Engl J
Med. 351:2704–2714. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hewitson P, Glasziou P, Irwig L, Towler B
and Watson E: Screening for colorectal cancer using the faecal
occult blood test, Hemoccult. Cochrane Database Syst Rev.
CD0012162007. View Article : Google Scholar
|
|
47
|
Cotton PB, Durkalski VL, Benoit PC, et al:
Computed tomographic colonography (virtual colonoscopy): a
multicenter comparison with standard colonoscopy for detection of
colorectal neoplasia. JAMA. 291:1713–1719. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mulhall BP, Veerappan GR and Jackson JL:
Meta-analysis: computed tomographic colonography. Ann Intern Med.
142:635–650. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Johnson CD, Chen MH, Toledano AY, et al:
Accuracy of CT colonography for detection of large adenomas and
cancers. New Engl J Med. 359:1207–1217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Coady-Fariborzian L, Angel LP and
Procaccino JA: Perforated colon secondary to virtual colonoscopy:
report of a case. Dis Colon Rectum. 47:1247–1249. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kanaoka S, Yoshida K, Miura N, Sugimura H
and Kajimura M: Potential usefulness of detecting cyclooxygenase 2
messenger RNA in feces for colorectal cancer screening.
Gastroenterology. 127:422–427. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ahlquist DA, Sargent DJ, Loprinzi CL, et
al: Stool DNA and occult blood testing for screen detection of
colorectal neoplasia. Ann Intern Med. 149:441–450. W4812008.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Brand RE, Ross ME and Shuber AP:
Reproducibility of a multitarget stool-based DNA assay for
colorectal cancer detection. Am J Gastroenterol. 99:1338–1341.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Calistri D, Rengucci C, Bocchini R,
Saragoni L, Zoli W and Amadori D: Fecal multiple molecular tests to
detect colorectal cancer in stool. Clin Gastroenterol Hepatol.
1:377–383. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yu YJ, Majumdar AP, Nechvatal JM, et al:
Exfoliated cells in stool: a source for reverse
transcription-PCR-based analysis of biomarkers of gastrointestinal
cancer. Cancer Epidemiol Biomarkers Prev. 17:455–458.
2008.PubMed/NCBI
|
|
56
|
Zauber AG, Levin TR, Jaffe CC, Galen BA,
Ransohoff DF and Brown ML: Implications of new colorectal cancer
screening technologies for primary care practice. Med Care.
46:S138–S146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Aslam MI, Taylor K, Pringle JH and Jameson
JS: MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg.
96:702–710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ahmed FE, Jeffries CD, Vos PW, et al:
Diagnostic microRNA markers for screening sporadic human colon
cancer and active ulcerative colitis in stool and tissue. Cancer
Genomics Proteomics. 6:281–295. 2009.PubMed/NCBI
|
|
59
|
Link A, Balaguer F, Shen Y, et al: Fecal
MicroRNAs as novel biomarkers for colon cancer screening. Cancer
Epidemiol Biomarkers Prev. 19:1766–1774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kalimutho M, Del Vecchio Blanco G, Di
Cecilia S, et al: Differential expression of miR-144* as
a novel fecal-based diagnostic marker for colorectal cancer. J
Gastroenterol. 46:1391–1402. 2011.
|
|
61
|
Koga Y, Yasunaga M, Takahashi A, et al:
MicroRNA expression profiling of exfoliated colonocytes isolated
from feces for colorectal cancer screening. Cancer Prev Res.
3:1435–1442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li JM, Zhao RH, Li ST, et al:
Down-regulation of fecal miR-143 and miR-145 as potential markers
for colorectal cancer. Saudi Med J. 33:24–29. 2012.PubMed/NCBI
|
|
63
|
Kalimutho M, Di Cecilia S, Blanco GD, et
al: Epigenetically silenced miR-34b/c as a novel faecal-based
screening marker for colorectal cancer. Br J Cancer. 104:1770–1778.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Greenlee RT, Hill-Harmon MB, Murray T and
Thun M: Cancer statistics, 2001. CA Cancer J Clin. 51:15–36. 2001.
View Article : Google Scholar
|
|
65
|
Xing LX, Todd NW, Yu L, Fang HB and Jiang
F: Early detection of squamous cell lung cancer in sputum by a
panel of microRNA markers. Modern Pathol. 23:1157–1164. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Akira M: High-resolution CT in the
evaluation of occupational and environmental disease. Radiol Clin
North Am. 40:43–59. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bastarrika G, Garcia-Velloso MJ, Lozano
MD, et al: Early lung cancer detection using spiral computed
tomography and positron emission tomography. Am J Respir Crit Care
Med. 171:1378–1383. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gohagan JK, Marcus PM, Fagerstrom RM, et
al: Final results of the Lung Screening Study, a randomized
feasibility study of spiral CT versus chest X-ray screening for
lung cancer. Lung Cancer. 47:9–15. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Swensen SJ, Jett JR, Hartman TE, et al:
Lung cancer screening with CT: Mayo Clinic experience. Radiology.
226:756–761. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Koga H, Eguchi K, Shinkai T, et al:
Preliminary evaluation of the new tumor marker, CYFRA 21-1, in lung
cancer patients. Jpn J Clin Oncol. 24:263–268. 1994.PubMed/NCBI
|
|
71
|
Sun S, Schiller JH and Gazdar AF: Lung
cancer in never smokers: a different disease. Nat Rev Cancer.
7:778–790. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hirsch FR, Franklin WA, Gazdar AF and Bunn
PA Jr: Early detection of lung cancer: clinical perspectives of
recent advances in biology and radiology. Clin Cancer Res. 7:5–22.
2001.PubMed/NCBI
|
|
73
|
Hassanein M, Callison JC, Callaway-Lane C,
Aldrich MC, Grogan EL and Massion PP: The state of molecular
biomarkers for the early detection of lung cancer. Cancer Prev Res.
5:992–1006. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Belinsky SA: Gene-promoter
hypermethylation as a biomarker in lung cancer. Nat Rev Cancer.
4:707–717. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Castro M, Grau L, Puerta P, et al:
Multiplexed methylation profiles of tumor suppressor genes and
clinical outcome in lung cancer. J Transl Med. 8:862010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Halling KC, Rickman OB, Kipp BR, Harwood
AR, Doerr CH and Jett JR: A comparison of cytology and fluorescence
in situ hybridization for the detection of lung cancer in
bronchoscopic specimens. Chest. 130:694–701. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Beane J, Sebastiani P, Whitfield TH, et
al: A prediction model for lung cancer diagnosis that integrates
genomic and clinical features. Cancer Prev Res. 1:56–64. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Greenberg AK, Rimal B, Felner K, et al:
S-adenosylmethionine as a biomarker for the early detection of lung
cancer. Chest. 132:1247–1252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Showe MK, Vachani A, Kossenkov AV, et al:
Gene expression profiles in peripheral blood mononuclear cells can
distinguish patients with non-small cell lung cancer from patients
with nonmalignant lung disease. Cancer Res. 69:9202–9210. 2009.
View Article : Google Scholar
|
|
81
|
Lai CY, Yu SL, Hsieh MH, et al: MicroRNA
expression aberration as potential peripheral blood biomarkers for
schizophrenia. PLoS One. 6:e216352011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Peck K, Sher YP, Shih JY, Roffler SR, Wu
CW and Yang PC: Detection and quantitation of circulating cancer
cells in the peripheral blood of lung cancer patients. Cancer Res.
58:2761–2765. 1998.PubMed/NCBI
|
|
83
|
van der Drift MA, Prinsen CFM, Hol BEA, et
al: Can free DNA be detected in sputum of lung cancer patients?
Lung Cancer. 61:385–390. 2008.PubMed/NCBI
|
|
84
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Neragi-Miandoab S: Malignant pleural
effusion, current and evolving approaches for its diagnosis and
management. Lung Cancer. 54:1–9. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang T, Lv M, Shen S, et al: Cell-free
microRNA expression profiles in malignant effusion associated with
patient survival in non-small cell lung cancer. PLoS One.
7:e432682012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lee JH, Hong YS, Ryu JS and Chang JH: p53
and FHIT mutations and microsatellite alterations in
malignancy-associated pleural effusion. Lung Cancer. 44:33–42.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Spector M and Pollak JS: Management of
malignant pleural effusions. Semin Respir Crit Care Med.
29:405–413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Topolcan O, Holubec L, Polivkova V, et al:
Tumor markers in pleural effusions. Anticancer Res. 27:1921–1924.
2007.PubMed/NCBI
|
|
90
|
Katayama H, Hiraki A, Aoe K, et al:
Aberrant promoter methylation in pleural fluid DNA for diagnosis of
malignant pleural effusion. Int J Cancer. 120:2191–2195. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang T, Qian X, Wang Z, et al: Detection
of cell-free BIRC5 mRNA in effusions and its potential diagnostic
value for differentiating malignant and benign effusions. Int J
Cancer. 125:1921–1925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Nair VS, Maeda LS and Ioannidis JPA:
Clinical outcome prediction by microRNAs in human cancer: a
systematic review. J Natl Cancer Inst. 104:528–540. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
94
|
Snowdon J, Boag S, Feilotter H, Izard J
and Siemens DR: A pilot study of urinary microRNA as a biomarker
for urothelial cancer. Can Urol Assoc J. 7:28–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yamada Y, Enokida H, Kojima S, et al:
MiR-96 and miR-183 detection in urine serve as potential tumor
markers of urothelial carcinoma: correlation with stage and grade,
and comparison with urinary cytology. Cancer Sci. 102:522–529.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Ratliff TL: Urine markers for bladder
cancer surveillance: a systematic review. J Urol. 174:2065–2066.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Sarosdy MF, Hudson MA, Ellis WJ, et al:
Improved detection of recurrent bladder cancer using the Bard BTA
stat Test. Urology. 50:349–353. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Carpinito GA, Stadler WM, Briggman JV, et
al: Urinary nuclear matrix protein as a marker for transitional
cell carcinoma of the urinary tract. J Urol. 156:1280–1285. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Fradet Y and Lockhard C: Performance
characteristics of a new monoclonal antibody test for bladder
cancer: ImmunoCyt™. Can J Urol. 4:400–405. 1997.PubMed/NCBI
|
|
100
|
Greenlee RT, Murray T, Bolden S and Wingo
PA: Cancer statistics, 2000. CA Cancer J Clin. 50:7–33. 2000.
View Article : Google Scholar
|
|
101
|
Wang G, Chan ESY, Kwan BCH, et al:
Expression of microRNAs in the urine of patients with bladder
cancer. Clin Genitourin Cancer. 10:106–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Roos PH and Jakubowski N: Methods for the
discovery of low-abundance biomarkers for urinary bladder cancer in
biological fluids. Bioanalysis. 2:295–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Puerta-Gil P, Garcia-Baquero R, Jia AY, et
al: miR-143, miR-222, and miR-452 are useful as tumor
stratification and noninvasive diagnostic biomarkers for bladder
cancer. Am J Pathol. 180:1808–1815. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Hanke M, Hoefig K, Merz H, et al: A robust
methodology to study urine microRNA as tumor marker: microRNA-126
and microRNA-182 are related to urinary bladder cancer. Urol Oncol.
28:655–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Veerla S, Lindgren D, Kvist A, et al:
MiRNA expression in urothelial carcinomas: important roles of
miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and
metastasis, and frequent homozygous losses of miR-31. Int J Cancer.
124:2236–2242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Han Y, Chen J, Zhao X, et al: MicroRNA
expression signatures of bladder cancer revealed by deep
sequencing. PLoS One. 6:e182862011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Noguchi S, Mori T, Hoshino Y, et al:
MicroRNA-143 functions as a tumor suppressor in human bladder
cancer T24 cells. Cancer Lett. 307:211–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kassouf W, Black PC, Tuziak T, et al:
Distinctive expression pattern of ErbB family receptors signifies
an aggressive variant of bladder cancer. J Urol. 179:353–358. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Junttila TT, Laato M, Vahlberg T, et al:
Identification of patients with transitional cell carcinoma of the
bladder overexpressing ErbB2, ErbB3, or specific ErbB4 isoforms:
real-time reverse transcription-PCR analysis in estimation of ErbB
receptor status from cancer patients. Clin Cancer Res. 9:5346–5357.
2003.
|
|
110
|
Cissell KA and Deo SK: Trends in microRNA
detection. Anal Bioanal Chem. 394:1109–1116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
de Planell-Saguer M and Rodicio MC:
Analytical aspects of microRNA in diagnostics: a review. Anal Chim
Acta. 699:134–152. 2011.PubMed/NCBI
|
|
112
|
Ach RA, Wang H and Curry B: Measuring
microRNAs: comparisons of microarray and quantitative PCR
measurements, and of different total RNA prep methods. BMC
Biotechnol. 8:692008. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
de Planell-Saguer M and Rodicio MC:
Analytical aspects of microRNA in diagnostics: a review. Anal Chim
Acta. 699:134–152. 2011.PubMed/NCBI
|
|
114
|
Sarver AL: Toward understanding the
informatics and statistical aspects of micro-RNA profiling. J
Cardiovasc Transl Res. 3:204–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Valoczi A, Hornyik C, Varga N, Burgyan J,
Kauppinen S and Havelda Z: Sensitive and specific detection of
microRNAs by northern blot analysis using LNA-modified
oligonucleotide probes. Nucleic Acids Res. 32:e1752004. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Pall GS, Codony-Servat C, Byrne J, Ritchie
L and Hamilton A: Carbodiimide-mediated cross-linking of RNA to
nylon membranes improves the detection of siRNA, miRNA and piRNA by
northern blot. Nucleic Acids Res. 35:e602007. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Varallyay E, Burgyan J and Havelda Z:
MicroRNA detection by northern blotting using locked nucleic acid
probes. Nat Protoc. 3:190–196. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Chamnongpol S, Maroney PA and Nilsen TW: A
rapid, quantitative assay for direct detection of microRNAs and
other small RNAs using splinted ligation. Methods Mol Biol.
667:3–17. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Cissell KA, Rahimi Y, Shrestha S, Hunt EA
and Deo SK: Bioluminescence-based detection of microRNA, miR21 in
breast cancer cells. Anal Chem. 80:2319–2325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Weaver S, Dube S, Mir A, et al: Taking
qPCR to a higher level: analysis of CNV reveals the power of high
throughput qPCR to enhance quantitative resolution. Methods.
50:271–276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Raymond CK, Roberts BS, Garrett-Engele P,
Lim LP and Johnson JM: Simple, quantitative primer-extension PCR
assay for direct monitoring of microRNAs and short-interfering
RNAs. RNA. 11:1737–1744. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Neely LA, Patel S, Garver J, et al: A
single-molecule method for the quantitation of microRNA gene
expression. Nat Methods. 3:41–46. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kloosterman WP, Wienholds E, de Bruijn E,
Kauppinen S and Plasterk RHA: In situ detection of miRNAs in animal
embryos using LNA-modified oligonucteotide probes. Nat Methods.
3:27–29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
de Planell-Saguer M, Rodicio MC and
Mourelatos Z: Rapid in situ codetection of noncoding RNAs and
proteins in cells and formalin-fixed paraffin-embedded tissue
sections without protease treatment. Nat Protoc. 5:1061–1073.
2010.PubMed/NCBI
|
|
126
|
Obernosterer G, Martinez J and Alenius M:
Locked nucleic acid-based in situ detection of microRNAs in mouse
tissue sections. Nat Protoc. 2:1508–1514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Nelson PT, Baldwin DA, Kloosterman WP,
Kauppinen S, Plasterk RHA and Mourelatos Z: RAKE and LNA-ISH reveal
microRNA expression and localization in archival human brain. RNA.
12:187–191. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Liu CG, Calin GA, Volinia S and Croce CM:
MicroRNA expression profiling using microarrays. Nat Protoc.
3:563–578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Li W and Ruan KC: MicroRNA detection by
microarray. Anal Bioanal Chem. 394:1117–1124. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Nelson PT, Baldwin DA, Scearce LM,
Oberholtzer JC, Tobias JW and Mourelatos Z: Microarray-based,
high-throughput gene expression profiling of microRNAs. Nat
Methods. 1:155–161. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Takada S and Mano H: Profiling of microRNA
expression by mRAP. Nat Protoc. 2:3136–3145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Weber JA, Baxter DH, Zhang SL, et al: The
microRNA spectrum in 12 body fluids. Clin Chem. 56:1733–1741. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Kanaan Z, Rai SN, Eichenberger MR, et al:
Plasma miR-21: a potential diagnostic marker of colorectal cancer.
Ann Surg. 256:544–551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li BS, Zhao YL, Guo G, et al: Plasma
microRNAs, miR-223, miR-21 and miR-218, as novel potential
biomarkers for gastric cancer detection. PLoS One. 7:e416292012.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang X, Cui L, Ye G, et al: Gastric juice
microRNA-421 is a new biomarker for screening gastric cancer.
Tumour Biol. 33:2349–2355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Janakiram NB and Rao CV: Molecular markers
and targets for colorectal cancer prevention. Acta Pharmacol Sin.
29:1–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Dong Y, Wu WK, Wu CW, Sung JJ, Yu J and Ng
SS: MicroRNA dysregulation in colorectal cancer: a clinical
perspective. Br J Cancer. 104:893–898. 2011. View Article : Google Scholar : PubMed/NCBI
|