Introduction
microRNAs (miRNAs) constitute a class of 20–22-nt
noncoding single-strand RNA that are initially synthesized by RNA
polymerase II as long primary transcripts, which are subsequently
capped and polyadenylated. These precursors of miRNAs, pri-miRNAs,
are cleaved by Drosha into pre-miRNAs and transported out of the
nucleus by exportin-5. Pre-miRNAs are further processed by Dicer in
the cytoplasm to yield mature miRNAs. Through base pairing to the
3′ untranslated region (3′UTR) of mRNA, miRNAs negatively regulate
gene expression post-transcriptionally to induce suppression of
translation or degradation of multiple mRNAs. They are thus crucial
players that participate in numerous key cellular processes such as
cell growth, differentiation and death (1–3). In
malignancy, functional characterization has revealed a role for
miRNAs as oncogenes or tumor-suppressor genes (4). The contribution of miRNAs to the
malignant progression of human tumors was recently investigated.
Malignant tumor progression shares several characteristics with the
process of epithelial-to-mesenchymal transition (EMT), which
facilitates tissue remodeling during embryonic development and is
considered an essential early step in tumor metastasis. In EMT,
epithelial tumor cells are stimulated by extracellular cytokines to
lose their epithelial polarity and gain mesenchymal phenotypes with
increased migratory and invasive capabilities (5,6). One
of the molecular hallmarks driving this transition is the
functional loss of E-cadherin, which is a cell adhesion protein and
a major constituent of adherens junctions, and is thought to be a
suppressor of migration/invasion during carcinoma progression
(7,8). Recent studies have indicated that the
expression of the miRNA-200 family (miR-200a, miR-200b, miR-200c,
miR-141 and miR-429) is markedly downregulated in cells that have
undergone EMT. By serving as a powerful regulator of the EMT,
miR-200c can maintain the epithelial phenotype of tissues by
suppressing expression of the E-cadherin transcription factor ZEB1
in some types of cancer (9–13).
Renal cell carcinoma (RCC) is the third most common
urological cancer after prostate and bladder cancer, and it
accounts for approximately 3% of adult malignancies and 90–95% of
neoplasms arising from the kidney (14,15).
Previous studies have shown that almost 20–30% of patients have
distant metastasis when diagnosed and that metastasis is associated
with poor prognosis. The mechanisms underlying metastasis are
multifaceted and intricate. It is therefore important to explore
metastatic mechanisms in RCC as these may provide new targets for
treating metastasis. In this study, we investigated the possible
metastatic mechanisms of action of miR-200c in 2 RCC cell lines:
the 786-0 cancer cell line, derived from primary human RCC in a
58-year-old male patient, and the SN12-PM6 cancer cell line,
obtained from lung metastases produced in nude mice (16,17).
Materials and methods
Clinical specimens
All primary RCC tissues and normal renal tissues
were surgically resected at Wuhan Union Hospital. Tissue samples
were immediately frozen in liquid nitrogen and then stored in a
deep freezer at −80°C. Histopathological diagnoses were performed
according to the World Health Organization (WHO) classification
system. Informed consent was obtained from each patient, and the
use of tissue samples for all experiments was approved by the
Clinical Research Ethics Committee of Wuhan Union Hospital.
miRNA microarray and analysis
A miRNA microarray platform was used to determine
the expression profiles of miRNA in 5 paired RCCs and normal renal
tissues. The array used was the commercially available G4471A Human
miRNA Microarray (Agilent Technologies, Santa Clara, CA, USA),
which consists of 961 probes for 851 human miRNAs, and is based on
Sanger miRBase release 12.0. The arrays were washed and scanned
using a laser confocal scanner (G2565BA; Agilent Technologies)
according to the manufacturer’s instructions. The fluorescence
intensity was calculated using the Feature Extraction software
(Agilent Technologies). Differentially expressed miRNAs were
identified using a filter based on a 2-fold change in expression,
combined with ANOVA analysis (P<0.01).
Lentivirus production and
transduction
We used the lentiviral vector pGCSIL-GFP (GeneChem
Co., Ltd., Shanghai, China) to construct the hsa-miR-200c
lentiviral expression vector pGCSIL-GFP-hsa-miR-200c as well as the
lentiviral vector pGCSIL-GFP-negative as a negative control. For
6-well transduction in SN12-PM6 and 786-0 cells, cells were plated
at a density of 150,000/well in a 6-well dish. When the cells were
50% confluent, they were transduced with recombinant lentivirus
vectors at a multiplicity of infection (MOI) of 30 and 10,
respectively, and supplemented with 5 μg/ml polybrene from GeneChem
Co., Ltd. The cells were then collected for western blot analysis,
quantitative RT-PCR (qRT-PCR), and migration and invasion assays.
The SN12-PM6 cell line stably expressing miR-200c was termed
SN12-PM6 miR-200c; the negative control cell line was termed
SN12-PM6 miR-Ctr. We also generated 786-0 miR-200c and 786-0
miR-Ctr cells.
Cell culture and transfection
All cell lines were maintained in DMEM medium
(Invitrogen, Carlsbad, CA, USA) supplemented with 10%
heat-inactivated fetal bovine serum (Gibco-BRL, Grand Island, NY,
USA), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a
humidified atmosphere containing 5% CO2. Lipofectamine™
2000 (Invitrogen) was used to transfect SN12-PM6 miR-200c and 786-0
miR-200c with the antagomiR-200c (RiboBio Co., Ltd., Guangzhou,
China) according to the manufacturer’s instructions.
Transwell migration and invasion
assays
For transwell migration assays, 20,000 cells were
plated in the top chamber onto a non-coated membrane (24-well
insert; pore size, 8 μm; Corning-Costar, Corning, NY, USA). For
invasion assays, 20,000 cells were plated in the top chamber onto a
Matrigel-coated membrane. Each well was freshly coated with 60 μg
of Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) prior to the
invasion assay. In both assays, cells were plated in medium without
serum or growth factors, and medium supplemented with serum was
used as a chemoattractant in the lower chamber. The cells were
incubated for 24 h in the migration and the invasion assay, and
cells that did not migrate or invade through the pores were removed
with a cotton swab. Cells on the lower surface of the membrane were
fixed with methanol and stained with crystal violet. The cells
migrating or invading through the membrane were counted with a
Nikon Eclipse TE2000-S (Nikon, Japan) at ×100 magnification in 4
random fields/well.
Quantitative RT-PCR analysis
Total RNA was extracted using TRIzol (Invitrogen),
and 1-μg RNA samples were reverse transcribed to cDNA by using
reverse transcriptase M-MLV (Invitrogen). qRT-PCR was performed
using the SYBR-Green PCR master mix (Invitrogen) on a Roche
LightCycler 480 System (Roche Diagnostics, Germany). For analysis
of miRNA expression by qRT-PCR, reverse transcription and PCR were
conducted using a Bulge-Loop™ miRNA qPCR Primer Set for
hsa-miR-200c and U6 snRNA (RiboBio Co., Ltd.) according to the
manufacturer’s instructions. Other primers used in qRT-PCR are
described in Table I. GAPDH and U6
snRNA were used as endogenous controls.
 | Table IPrimers used for quantitative
RT-PCR. |
Table I
Primers used for quantitative
RT-PCR.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| ZEB1 |
TGCTCCCTGTGCAGTTACACCTT |
CCAGACTGCGTCACATGTCTTTGA |
| E-cadherin |
GTCCTGGGCAGAGTGAATTT |
CATCTGTGCCCACTTTGAAT |
| GAPDH |
GAGTCAACGGATTTGGTCGT |
GACAAGCTTCCCGTTCTCAG |
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer
(Beyotime, Shanghai, China) containing a cocktail of protease
inhibitors and phosphatase inhibitors (Roche Applied Science,
Indianapolis, IN, USA). Equal amounts of protein sample (40–60 μg)
were separated by 10% SDS-PAGE and transferred to a PVDF membrane
(Millipore, Bedford, MA, USA) using the Invitrogen semidry transfer
system and then incubated with the specific primary antibody
overnight at 4°C. The membranes were then washed and subsequently
incubated with a secondary antibody conjugated to horseradish
peroxidase (HRP). The protein detected was then visualized using
enhanced chemiluminescence. The following antibodies were used for
western blotting: anti-β-actin (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA), anti-E-cadherin and anti-ZEB1 (both from
Bioworld, Minneapolis, MN, USA), anti-Akt and anti-p-Akt (both from
Cell Signaling Technology, Beverly, MA, USA). The blotted proteins
were detected and quantified using ChemiDoc-XRS+ (Bio-Rad,
USA).
Statistical analysis
Data from different experiments are presented in
terms of mean ± standard deviation (SD) values. Differences among
different groups with respect to the number of cancer cells in the
invasion and migration assays were compared using the
paired-samples t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Downregulated miRNAs in RCC and normal
renal tissues
A comprehensive miRNA expression profile was
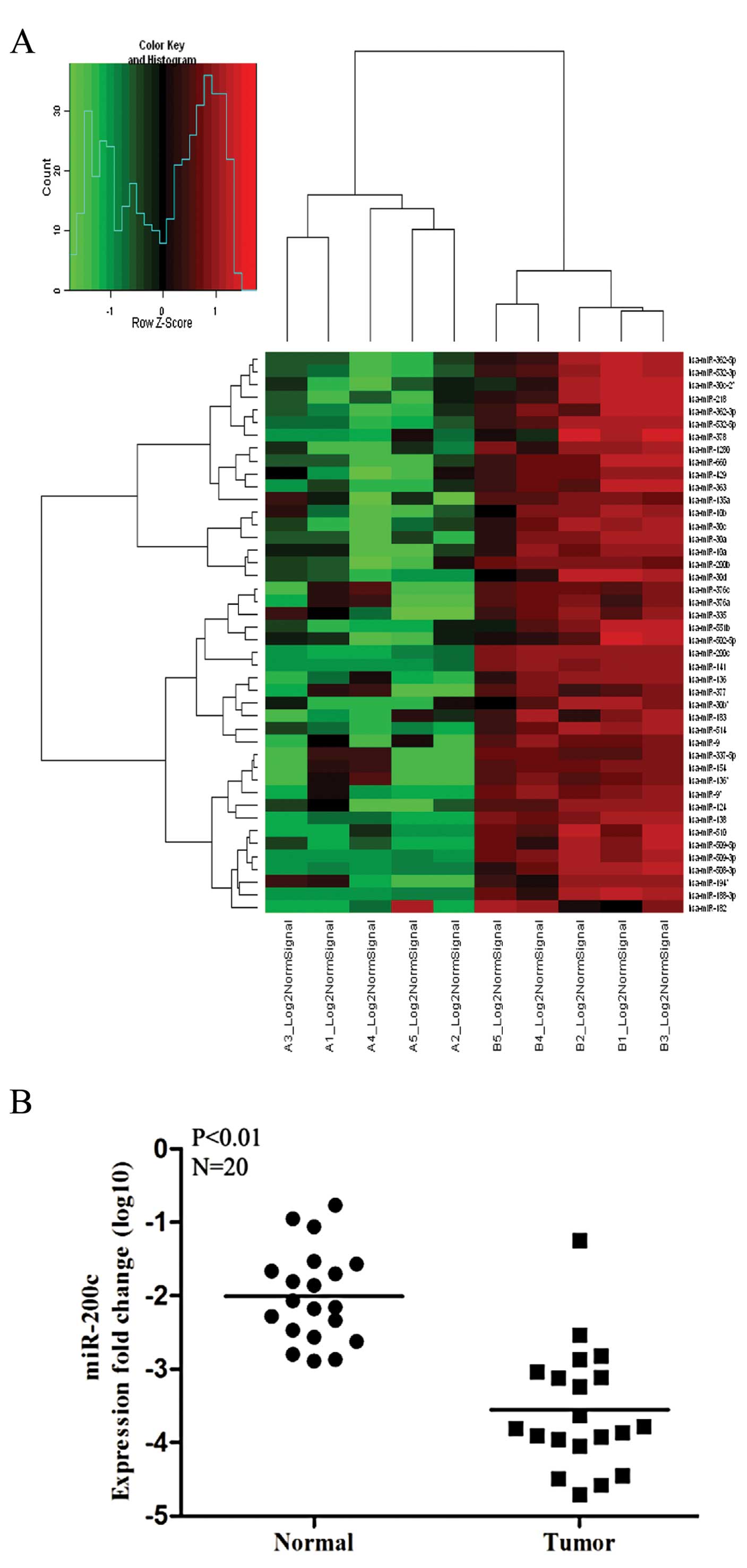
obtained for the 5 paired RCC and normal renal tissues. Fig. 1A shows miRNAs that were
downregulated in tumors relative to normal tissues: 12 miRNAs were
downregulated >4-fold in tumor tissues, as listed in Table II. miR-200c was one of the most
strongly downregulated miRNAs in tumor tissues, which was expected,
as it had already been shown to inhibit metastasis in some types of
tumors. We further examined the expression of miR-200c in 20 more
pairs of RCC and normal human specimens to validate the microarray
data and found that the data for 19 of 20 pairs were consistent
with the microarray data in that miR-200c expression was
significantly lower in RCCs than in normal kidney tissues (Fig. 1B). Therefore, miR-200c was selected
for further study.
 | Table IIDownregulated miRNAs in tumors. |
Table II
Downregulated miRNAs in tumors.
| miRNA | P-value | Fold-change |
|---|
| hsa-miR-141 | 4.11E-08 | 103.990700 |
| hsa-miR-200c | 4.35E-06 | 99.787830 |
| hsa-miR-138 | 1.14E-06 | 22.829310 |
| hsa-miR-514 | 5.95E-04 | 18.366480 |
| hsa-miR-509-3p | 3.74E-04 | 8.679166 |
|
hsa-miR-9* | 9.41E-04 | 8.520466 |
| hsa-miR-508-3p | 0.003473 | 7.809710 |
| hsa-miR-124 | 0.004609 | 7.624603 |
| hsa-miR-510 | 8.37E-05 | 6.083151 |
| hsa-miR-218 | 0.001076 | 5.091851 |
| hsa-miR-509-5p | 0.001346 | 4.811123 |
| hsa-miR-363 | 6.94E-04 | 4.533517 |
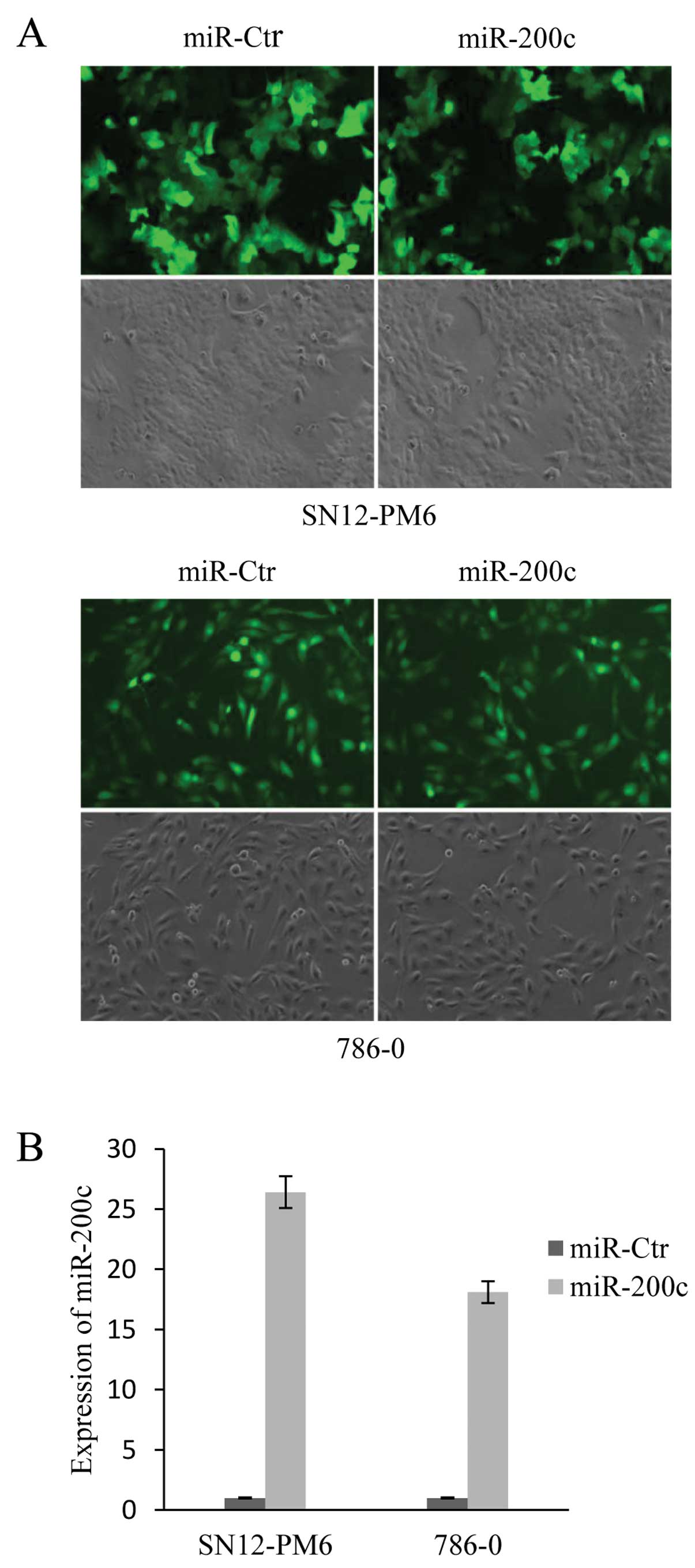
Establishment of stably transduced RCC
cell lines
To investigate the effect of miR-200c in tumor
cells, SN12-PM6 and 786-0 cells were transduced with
pGCSIL-GFP-hsa-miR-200c to establish cell lines stably expressing
miR-200c. The transduction efficiency was determined by counting
fluorescent cells and total cells from 6 random fields for each
condition. The transduction efficiency was ~88% in SN12-PM6 cells
and ~96% in 786-0 cells (Fig. 2A).
qRT-PCR indicated that compared to the control, SN12-PM6 miR-200c
cells and 786-0 miR-200c cells had 26.4-fold and 18.1-fold higher
miR-200c expression, respectively (Fig.
2B).
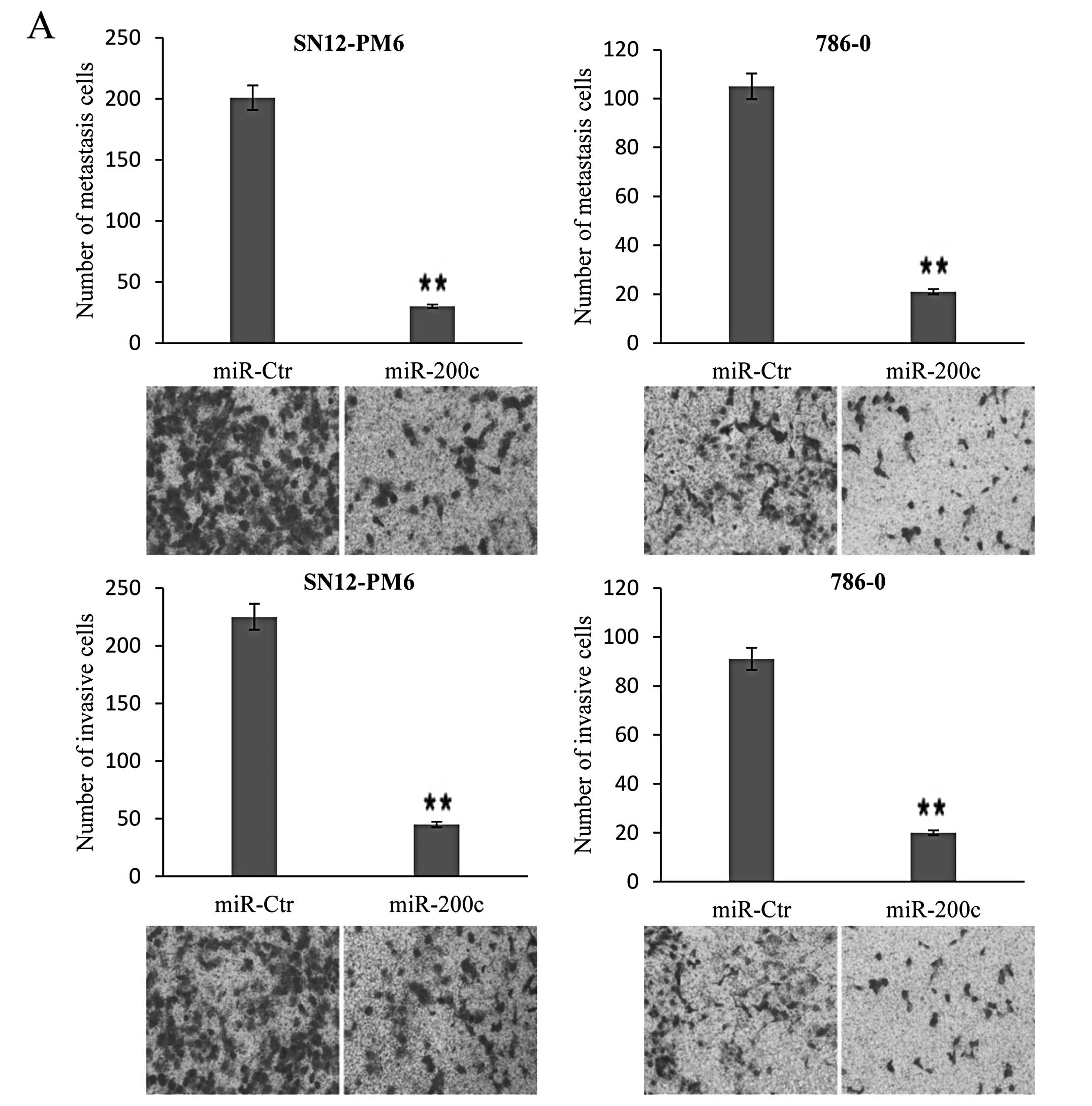
miR-200c inhibits migration and invasion
in SN12-PM6 and 786-0 cancer cells
To validate the involvement of miR-200c
dysregulation in migration and invasion, functional analysis was
performed to test the effects of miR-200c. In the migration test,
the mean number of SN12-PM6 cells expressing miR-200c per field was
30, which was significantly different from that for the control
group. The mean number of 786-0 cells expressing miR-200c per field
was 21, which was also significantly different from the control
group. In the invasion test, the mean number of SN12-PM6 and 786-0
cells expressing miR-200c per field was 45 and 20, respectively,
which was significantly different from the number for the control
group (Fig. 3A). To further explore
the role of miR-200c and to determine whether inhibition of
miR-200c could reverse the inhibition of cellular migration and
invasion produced by overexpression, loss-of-function analyses were
performed. We transfected cells stably expressing miR-200c with
antagomiR-200c, which resulted in significant reduction of miR-200c
levels in SN12-PM6 miR-200c and 786-0 miR-200c cells. In the
migration and invasion tests, the mean number of
inhibitor-transfected SN12-PM6 cells expressing miR-200c per field
was 188 and 176, and the mean number of inhibitor-transfected 786-0
cells expressing miR-200c per field was 178 and 156, respectively,
which was significantly different from the control group (Fig. 3B). These findings indicate that the
migration and invasion ability of SN12-PM6 and 786-0 cells can be
negatively modulated by miR-200c.
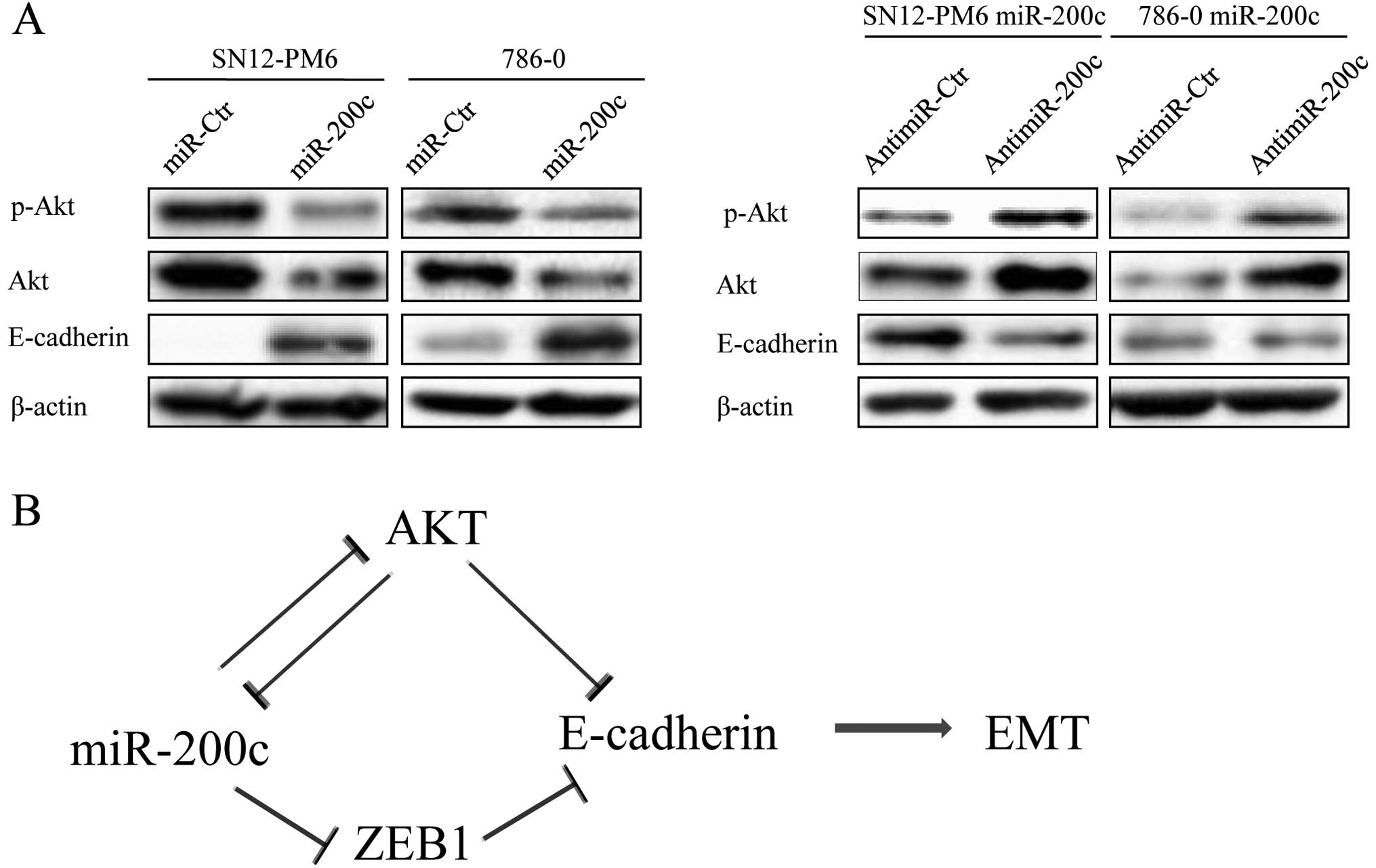
Expression of ZEB1 is negatively
regulated by miR-200c in RCC cell lines
The E-cadherin transcriptional repressor ZEB1 has
previously been implicated in EMT and tumor metastasis. EMT occurs
during tumor progression and confers invasive and metastatic
properties to cancer cells. ZEB1 has been proposed as a putative
target of miR-200c on the basis of in silico miRNA target
prediction programs (TargetScan, PicTar, miRanda) and studies in
other types of cancer (18,19). Our results also showed that miR-200c
levels were markedly elevated in SN12 miR-200c and 786-0 miR-200c
cells, whereas the mRNA and protein level of ZEB1 decreased.
Conversely, inhibition of miR-200c expression resulted in
upregulation of ZEB1. These observations indicate that ZEB1 is
negatively regulated by miR-200c at the posttranscriptional level.
To further examine whether miR-200c overexpression-induced
inhibition of EMT-related genes actually inhibits the EMT
phenotype, we evaluated the expression of E-cadherin.
Overexpression of miR-200c significantly increased E-cadherin mRNA
and protein expression, whereas anti-miR-200c produced the opposite
result (Fig. 4A and B).
Next, we analyzed whether miR-200c, ZEB1 and
E-cadherin were correlated in RCC cells and human cancer tissues.
We measured the expression of miR-200c, ZEB1, and E-cadherin in
SN12-PM6 cells, 786-0 cells, and 20 pairs of human RCC tissues by
qRT-PCR. The expression of miR-200c was inversely correlated with
that of ZEB1 but positively correlated with that of E-cadherin
(Fig. 4C).
miR-200c is involved in the regulation of
p-Akt and Akt protein expression
Akt is involved in a number of basic cellular
processes, including EMT. Akt strongly represses transcription of
the E-cadherin gene and internalization/sequestration of E-cadherin
in perinuclear organelles to induce EMT (20,21).
We found that overexpression of miR-200c in SN12-PM6 and 786-0
cells was able to inhibit the protein expression of p-Akt and Akt
and that blockade of miR-200c could reverse this result (Fig. 5A).
Discussion
Previous studies have linked the miR-200 family with
the epithelial phenotype and the ZEB family. miR-200c is a member
of the miR-200 family; this family has been associated with
metastasis in several types of tumors, particularly through
negative correlation with ZEB1, which inhibits EMT (9,22,23).
In breast cancer, melanoma, and pancreatic cancer, miR-200c
suppresses tumor migration and invasion (24–26).
Nakada et al(27) reported
that overexpression of miR-141 and miR-200c caused downregulation
of ZFHX1B and upregulation of E-cadherin in 2 renal carcinoma cell
lines. However, the functional involvement of miR-200c in EMT and
tumor migration, as well as direct targeting of ZEB1 by miR-200c,
has not been investigated in renal cell carcinoma (RCC) studies. In
our tissue microarray, miR-200c in tumor tissue was downregulated
significantly relative to other miRNAs. Therefore, we selected
miR-200c as our research target.
First, we performed functional analysis in SN12-PM6
and 786-0 cells stably overexpressing miR-200c. EMT is an important
process in which epithelial cells acquire mesenchymal
fibroblast-like properties and show reduced intercellular adhesion
and increased motility during development. Accumulating evidence
points to a critical role of EMT during tumor progression and
malignant transformation, as it endows cancer cells with invasive
and metastatic properties (8,28,29).
In our migration and invasion test, enforced expression of miR-200c
in SN12-PM6 and 786-0 cells inhibited cell mobility and invasion
activity. Blockade of expression of miR-200c in SN12-PM6 miR-200c
and 786-0 miR-200c cells increased their invasion and migration
abilities. We demonstrated that modulation of miR-200c expression
can alter the invasion and migration abilities of SN12-PM6 and
786-0 cells in metastasis assays, indicating that miR-200c is of
functional significance in RCC cells.
We then identified miR-200c as a suppressor of EMT
through direct targeting of ZEB1, which is a well-known
transcriptional repressor of E-cadherin. miRNA target prediction
programs and previous studies have indicated that ZEB1 has 2 target
sites for miR-200c (Fig. 4D), as
confirmed by luciferase reporter assays (8,19,30).
ZEB1 is a crucial inducer of EMT, which has been shown to promote
tumor invasion and migration through E-cadherin gene silencing
(9,12). In our present study, we analyzed
whether miR-200c played a role in the regulation of metastasis in
these 2 renal carcinoma cells by targeting ZEB1. Using quantitative
RT-PCR (qRT-PCR), we observed that stable overexpression of
miR-200c by an hsa-miR-200c lentivirus in SN12-PM6 and 786-0 cells
led to decreased expression of ZEB1 and increased expression of
E-cadherin, whereas blockade of miR-200c in SN12-PM6 miR-200c and
786-0 miR-200c cells led to restoration of ZEB1 expression and
decreased E-cadherin expression. These results were observed at the
protein level as well as the mRNA level. We also found that ZEB1
expression was inversely correlated with miR-200c expression in RCC
cells and human RCC specimens. These data demonstrate that
increased expression of miR-200c results in negative regulation of
its gene target ZEB1, which regulates E-cadherin expression to
trigger EMT in RCC cells.
Collectively, these findings suggest a potential
tumor suppressor role for miR-200c, which is downregulated in human
RCC, thereby leading to EMT and tumor cell invasion and metastasis.
Loss of miR-200c is associated with an aggressive cancer cell
phenotype. According to previous research and our data, Akt is
frequently activated in human epithelial cancer and the PI3K/Akt
pathway plays a pivotal role in RCC pathogenesis (31,32).
Additionally, Akt has been found to regulate E-cadherin mRNA and
protein and to induce EMT (20),
and miR-200c regulates the expression of E-cadherin by targeting
ZEB1 in some cancer cell lines, including RCC cell lines (12). Finally, miR-200c is downregulated in
human RCC specimens and regulates the protein level of Akt in RCC
cell lines. We therefore consider that RCC metastasis may be under
the control of the Akt-miR-200c-E-cadherin axis. A summary diagram
that outlines this hypothesis is shown in Fig. 5B. Our future work will focus on the
identification and functional characterization of additional direct
miR-200c downstream targets and metastasis promoter proteins, as
well as the specific mechanism underlying the regulation of
miR-200c by Akt. This will improve our understanding of the events
contributing to EMT and metastasis, leading to the development of
novel therapeutic strategies for RCC.
Acknowledgements
This research was supported by National Natural
Science Foundation of China (grant no. 30572139, 30872924 and
81072095), Program for New Century Excellent Talents in University
from Department of Education of China (NCET-08-0223) and the
National High Technology Research and Development Program of China
(863 Program) (2012AA021101) to X.Z, and supported by National
Natural Science Foundation of China (grant no. 2010MS011 and
31070142) to H.Y.
References
|
1
|
Bagga S, Bracht J, Hunter S, et al:
Regulation by let-7 and lin-4 miRNAs results in target mRNA
degradation. Cell. 122:553–563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lim LP, Lau NC, Garrett-Engele P, et al:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ambros V: microRNAs: tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bracken CP, Gregory PA, Khew-Goodall Y and
Goodall GJ: The role of microRNAs in metastasis and
epithelial-mesenchymal transition. Cell Mol Life Sci. 66:1682–1699.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: at the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bracken CP, Gregory PA, Kolesnikoff N, et
al: A double-negative feedback loop between ZEB1-SIP1 and the
microRNA-200 family regulates epithelial-mesenchymal transition.
Cancer Res. 68:7846–7854. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hur K, Toiyama Y, Takahashi M, et al:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. Jul 10–2012.(Epub ahead
of print).
|
|
9
|
Gregory PA, Bert AG, Paterson EL, et al:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Christoffersen NR, Silahtaroglu A, Orom
UA, Kauppinen S and Lund AH: miR-200b mediates post-transcriptional
repression of ZFHX1B. RNA. 13:1172–1178. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hurteau GJ, Carlson JA, Spivack SD and
Brock GJ: Overexpression of the microRNA hsa-miR-200c leads to
reduced expression of transcription factor 8 and increased
expression of E-cadherin. Cancer Res. 67:7972–7976. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mongroo PS and Rustgi AK: The role of the
miR-200 family in epithelial-mesenchymal transition. Cancer Biol
Ther. 10:219–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gottardo F, Liu CG, Ferracin M, et al:
Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
White NM and Yousef GM: MicroRNAs:
exploring a new dimension in the pathogenesis of kidney cancer. BMC
Med. 8:652010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saiki I, Naito S, Yoneda J, Azuma I, Price
JE and Fidler IJ: Characterization of the invasive and metastatic
phenotype in human renal cell carcinoma. Clin Exp Metastasis.
9:551–566. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Williams RD, Elliott AY, Stein N and
Fraley EE: In vitro cultivation of human renal cell cancer. II
Characterization of cell lines. In Vitro. 14:779–786. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen ML, Liang LS and Wang XK: miR-200c
inhibits invasion and migration in human colon cancer cells
SW480/620 by targeting ZEB1. Clin Exp Metastasis. 29:457–469. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Burk U, Schubert J, Wellner U, et al: A
reciprocal repression between ZEB1 and members of the miR-200
family promotes EMT and invasion in cancer cells. EMBO Rep.
9:582–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grille SJ, Bellacosa A, Upson J, et al:
The protein kinase Akt induces epithelial mesenchymal transition
and promotes enhanced motility and invasiveness of squamous cell
carcinoma lines. Cancer Res. 63:2172–2178. 2003.PubMed/NCBI
|
|
21
|
Larue L and Bellacosa A:
Epithelial-mesenchymal transition in development and cancer: role
of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene.
24:7443–7454. 2005.
|
|
22
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hurteau GJ, Carlson JA, Roos E and Brock
GJ: Stable expression of miR-200c alone is sufficient to regulate
TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle.
8:2064–2069. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ahmad A, Aboukameel A, Kong D, et al:
Phosphoglucose isomerase/autocrine motility factor mediates
epithelial-mesenchymal transition regulated by miR-200 in breast
cancer cells. Cancer Res. 71:3400–3409. 2011. View Article : Google Scholar
|
|
25
|
Elson-Schwab I, Lorentzen A and Marshall
CJ: MicroRNA-200 family members differentially regulate
morphological plasticity and mode of melanoma cell invasion. PLoS
One. 5:e131762010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu J, Ohuchida K, Mizumoto K, et al:
MicroRNA, hsa-miR-200c, is an independent prognostic factor in
pancreatic cancer and its upregulation inhibits pancreatic cancer
invasion but increases cell proliferation. Mol Cancer. 9:1692010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakada C, Matsuura K, Tsukamoto Y, et al:
Genome-wide microRNA expression profiling in renal cell carcinoma:
significant down-regulation of miR-141 and miR-200c. J Pathol.
216:418–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kang Y and Massague J:
Epithelial-mesenchymal transitions: twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thiery JP and Morgan M: Breast cancer
progression with a Twist. Nat Med. 10:777–778. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
TargetScanHuman release 6.2. Available at:
http://www.targetscan.org.
Accessed Sep 1 2012
|
|
31
|
Porta C and Figlin RA:
Phosphatidylinositol-3-kinase/Akt signaling pathway and kidney
cancer, and the therapeutic potential of
phosphatidylinositol-3-kinase/Akt inhibitors. J Urol.
182:2569–2577. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Testa JR and Bellacosa A: AKT plays a
central role in tumorigenesis. Proc Natl Acad Sci USA.
98:10983–10985. 2001. View Article : Google Scholar : PubMed/NCBI
|